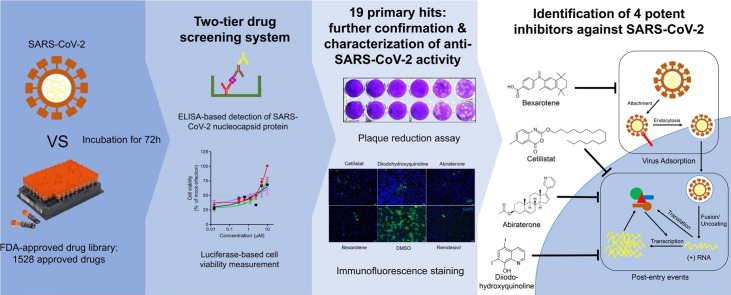
Graphical abstract
Chemical compounds studied in this article: Abiraterone acetate (PubChem CID: 9821849), Bexarotene (PubChem CID: 82146), Cetilistat (PubChem CID: 9952916), Diiodohydroxyquinoline (PubChem CID: 3728)
Keywords: Antiviral, Coronavirus, COVID-19, Library, SARS-CoV-2, Treatment, Cetilistat, Diiodohydroxyquinoline, Abiraterone, Bexarotene
Abstract
Coronavirus Disease 2019 (COVID-19) caused by the emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is associated with a crude case fatality rate of about 0.5–10 % depending on locality. A few clinically approved drugs, such as remdesivir, chloroquine, hydroxychloroquine, nafamostat, camostat, and ivermectin, exhibited anti-SARS-CoV-2 activity in vitro and/or in a small number of patients. However, their clinical use may be limited by anti-SARS-CoV-2 50 % maximal effective concentrations (EC50) that exceeded their achievable peak serum concentrations (Cmax), side effects, and/or availability. To find more immediately available COVID-19 antivirals, we established a two-tier drug screening system that combines SARS-CoV-2 enzyme-linked immunosorbent assay and cell viability assay, and applied it to screen a library consisting 1528 FDA-approved drugs. Cetilistat (anti-pancreatic lipase), diiodohydroxyquinoline (anti-parasitic), abiraterone acetate (synthetic androstane steroid), and bexarotene (antineoplastic retinoid) exhibited potent in vitro anti-SARS-CoV-2 activity (EC50 1.13–2.01 μM). Bexarotene demonstrated the highest Cmax:EC50 ratio (1.69) which was higher than those of chloroquine, hydroxychloroquine, and ivermectin. These results demonstrated the efficacy of the two-tier screening system and identified potential COVID-19 treatments which can achieve effective levels if given by inhalation or systemically depending on their pharmacokinetics.
1. Introduction
Coronaviruses are positive sense, single stranded, enveloped RNA viruses that have repeatedly crossed species barriers to cause disease in human and animals [1]. In the past two decades, three novel human-pathogenic coronaviruses have emerged to cause epidemics of severe respiratory infection among human, including severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, Middle East respiratory syndrome coronavirus (MERS-CoV) since 2012, and most recently SARS-CoV-2 since December 2019 [[2], [3], [4]]. Within just 4 months, the number of patients with SARS-CoV-2 infection, or Coronavirus Disease 2019 (COVID-19), has exceeded the total number of cases of SARS and MERS by nearly 100 times, with more than 1.2 million confirmed cases and over 60,000 deaths globally [5]. The clinical severity of COVID-19 ranges from asymptomatic infection to fatal disease. The disease is usually mild in children, but severe infection in immunocompromised and elderly patients may be associated with a crude case fatality rate of about 15 % [[6], [7], [8]]. Patient with severe COVID-19 may develop acute respiratory distress syndrome, multiorgan dysfunction syndrome, and other extrapulmonary manifestations such as lymphopenia, diarrhea, confusion, deranged liver and renal function tests, and elevated d-dimer, fibrinogen, lactate dehydrogenase, and inflammatory marker levels [9,10]. A major reason for the poor clinical outcome of COVID-19 patients and difficulty in controlling the expansion of the pandemic is the lack of effective vaccine or antiviral for treatment and prophylaxis.
Similar to other emerging viral infections, the de novo development of antiviral drugs would inevitably lag behind the rapid progression of the epidemic [11]. Drug repurposing is therefore a feasible strategy to quickly identify clinically approved drugs with known pharmacological properties and safety profiles that can be immediately used in clinical trial settings. A number of existing drugs, such as remedsivir, chloroquine, hydroxychloroquine, nafamostat, camostat, and ivermectin, have been reported to exhibit anti-SARS-CoV-2 activity in vitro and/or in a very small number of patients [[12], [13], [14], [15]]. Remdesivir is a nucleotide analogue with broad-spectrum antiviral activities including against SARS-CoV-2 [12]. Chloroquine and hydroxychloroquine are mildly immunosuppressive drugs used in the treatment of autoimmune diseases and malaria that exhibited 50 % maximal effective concentration (EC50) at or above the peak serum concentration (Cmax) achievable with standard dosing in human [12,16]. A recent non-randomized small-scale clinical study showed that hydroxychloroquine with or without azithromycin significantly reduced the viral load and duration of virus shedding in 20 COVID-19 patients [13]. Nafamostat and camostat are a serine protease inhibitor used in the treatment of chronic pancreatitis and reflux esophagitis [14]. Ivermectin is a macrocyclic lactone used in the treatment of various parasitic infections [15]. However, data from well-designed randomized controlled trials for these drugs are not yet available. Therefore, there is an urgent need to search for additional drug compounds with anti-SARS-CoV-2 activity among clinically approved drugs. In this study, we first established a robust two-tier drug screening system by combining SARS-CoV-2 enzyme-linked immunosorbent assay with cell viability assay, and then applied it to screen an FDA-approved drug compound library. We successfully identified a number of drug compounds with anti-SARS-CoV-2 activity, including bexarotene which has broad-spectrum anti-coronaviral activity and a higher Cmax to EC50 ratio than most other reported potential anti-SARS-CoV-2 agents.
2. Materials and methods
2.1. Viruses, cell lines, and drug compounds
SARS-CoV-2 HKU-001a (GenBank accession number: MT230904) was isolated from the nasopharyngeal aspirate specimen of a laboratory-confirmed COVID-19 patient in Hong Kong [17]. MERS-CoV EMC/2012 strain (GenBank accession number: NC_019843.3) was kindly provided by Ron Rouchier (Erasmus Medical Center, Rotterdam, the Netherlands) [18]. The viruses were propagated in VeroE6 cells and kept at −80 °C in aliquots until use. Plaque forming unit (PFU) and TCID50 assays were performed to titrate the cultured SARS-CoV-2. VeroE6 (ATCC® CRL-1586™) and Caco2 cells (ATCC® HTB-37™) were purchased from ATCC and maintained in Dulbecco’s modified eagle medium (DMEM, Gibco, CA, USA) culture medium supplemented with 10 % heat-inactivated fetal bovine serum (FBS, Gibco), 50 U/mL penicillin, and 50 μg/ml streptomycin as previously described [17]. All experiments involving live SARS-CoV-2 and MERS-CoV followed the approved standard operating procedures of the Biosafety Level 3 facility at the Department of Microbiology, The University of Hong Kong, as previously described [19,20]. The FDA-approved drug library (Cat# HY-L022) and all the tested drug compounds were purchased from MedChem Express (Monmouth Junction, NJ, USA).
2.2. Cell viability assay and CPE inhibition assay
The CellTiterGlo® luminescent assay (Promega Corporation, Madison, WI, USA) was performed to detect the cytotoxicity of the selected drug compounds as previously described [21]. Briefly, VeroE6 cells (4 × 104 cells/well) were incubated with different concentrations of the individual compound for 48 h, followed by the addition of substrate and measurement of luminance 10 min later. The CC50 of the drug compounds were calculated by Sigma plot (SPSS) in an Excel add-in ED50V10. The CPE inhibition assay was performed as previously described with slight modifications to evaluate the individual drug compounds’ cell protection effects against SARS-CoV-2 infection [21]. Briefly, VeroE6 cells seeded in 96-well plates were infected with SARS-CoV-2 for 1 h with 0.01 multiplicity of infection (MOI), followed by washing with phosphate-buffered saline (PBS) and replacement of fresh DMEM medium containing serially diluted drug compounds and 0.1 % DMSO as negative control. The cell viability of each well was determined at 3 days post-infection (dpi) by the CellTiterGlo® luminescent assay.
2.3. ELISA
ELISA was performed to determine the amount of viral N protein expression in the culture supernatant of SARS-CoV-2-infected cells using a similar approach as previously described [22]. Briefly, 500 ng/well of mouse-anti-SARS-CoV-N monoclonal antibody (clone 14E7A11A8) was coated in 96-well ELISA plates for overnight incubation at 4 °C, followed by blocking with 2.5 % FBS plus 2.5 % FBS in PBS with Tween 20 (Sigma-Aldrich, St. Louis, MO, USA) for 2 h at 37 °C. After washing, the infectious culture supernatants were transferred to the ELISA plates accordingly (50 μL, incubated at room temperature for 2 h), followed by intensive wash and addition of another 50 μL/well rabbit anti-SARS-CoV-N polyclonal antibody (1:4000), the secondary goat-anti-rabbit horseradish peroxidase (HRP) antibody (1:1500, Invitrogen, Carlsbad, CA, USA), the 3,3′,5,5′-tetramethylbenzidine (TMB) solution (Invitrogen), and the stop solution (0.1 M HCl). Subsequently, the optical density of each well was read at 450 nm (OD450) using VICTOR 3 multi-label plate reader (PerkinElmer, Inc., Waltham, MA, USA). The rabbit anti-SARS-CoV-N monoclonal antibody showed good cross-reactivity against SARS-CoV-2-N [17,23].
2.4. FDA drug compound library screening
To identify anti-SARS-CoV-2 inhibitors, confluent VeroE6 cells in 96-well culture plates (4 × 104 cells/well) in triplicates were infected with SARS-CoV-2 at 0.004 MOI. One hour after virus adsorption, the inoculum was removed and then drug-containing medium (10μM) was added. Next, after 72 h, 20 μL of cell culture supernatant was diluted into 80 μL of PBS before adding to the antibody-coated ELISA plate for SARS-CoV-2-N detection, while another 20 μL/well of CellTiterGlo® substrate was added to the original cell culture plates for cell viability evaluation by CPE inhibition assay.
2.5. Viral load reduction assay
Viral load reduction assay was performed on VeroE6 and Caco2 cells, as described previously with modifications [24,25]. Supernatant samples from the infected cells were harvested at different time-points for qRT-PCR analysis of virus replication. Briefly, 100 μl of viral supernatant was lyzed with 400 μl of AVL buffer and then extracted for total RNA with the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany). Real-time one-step qRT-PCR was used for quantitation of SARS-CoV-2 and SARS-CoV viral load using the QuantiNova Probe RT-PCR kit (Qiagen) with a LightCycler 480 Real-Time PCR System (Roche) as previously described [26]. Each 20 μl reaction mixture contained 10 μl of 2×QuantiNova Probe RT-PCR Master Mix, 1.2 μl of RNase-free water, 0.2 μl of QuantiNova Probe RT-Mix, 1.6 μl each of 10 μM forward and reverse primer, 0.4 μl of 10 μM probe, and 5 μl of extracted RNA as the template. Reactions were incubated at 45 °C for 10 min for reverse transcription, 95 °C for 5 min for denaturation, followed by 45 cycles of 95 °C for 5 s and 55 °C for 30 s. Signal detection and measurement were taken in each cycle after the annealing step. The cycling profile ended with a cooling step at 40 °C for 30 s. The primers and probe sequences were against the RNA-dependent RNA polymerase/Helicase (RdRP/Hel) gene region of SARS-CoV-2: Forward primer: 5′ CGCATACAGTCTTRCAGGCT-3′; Reverse primer: 5′-GTGTGATGTTGAWATGACATGGTC-3′; specific probe: 5′-FAM TTAAGATGTGGTGCTTGCATACGTAGAC-IABkFQ-3′ [26].
2.6. Plaque reduction assay
Plaque reduction assay was performed to plot the 50 % antiviral effective dose (EC50) as we previously described with slight modifications [24,27]. Briefly, VeroE6 cells were seeded at 4 × 105 cells/well in 12-well tissue culture plates on the day before carrying out the assay. After 24 h of incubation, 50 plaque-forming units (PFU) of SARS-CoV-2 were added to the cell monolayer with or without the addition of drug compounds and the plates were further incubated for 1 h at 37 °C in 5% CO2 before removal of unbound viral particles by aspiration of the media and washing once with DMEM. Monolayers were then overlaid with media containing 1% low melting agarose (Cambrex Corporation, New Jersey, USA) in DMEM and appropriate concentrations of individual compound, inverted and incubated as above for another 72 h. The wells were then fixed with 10 % formaldehyde (BDH, Merck, Darmstadt, Germany) overnight. After removal of the agarose plugs, the monolayers were stained with 0.7 % crystal violet (BDH, Merck) and the plaques counted. The percentage of plaque inhibition relative to the control (i.e. without the addition of compound) wells were determined for each drug compound concentration. The EC50 was calculated using Sigma plot (SPSS) in an Excel add-in ED50V10. The plaque reduction assay experiments were performed in triplicate and repeated twice for confirmation.
2.7. Time-of-drug-addition assay
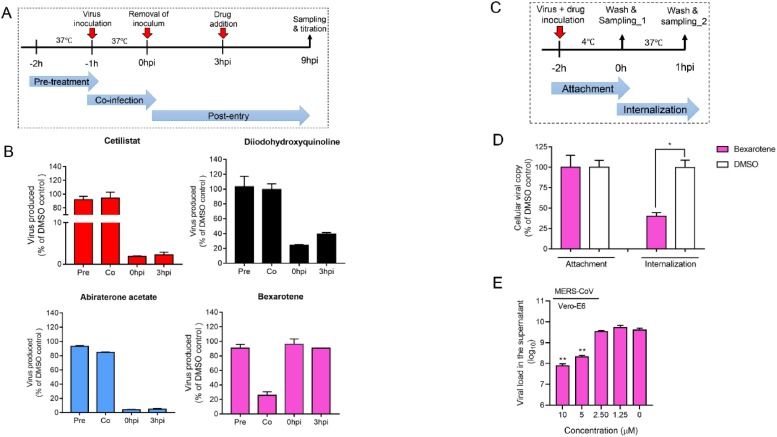
Time-of-drug-addition assay was performed for the selected compound as previously described with slight modifications [25]. Briefly, VeroE6 cells were seeded in 24-well plates (2 × 105 cells/well). The cells were inoculated with SARS-CoV-2 (MOI = 0.500) and then incubated for 1 h for virus internalization. Drugs were added at different time points of the virus replication cycle, including: pre-treatment (From -2 h post infection (hpi) to -1 hpi), co-infection with virus (-1 to 0 hpi) or post-infection (0 to +1 hpi). Dimethyl sulfoxide (0.5 %) was included as a negative control. Other details were depicted as schematic representations in Fig. 6.
Fig. 6.
Anti-SARS-CoV-2 modes of action of the four selected drug compounds. (A) Time-of-drug-addition assay was performed to determine the steps of the viral replication cycle targeted by each of the four identified drug compounds. The assay was conducted at 37 °C. (B) Viral load in the treated VeroE6 cell culture supernatants normalized by DMSO as control. (C) Virus entry assay with or without bexarotene addition. Viral attachment was performed at 4℃ and then shifted to 37℃ to enable virus internalization. (D) Intracellular viral load in the treated VeroE6 cells normalized by DMSO as control. (E) Viral load reduction assay showing the dose-dependent anti-MERS-CoV activity of bexarotene (10μM). VeroE6 cells were infected with MERS-CoV (MOI = 0.01) and treated with bexarotene, and the culture supernatant was then collected at 48 h post-inoculation for viral load quantitation by qRT-PCR. *P indicates < 0.05 and ** indicates P < 0.01 (Student’s t-test). All the experiments were performed in triplicate and replicated twice. The results are shown as mean ± standard deviations.
2.8. Immunofluorescence staining
Antigen expression in SARS-CoV-2-infected cells was detected with an in-house rabbit antiserum against SARS-CoV-N protein, which cross-reacted with the SARS-CoV-2-N protein due to their high amino acid homologies [17,23]. Cell nuclei were labelled with the DAPI nucleic acid stain from Thermo Fisher Scientific (Waltham, MA, USA). The Alexa Fluor secondary antibody was obtained from Thermo Fisher Scientific. Mounting was performed with the Diamond Prolong Antifade mountant from Thermo Fisher Scientific.
3. Results
3.1. Characterization of SARS-CoV-2 replication kinetics
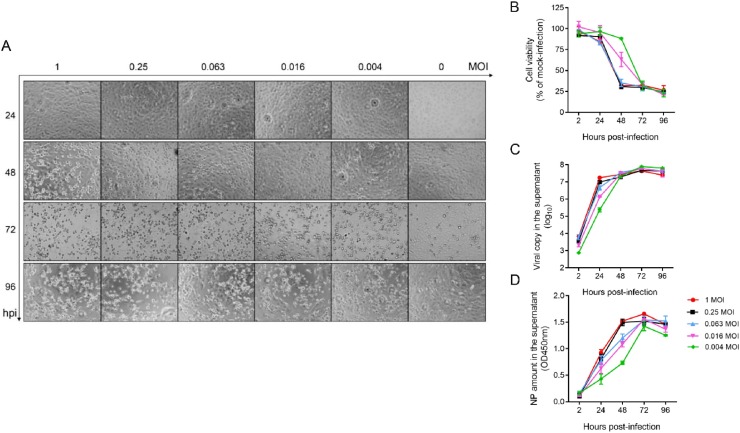
To develop a drug screening system for SARS-CoV-2, we first characterized the viral replication kinetics of SARS-CoV-2 in VeroE6 cells and compared the signal dynamic range utilizing three approaches. These approaches included luminescent cell viability assay with CellTiterGlo® for evaluation of virus induced-cytopathic effects (CPE), viral load quantitation assay with quantitative RT-PCR (qRT-PCR) targeting the viral RNA-dependent RNA polymerase (RdRp)/ helicase (Hel) genes, and viral nucleocapsid (N) protein expression with enzyme-linked immunosorbent assay (ELISA). VeroE6 cells were used because they robustly support SARS-CoV-2 replication [17]. Time-dependent observation of CPE development was recorded using different multiplicities of infection (MOI) (1.000, 0.250, 0.063, 0.016, and 0.004). SARS-CoV-2 infection (MOI = 1.000) caused significant CPE in VeroE6 cells as early as 48 h post-infection (hpi) (Fig. 1 A). At 72 hpi, prominent CPE were detected in the virus-infected VeroE6 cells even with the lowest MOI of 0.004, while the morphology of the non-infected VeroE6 cells remained intact. This observation was in line with the cell viability determined by using the CellTiter-Glo® substrate, which is a reagent that generates a luminescent signal directly proportional to the amount of adenosine triphosphate (ATP) present and is proportional to the number of metabolically active cells. As shown in Fig. 1B, there was approximately 75 % reduction in cell viability at 48 hpi with MOI of 1.000, 0.250, or 0.063 and at 72 hpi with MOI of 0.016 or 0.004. In the qRT-PCR assay, viral RNA load in the culture supernatant markedly increased by ≥2 logs gene copies per reaction within 24 hpi (∼2 logs for 0.004 MOI, ∼3 logs for 0.016 MOI, and ∼3.5 logs for 0.063, 0.250, and 1.000 MOI) (Fig. 1C). Regardless of the MOI, the viral RNA load plateaued at 48 hpi. The viral N protein expression followed a delayed but similar pattern as that of the viral RNA load, with the peak viral antigen expression being observed at 72 hpi (Fig. 1D).
Fig. 1.
Development of a two-tier system for anti-SARS-CoV-2 drug compound screening. VeroE6 cells seeded in a 96-well plate were infected with SARS-CoV-2 of various multiplicities of infection (MOIs)) as indicated, followed by phosphate buffered saline (PBS) wash and replacement of fresh Dulbecco’s modified eagle medium (DMEM). Various time points of data collection were performed for (A) Cytopathic effects (CPE) observed by a bright-field at 20× magnification. (B) Cell viability of each treatment group normalized with that of the mock-infected cells. (C) Cell culture supernatant was collected at the indicated time points with viral copy number determined by quantitative RT-PCR (qRT-PCR). (D) The supernatant was concomitantly applied for ELISA to measure the SARS-CoV-2-nucleoprotein (NP) protein amount. The experiments were carried out in triplicate. The results are shown as mean ± standard deviation.
3.2. Establishment of a two-tier drug screening system for rapid identification of antivirals against SARS-CoV-2 from an FDA-approved drug compound library
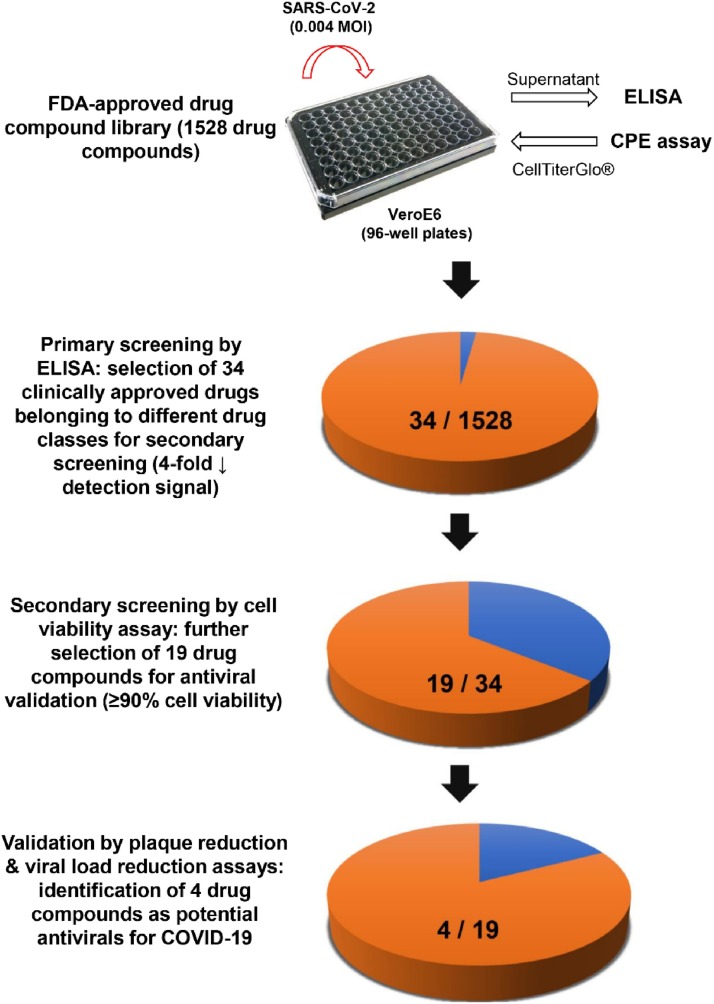
The signal window is determined by the upper and lower boundaries of the assay readout and is considered a major parameter for the overall assay quality measurement. The qRT-PCR assay exhibited the highest signal window (∼4 log difference between SARS-CoV-2-infected and non-infected samples) but was laborious, and was thus inefficient as a high-throughput drug compound screening assay. On the other hand, the ELISA (∼6 folds) and cell viability assay (∼4 folds) also exhibited reasonably high signal differences between infected cells and baseline at 72 hpi and provided a wide signal window. Therefore, we exploited the combination of ELISA and cell viability assay as a two-tier drug screening system for finding potential SARS-CoV-2 antivirals. This combinational screening system is rapid, non-labor-intensive, and can easily exclude false-positive results due to cytotoxicity. Using this newly established system, we screened an FDA-approved drug library containing 1528 drug compounds under the optimized conditions of 0.004 MOI at 72 hpi with each drug concentration fixed at 10μM. At 72 hpi, 20 μL/well of cell culture supernatant was harvested for SARS-CoV-2-N detection before addition of another 40 μL/well CellTiter-Glo® substrate for determination of cell viability (Fig. 2 ). Among the 1528 drug compounds, we identified 34 primary hit drug compounds with ≥4-fold reduction of detection signal in ELISA. In the secondary screen by cell viability assay, 19 of these 34 drug compounds showed <10 % CPE at 72 hpi (Table 1 ). Notably, hydroxychloroquine, nafamostat, and ivermectin which were previously shown to exhibit anti-SARS-CoV-2 activity were also identified in our primary screen with ELISA, but they showed >10 % CPE at 72 hpi and were therefore excluded from the 19 drug compound list. Camostat was not detected in the primary screen but this might be related to its EC50 being higher than 10μM which was used in our primary screen. Remdesivir was not included in the library and was therefore not identified.
Fig. 2.
Schematic representation of the study design. Using the newly established two-tier drug compound screening system, an FDA-approved drug compound library consisting 1528 drug compounds was screened for potential anti-SARS-CoV-2 agents. Primary screening by ELISA identified 34 drug compounds with 4-fold reduction in the detection signal. Secondary screening by cell viability assay further selected 19 of the 34 drug compounds that exhibited ≥90 % cell viability. Four drug compounds were then prioritized for cytotoxicity and antiviral activity evaluation by plaque reduction and viral load reduction assays.
Table 1.
Primary hit drug compounds identified from an FDA-approved drug library with a two-tier drug screening platform for SARS-CoV-2.
| Compound | Drug class | Main clinical use(s) |
|---|---|---|
| Abiraterone (acetate) | Synthetic androstane steroid | Prostate cancer |
| Asenapine (hydrochloride) | Atypical antipsychotic | Schizophrenia |
| Azacytidine | Nucleoside analogue | Myelodysplastic syndrome and acute myeloid leukemia |
| Bexarotene | Retinoid (retinoid X receptor activator) | Cutaneous T cell lymphoma |
| Candesartan (Cilexetil) | Angiotensin receptor blocker | Hypertension and congestive heart failure |
| Cetilistat | Pancreatic lipase inhibitor | Obesity |
| Chloroquine (diphosphate) | 4-Aminoquinoline | Malaria and amoebic liver abscess |
| Ciclesonide | Glucocorticoid | Asthma and allergic rhinitis |
| Diethylstilbestrol | Nonsteroidal estrogen | Prostate cancer |
| Diiodohydroxyquinoline | Quinoline derivative | Amoebiasis |
| Fluoxetine (hydrochloride) | Selective serotonin reuptake inhibitor | Depression |
| Micafungin (sodium) | Echinocandin | Fungal infection |
| Permethrin | Insecticide | Scabies and lice |
| Pimavanserin | Atypical antipsychotic | Parkinson’s disease psychosis |
| Raloxifene (hydrochloride) | Selective estrogen receptor modulator | Osteoporosis |
| Sofalcone | Synthetic sophoradin analogue | Gastrointestinal tract mucosal protection |
| Tamoxifen (Citrate) | Selective estrogen receptor modulator | Breast cancer, infertility, and gynecomastia |
| Tilorone (dihydrochloride) | Interferon inducer | Antiviral |
| Tocofersolan | Synthetic vitamin E | Vitamin E deficiency |
3.3. In vitro antiviral evaluation of the selected drug compounds
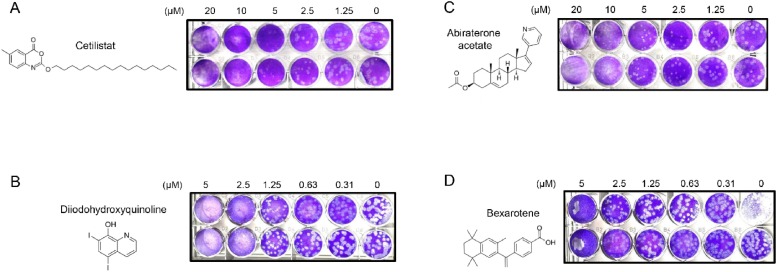
Next, these 19 primary hit drug compounds were subjected to viral load reduction assay for prioritization based on their dose-dependent anti-SARS-CoV-2 effects. Four drug compounds, namely, cetilistat, diiodohydroxyquinoline, abiraterone acetate, and bexarotene were identified as the most potent anti-SARS-CoV-2 drug compounds using the cut-off of 90 % effective concentrations (EC90) <10 μM after treating VeroE6 cells with 0.01 MOI of SARS-CoV-2 for 48 h. Using plaque reduction assay, the EC50 of the drug compounds were determined to be 1.13 μM (cetilistat), 1.38μM (diiodohydroxyquinoline), 1.94 μM (abiraterone acetate), and 2.01 μM (bexarotene) (Fig. 3 A–D). At 48 hpi, the 50 % cytotoxic concentrations (CC50) of cetilistat, diiodohydroxyquinoline, abiraterone acetate, and bexarotene were >100 μM, >100 μM, 92.35 μM, and 38.21 μM, respectively (Table 2 ). The selectivity index of these four drug compounds were >88.50 (cetilistat), >72.46 (diiodohydroxyquinoline), 47.60 (abiraterone acetate), and 19.01 (bexarotene), respectively.
Fig. 3.
Identification of four FDA-approved drug compounds with potent anti-SARS-CoV-2 activity. Chemical structures of the selected compounds and photos of plaque reduction assay are shown: (A) cetilistat, (B) diiodohydroxyquinoline, (C) abiraterone acetate, and (D) bexarotene.
Table 2.
Antiviral activity and cytotoxicity of the four most potent drug compounds.
| Compound | CC50 (μM) (CellTiterGlo®)a |
EC90 (μM) (viral load reduction assay) |
EC50 (μM) (plaque reduction assay) |
Select index (CC50/EC50) |
|---|---|---|---|---|
| Cetilistat | >100.00 | 2.90 | 1.13 | >88.50 |
| Diiodohydroxyquinoline | >100.00 | 4.50 | 1.38 | >72.46 |
| Abiraterone (acetate) | 92.35 | 8.40 | 1.94 | 47.60 |
| Bexarotene | 38.21 | 9.40 | 2.01 | 19.01 |
Abbreviations: CC50, 50 % cytotoxic concentration; EC50, 50 % maximal effective concentration; EC90, 90 % maximal effective concentration.
>100 indicates the highest drug concentration tested in the cytotoxicity assay was 100μM.
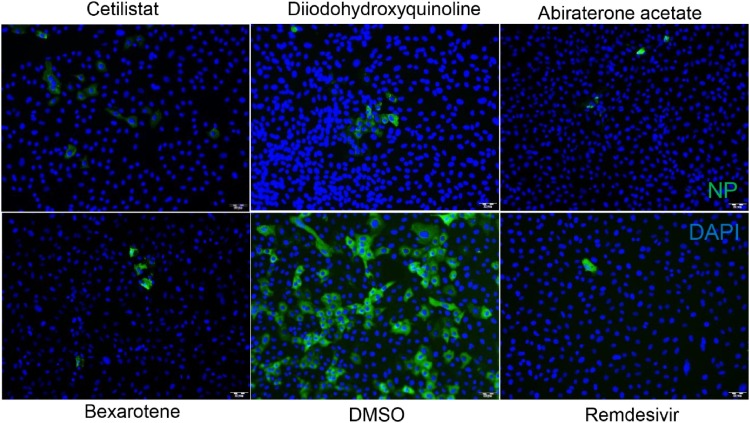
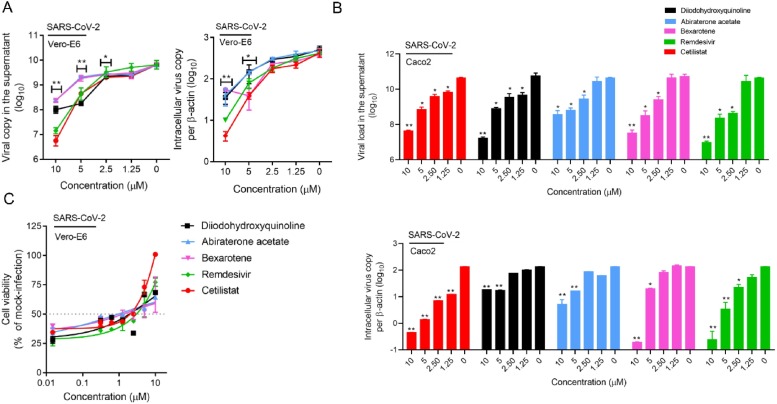
To more clearly demonstrate the anti-SARS-CoV-2 activities of these four identified drug compounds, we included remdesivir as a comparator drug. Similar to remdesivir, treatment with any one of the four drug compounds below the non-toxic drug concentration of 10μM, markedly suppressed SARS-CoV-2-N protein expression (Fig. 4 ). As shown in Fig. 5 A, all four drug compounds exhibited dose-dependent viral load reduction with magnitudes of 1 log to 3 logs. At 10μM of drug concentration, treatment with cetilistat exhibited about 3 logs reduction at 48 hpi, as comparable level achieved by remdesivir which demonstrated an EC50 of 1.04μM in our study. Similar levels of dose-dependent viral load reduction were observed in SARS-CoV-2-infected Caco2 (human colorectal adenocarcinoma) cell culture supernatants and cell lysates (Fig. 5B). In addition to viral load reduction, treatment with any one of these four drug compounds provided cell protection effects. In the CPE inhibition assay, treatment with 10μM of cetilistat completely inhibited CPE development in VeroE6 cells at 72 hpi, while diiodohydroxyquinoline, abiraterone acetate, and bexarotene achieved up to ∼70 % CPE inhibition (Fig. 5C). Overall, these results demonstrated that cetilistat, diiodohydroxyquinoline, abiraterone acetate, and bexarotene inhibited the viral load, viral antigen expression, and infectious viral particle production of SARS-CoV-2, and protected SARS-CoV-2-induced cell damages at comparable levels of remdesivir.
Fig. 4.
Immunofluorescence staining showing the anti-SARS-CoV-2 effects of the four selected drug compounds. Fixation and staining were performed on SARS-CoV-2-infected (MOI = 0.100) VeroE6 cells after treatment with cetilistat, diiodohydroxyquinoline, abiraterone acetate, or bexarotene (10μM each) and incubated at 37 °C with 5% CO2 for 24 h. The SARS-CoV-2-N antigens and cell nuclei (DAPI) were stained in green and blue, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Fig. 5.
Characterization of the anti-SARS-CoV-2 activity of each of the four selected drug compounds in vitro. (A) VeroE6 and (B) Caco2 cells were infected with SARS-CoV-2 and treated with different concentrations of the selected drug compounds as indicated. The viral load under each condition was collected at 48 hpi for viral load reduction assay by qRT-PCR. Intracellular viral loads were normalized by human β-actin. (C) VeroE6 cells were infected with SARS-CoV-2 and treated with different concentrations of the selected drug compounds as indicated and evaluated by the CPE inhibition assay at 72 hpi. Remdesivir was used as a positive control in all of these experiments. One-way ANOVA was used to compare the treatment groups with the 0μM (negative control) group. *P indicates < 0.05 and ** indicates P < 0.01 (Student’s t-test). All the experiments were performed in triplicate and replicated twice. The results are shown as mean ± standard deviations.
3.4. Modes of action of cetilistat, diiodohydroxyquinoline, abiraterone acetate, and bexarotene against SARS-CoV-2
To investigate which steps of the SARS-CoV-2 replication cycle were interrupted by the selected drug compounds, we performed a time-of-drug-addition assay by treating virus-infected VeroE6 cells to each drug compound at different time points, followed by viral titer measurements at 9 hpi, when the first round of progeny virions were detectable in the cell culture supernatant. VeroE6 cells were infected by 0.500 MOI of SARS-CoV-2, before and after which four different treatments were carried out (Fig. 6 A). Addition of cetilistat, diiodohydroxyquinoline, or abiraterone acetate at 0 hpi and 3 hpi, but not pre-treatment with host cells or during virus adsorption (co-infection) significantly reduced viral replication, suggesting that these three drug compounds disrupted the post-entry events of the SARS-CoV-2 replication cycle (Fig. 6B). In contrast, bexarotene was only effective when co-infected with cells, suggesting that it interfered with virus entry either by blocking the host ACE2 receptor or the viral components required for cell attachment and entry. To further delineate the step of SARS-CoV-2 entry that is affected by bexarotene, VeroE6 cells were placed at 4 °C for 2 h which only permitted virus attachment to the cell surface and then at 37 °C for 1 h which enabled virus internalization (Fig. 6C). As shown in Fig. 6D, bexarotene did not block virus-host receptor binding, but significantly reduced viral load during virus internationalization. These results suggested that bexarotene interfered with SARS-CoV-2 internalization without blocking its binding to the host cell surface. Interestingly, bexarotene also demonstrated antiviral activity against the highly virulent MERS-CoV (EC50 of 2.12 μM), suggesting that it has the potential of being a “pan-coronavirus” antiviral (Fig. 6E).
4. Discussion
The newly established two-tier screening system in this study was robust and able to identify other drug compounds that were shown to exhibit anti-SARS-CoV-2 activity, including chloroquine, hydroxychloroquine, ivermectin, and nafamostat. This allowed us to rapidly and systematically screen a drug library consisting 1528 FDA-approved drug compounds and identify four drug compounds that showed anit-SARS-CoV-2 activities at low micromolar concentrations (EC50 = 1.13–2.01). Cetilistat is a pancreatic lipase inhibitor that blocks fat digestion and absorption used in the treatment of obesity [28]. Diiodohydroxylquinoline, also known as uidoquinol, is a quinolone derivative that is used as a luminal amebicide for the treatment of amoebiasis [29]. Cetilistat is rapidly hydrolyzed into its metabolites in the presence of bile and diiodohydroxylquinoline is poorly absorbed into circulatory system [28,30]. Importantly, about 15–20 % of COVID-19 patients develop gastrointestinal symptoms with some also having detectable viral RNA and even infectious virus particles [31]. In a SARS-CoV-2-infected hamster model, it was found that the animals’ intestines exhibited severe inflammation with detectable viral RNA and abundant viral nucleocapsid protein expression [23]. These findings showed that like in SARS, feces might also be a potential source of infection in COVID-19 [2,32]. Thus, besides being administered by inhalation, oral cetilisat and diiodohydroxylquinoline might have a role as topical luminal antivirals to reduce viral shedding in the gastrointestinal tract.
Both abiraterone acetate and bexarotene are non-chemotherapeutic antineoplastic drugs with limited immunosuppressive effects. Abiraterone acetate is used in combination with corticosteroid to treat refractory prostate cancer through androgen deprivation by inhibition of the androgen synthesizing enzyme CYP17A1 [33,34]. Bexarotene is a third-generation retinoid used in the treatment of cutaneous T cell lymphoma, non-small cell lung cancer, and breast cancer [[35], [36], [37], [38]]. We showed that abiraterone acetate and bexarotene potently inhibited SARS-CoV-2 replication in vitro with EC50 of 1.94 μM and 2.01 μM, respectively. Of particular interest is bexarotene which has a peak serum concentration (Cmax) of 3.39 μM after an oral dose of 300 mg/m2 [39]. The Cmax to EC50 ratio of bexarotene for SARS-CoV-2 (1.69) is higher than those achieved with standard dosing of chloroquine, hydroxychloroquine, and ivermectin (0.02–1.04). Moreover, the time required to achieve Cmax (Tmax) with standard oral dosing of bexarotene is short (2.5 h), which is important especially in COVID-19 patients who are acutely ill with rapid clinical deterioration [39]. We have previously shown that AM580 and tamibarotene, which belong to the same drug class as bexarotene, exhibited broad-spectrum antiviral activities against coronaviruses (MERS-CoV and SARS-CoV), influenza viruses, enterovirus A71, Zika virus, and adenovirus [21]. We therefore additionally tested the antiviral activity of bexarotene against the highly virulent MERS-CoV, and showed that it indeed also inhibited MERS-CoV with an EC50 of 2.12 μM. The potential of bexarotene and its related analogue compounds as “pan-coronavirus” agents should be further evaluated in suitable animal models for COVID-19 and other human-pathogenic coronaviruses.
In conclusion, the robust two-tier drug compound screening system established in this study represented a novel platform for conducting drug discovery programmes for COVID-19. The topical and/or systemic effects of cetilistat, diiodohydroxylquinoline, abiraterone acetate, and bexarotene should be further evaluated in suitable ex vivo human organ culture or organoids, animal models, and/or clinical trials.
Author contributions
SY, JFWC, and KYY had roles in the study design, data collection, data analysis, data interpretation, and writing of the manuscript. KKHC, CCYC, JOLT, RL, JC, KT, LLC, KW, JPC, ZWY, GL, HC and DYJ had roles in the experiments, data collection, data analysis, and/or data interpretation. All authors reviewed and approved the final version of the manuscript.
Declaration of Competing Interest
JFWC has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited, and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. The other authors declared no conflict of interests. The funding sources had no role in study design, data collection, analysis or interpretation or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgements
This study was partly supported by funding from Health@InnoHK (Centre for Virology, Vaccinology and Therapeutics), Innovation and Technology Commission, The Government of the Hong Kong Special Administrative Region of the People’s Republic of China; the National Program on Key Research Project of China (grant no. 2020YFA0707500 and 2020YFA0707504); the Theme-Based Research Scheme (T11/707/15) of the Research Grants Council, Hong Kong Special Administrative Region; and the High Level-Hospital Program, Health Commission of Guangdong Province, China; and donations of Richard Yu and Carol Yu, the Shaw Foundation of Hong Kong, Michael Seak-Kan Tong, May Tam Mak Mei Yin, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, and Kai Chong Tong. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Contributor Information
Jasper F.W. Chan, Email: jfwchan@hku.hk.
Kwok-Yung Yuen, Email: kyyuen@hku.hk.
References
- 1.Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization . 2020. Coronavirus Disease 2019 (COVID-19) Situation Report – 77.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200406-sitrep-77-covid-19.pdf?sfvrsn=21d1e632_2 [Google Scholar]
- 6.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai J., Xu J., Lin D., Yang Z., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Liu P. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020:ciaa198. doi: 10.1093/cid/ciaa198. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. pii: S1473-3099(20)30243-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. pii: S0092-8674(20)30229-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020 doi: 10.1016/j.antiviral.2020.104787. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020:ciaa237. doi: 10.1093/cid/ciaa237. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X. An observational study on the comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV: implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2013;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 19.Chan J.F., Chan K.H., Choi G.K., To K.K., Tse H., Cai J.P., Yeung M.L., Cheng V.C., Chen H., Che X.Y. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [accepted and in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S., Chu H., Chan J.F., Ye Z.W., Wen L., Yan B., Lai P.M., Tee K.M., Huang J., Chen D. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 2019;10:120. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Che X.Y., Qiu L.W., Liao Z.Y., Wang Y.D., Wen K., Pan Y.X., Hao W., Mei Y.B., Cheng V.C., Yuen K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020:ciaa325. doi: 10.1093/cid/ciaa325. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li C.P.T., Dai J., Mok F.K., Chen H. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan S., Chu H., Zhao H., Zhang K., Singh K., Chow B.K., Kao R.Y., Zhou J., Zheng B.J. Identification of a small-molecule inhibitor of influenza virus via disrupting the subunits interaction of the viral polymerase. Antiviral Res. 2016;125:34–42. doi: 10.1016/j.antiviral.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00310-20. pii: JCM.00310-20. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan S., Chu H., Zhang K., Ye J., Singh K., Kao R.Y., Chow B.K., Zhou J., Zheng B.J. A novel small-molecule compound disrupts influenza A virus PB2 cap-binding and inhibits viral replication. J. Antimicrob. Chemother. 2016;71:2489–2497. doi: 10.1093/jac/dkw194. [DOI] [PubMed] [Google Scholar]
- 28.Bryson A., de la Motte S., Dunk C. Reduction of dietary fat absorption by the novel gastrointestinal lipase inhibitor cetilistat in healthy volunteers. Br. J. Clin. Pharmacol. 2009;67:309–315. doi: 10.1111/j.1365-2125.2008.03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickards A.G. The treatment of amoebiasis with diodoquin. J. Trop. Med. Hyg. 1949;52:33–38. [PubMed] [Google Scholar]
- 30.Padwal R. Cetilistat, a new lipase inhibitor for the treatment of obesity. Curr. Opin. Investig. Drugs. 2008;9:414–421. [PubMed] [Google Scholar]
- 31.Cheung K.S., Hung I.F., Chan P.P., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W., Tam A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. pii: S0016-5085(20)30448-0. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung I.F., Cheng V.C., Wu A.K., Tang B.S., Chan K.H., Chu C.M., Wong M.M., Hui W.T., Poon L.L., Tse D.M. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duc I., Bonnet P., Duranti V., Cardinali S., Riviere A., De Giovanni A., Shields-Botella J., Barcelo G., Adje N., Carniato D. ) In vitro and in vivo models for the evaluation of potent inhibitors of male rat 17alpha-hydroxylase/C17,20-lyase. J. Steroid Biochem. Mol. Biol. 2003;84:537–542. doi: 10.1016/s0960-0760(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 34.Arasaratnam M., Crumbaker M., Bhatnagar A., McKay Mj, Molloy Mp, Gurney H. Inter- and intra-patient variability in pharmacokinetics of abiraterone acetate in metastatic prostate cancer. Cancer Chemother. Pharmacol. 2019;84:139–146. doi: 10.1007/s00280-019-03862-x. [DOI] [PubMed] [Google Scholar]
- 35.Boehm M.F., Zhang L., Zhi L., McClurg M.R., Berger E., Wagoner M., Mais D.E., Suto C.M. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J. Med. Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 36.Gniadecki R., Assaf C., Bagot M., Dummer R., Duvic M., Knobler R., Ranki A., Schwandt P., Whittaker S. The optimal use of bexarotene in cutaneous T-cell lymphoma. Br. J. Dermatol. 2007;157:433–440. doi: 10.1111/j.1365-2133.2007.07975.x. [DOI] [PubMed] [Google Scholar]
- 37.Dragnev K.H., Petty W.J., Shah S.J., Lewis L.D., Black C.C., Memoli V., Nugent W.C., Hermann T., Negro-Vilar A., Rigas J.R. A proof-of-principle clinical trial of bexarotene in patients with non-small cell lung cancer. Clin. Cancer Res. 2007;13:1794–1800. doi: 10.1158/1078-0432.CCR-06-1836. [DOI] [PubMed] [Google Scholar]
- 38.Esteva F.J., Glaspy J., Baidas S., Laufman L., Hutchins L., Dickler M., Tripathy D., Cohen R. Multicenter phase II study of oral bexarotene for patients with metastatic breast cancer. J. Clin. Oncol. 2003;21:999–1006. doi: 10.1200/JCO.2003.05.068. [DOI] [PubMed] [Google Scholar]
- 39.Liston D.R., Davis M. Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin. Cancer Res. 2017;23:3489–3498. doi: 10.1158/1078-0432.CCR-16-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]