Abstract
Presently, COVID-19 has posed a serious threat to researchers, scientists, health professionals, and administrations around the globe from its detection to its treatment. The whole world is witnessing a lockdown like situation because of COVID-19 pandemic. Persistent efforts are being made by the researchers to obtain the possible solutions to control this pandemic in their respective areas. One of the most common and effective methods applied by the researchers is the use of CT-Scans and X-rays to analyze the images of lungs for COVID-19. However, it requires several radiology specialists and time to manually inspect each report which is one of the challenging tasks in a pandemic. In this paper, we have proposed a deep learning neural network-based method nCOVnet, an alternative fast screening method that can be used for detecting the COVID-19 by analyzing the X-rays of patients which will look for visual indicators found in the chest radiography imaging of COVID-19 patients.
Keywords: COVID-19, Detection, X-Rays, Deep learning, Convolutional neural network (CNN), nCOVnet
1. Introduction
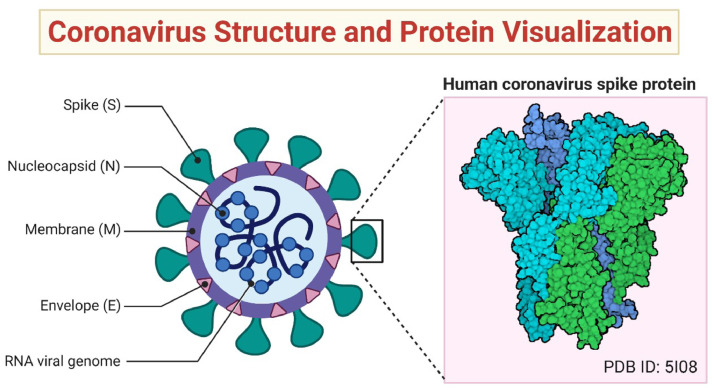
Research investigations show that around dozens of viruses exist in the family of the corona, but only seven types are dangerous to human beings [1]. It is noted that these viruses are transmitted in the human body via mammals like bats [2]. Initially, people of Wuhan city, China got infected by this virus in Dec, 2019, which causes an acute respiratory disease in humans, and has emerged as the latest worldwide epidemic. Now WHO has also declared it a pandemic known as COVID-19 [3], [4]. The structure of COVID-19 is shown in Fig. 1 .
Fig. 1.
The 2019-nCoV structure. Corona viruses belong in the family Coronaviridae and can cause disease in mammals and birds. The corona virus spike (S) protein mediates membrane fusion by binding to cellular receptors. (reprinted from [5] with permission under the terms of Creative Commons Attribution 4.0 International License.
In response to its sudden outbreak, the Research and Development wing of different research centers are actively participating in finding an effective diagnostic mechanism and vaccination for its treatment [6]. This area of research is not limited only up to medical or biotechnology, it involves the researchers of other fields like Artificial Intelligence, Data Science, Machine learning, to prevent and control this pandemic by providing their technical views and possible solutions [7], [8], [9], [10], [11].
1.1. Need of fast COVID-19 detection
The COVID-19 disease has become a raging cause of its worldwide mortality rate, the current death toll reaches up to 244K [12]. The novel corona-virus after entering through the human’s respiratory tract, affects the lungs of the person critically, causing conditions like and severe than the known ’Pneumonia’. The lungs become filled with fluid, get inflamed and develop patches called ‘Ground-Glass Opacity (GGO)’. Given that the symptoms of the disease are hard to recognise and there are limited testing kits available, we surely need to find other measures of its diagnosis [13]. Since a lot of efforts are being made for finding the effective medication of COVID-19 but till today the only effective way for protection is social distancing and lockdown of various cities in the country. However, the dark side of the lockdown is that it affects the GDP of the country and also have an adverse psychological effect on the health and mind of people. The number of afflicted sufferers because of COVID-19, are increasing exponentially around the globe. The majorly affected countries like the USA, Italy, and Spain have already surpassed China and will have an adverse catastrophic impact on the global economy. Richard Baldwin, a Professor of International Economics at the Graduate Institute in Geneva said “ǣThis virus is as economically contagious as it is medically contagious,” [14]. Therefore, it is mandatory to establish a health clinic system based on Artificial Intelligence (AI), that can be able to detect the cases fast and accurately to prevent this natural pandemic.
Deep Learning techniques have been on a rise since the last few years and have completely changed the scenario of many research fields. Especially, in medical field, image data set such as retina image, chest X-ray, and brain MRI provides promising results with an extended accuracy% by using the deep learning techniques [15], [16], [17]. As we know, X-ray machines provides inexpensive and faster results for scanning of various human organs in the hospitals. The interpretation of various X-rays images is usually performed manually by an expert radiologists. As a data scientist, if we train those captured images with the significance of deep learning that will be a great aid to medical experts for detecting the COVID-19 patients. This will help the developing countries where the X-ray facility is available but the availability of an expert is still a dream. To this advantage, we also aim to develop a deep neural network named ‘nCOVnet’ that can analyze the X-ray images of lungs and detect whether the person tests positive for the virus or not. Among various deep learning classifiers, in particular, the Convolutional Neural Networks (CNN) have been immensely effective in computer vision and medical image analysis tasks. The results of CNN have proven its cogency in mapping of image data to a precise and expected output. Since the lungs are the primary target of the virus, analysing their changes can give an explicit result of presence of the virus. The main contribution of this research work is to propose a CNN based model, which is able to train the images of corona virus infected lungs and those of healthy lungs. Proposed model is able to detect the COVID-19 cases at a faster speed by detecting the features of infected patients as hazy or shadowy patches in the X-ray images of lungs. The major research contributions of this paper can be summarized as follows:
-
•
We have proposed an algorithm nCOVnet to detect the COVID-19 patients using X-ray images.
-
•
We have focused on the observational analysis of lockdown is not only the solution and some artificial intelligence methods are extremely required to overcome from this solution.
-
•
We have presented the fast detection method using X-ray image analysis that would be a contribution to the society.
-
•
We have carefully analyzed the X-ray images of lungs for person in order to detect the COVID-19.
-
•
The obtained results have been evaluated by using three different parameters known as-confusion matrix, AUC of ROC, and Training Accuracy.
-
•
Proposed model correctly detect the COVID-19 positive patients with an accuracy of 97% whereas the overall accuracy of the proposed model is 88%.
Structure of the remaining paper is presented as. Section 2 focuses on the recent studies on X-ray image analysis using deep leaning based CNN Section 3 describes about the proposed methodology and data sets included to perform various experimentation’s. Section 4 elaborate the proposed nCOVnet model and its algorithm with detail description. Section 5 reports about obtained experimental results along with the performance of classifier. In Section 6 discusses the establishes the bridge between problem statement and outcomes came.
2. Deep learning in X-Ray image prognosis
Data scientists are significantly contributing in the research of medical data analysis that ultimately helps in the improvement of medical sciences. For this, deep learning/machine learning/data mining classifiers have been immensely applied in order to extract the relevant features from image data sets and classify them for disease diagnosis and prediction [10], [18], [19], [20], [21], [41], [42]. In COVID-19, human organ-lungs get infected and its diagnosis purely depends on the Lung X-ray. Therefore, it is quite evident to take the advantage of pre-research data from lung image analysis, which would be of great help to analyse the COVID-19 image data set and reach any sensible outcome with the deep learning approach. To detect the Pneumonia cases from chest X-rays, an effective CNN algorithm is applied with the 121-layered convolutional neural network. The model is trained on a data set containing over 100,000 images providing frontal-view of lung X-ray and describes 14 types of ailments. Rajpurkar et al. [22]. Similarly, in the current scenario of COVID-19, a CNN based approach has been applied to the X-ray images of the chest, since it causes similar symptoms to Pneumonia, verily, more severe. An automated COVID-19 method has been proposed by Alqudah et al. [23] they used machine learning classifiers–Support Vector Machine (SVM), random forest, K-Nearest Neighbor (KNN) with the CNN using soft-max, this helps in detection with a high accuracy rate of 98%. Singh and Gupta [18] applied Relu based deep learning method in identifying the malignant lung cancer from the image data set, their detection rate is 85.55%. Esteva et al. [24] focused on manifesting the classification of skin lesions, a single CNN layer is used, for diagnosis from diverse clinical images of skin. The model was tested to classify them binary with two critical cases– common cancer and deadly cancer. Further, its application also seen in tuberculosis data sets, Liu et al. [25] proposed a method to classify the Tuberculosis manifestations in the chest. The applied X-ray images data set are imbalanced. The approach took CNN and transfer learning into account and the method was stable for various CNN architectures with an excellent set of optimization solutions. However, it lacked considering region-level information while pre-processing of data. Singh and Gupta [18] applied Relu based deep learning method in identifying the malignant lung cancer from the image data set, their detection rate is 85.55%.
In the study conducted by Butt et al. [26] used ResNet23 and the classical ResNet-18. It was able to achieve an accuracy of 86.7%. The model was trained on CT Scans images. However, they are relatively more time consuming and are not available at many hospitals. Fanelli and Piazza [27] used the publicly available data set generated by John Hopkins University to detect the trends and forecast COVID-19 in difference scenarios. Through, Mean-field kinetics of the pandemic spreading they predicted the evolution of the COVID-19 outbreak and how the lockdown imposed by the government affected the situation. Apostolopoulos and Mpesiana [28] has applied Artificial Intelligence (AI) with different combinations of transfer learning and has achieved the best results with VGG19 with an accuracy of 98.75%. The used data set is also from Cohen’s GitHub, but they ignored the data leakage that may occur due to multiple images of the same patient and it may be a reason for the high accuracy. Li et al. [29], used AI with 3-D deep learning model for detecting COVID-19 patients on a data set containing 4356 CT Scans of 3322 patients. Chimmula and Zhang [30] built an automated model using deep learning and AI, specifically the LSTM networks (rather than the statistical methods), to forecast the trends and the possible cessation time of COVID-19 in different countries. All diseases have some patterns related to their spread, which are usually non-linear. Recognition of those patterns is the goal they could achieve with sequential networks. The model used time-series data for transmission prediction and could forecast Canada’s upcoming virus-trends and that several infection clusters could still persist until December 2020.
A wide variety of work has been done in research using the CNN, specifically in the medical image analysis field [31]. Various approaches collaborating engineering with health science have already been introduced that could reduce the wait time for the big process of diagnosis of some diseases. We have presented an extension of the existing techniques in the assessment of COVID-19 patients’ X-ray images data sets.
3. Methodology
3.1. Data set collection
The data set used in this work is an open source compiled by Cohen et al. [32] which currently consists of around 192 X-Ray images of COVID-19 positive patients and total 337 images. The repository of images is open for contributions and new images are added frequently. All the images are verified and annotated including the findings of the X-ray. Websites such as Radiopedia.org and Figure1.com were used to collect the images. In the data set we have Posterior Anterior (PA), Anterior Posterior (AP) and Anterior Posterior Supine (AP Supine) views of the lungs [10]. Here, we have selected only the PA views in the training and testing for the chest X-Rays. After selecting PA view, we have 142 X-ray images of COVID-19 positive patients. For normal healthy patients, we have used the Kaggle’s Chest X-Ray Images (Pneumonia) consisting of 5863 images. These images are divided into four different class values– ‘Normal’ ‘Bacterial Pneumonia’ and ‘Viral Pneumonia’. From this data set, we have selected around 142 images of normal X-rays. This data set is further converted into training and testing with the split of 70% data for training of proposed deep learning model and 30% data for testing purpose.
3.2. Data pre-processing
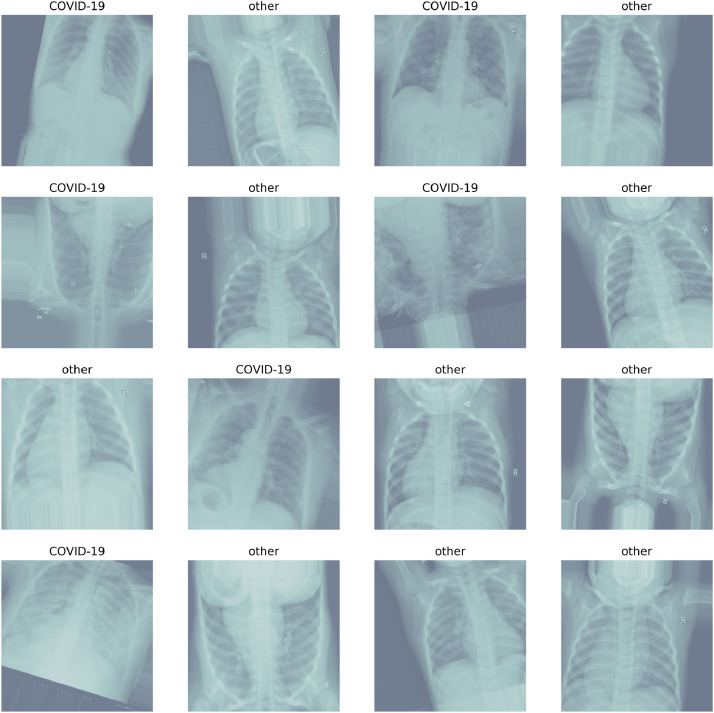
Since the data set is not uniform and the X-ray images are of different sizes therefore, we have converted all the images into the same size of 224 x 224 pixels. For this, RGB reordering has been applied and the final input to the proposed model is provided as 224 × 224 × 3 image. It has been noted that the data set is limited therefore, we have performed the data augmentation (refer to Fig. 2 ) with a rotation range of 20. The X-ray images were flipped horizontally and vertically so as to increase the diversity of data set significantly. Now, this data set is able to train on more images with the same data set.
Fig. 2.
Sample of the labelled X-rays after data augmentation taken from the combined data set of COVID-19 patients and normal patients.
3.3. Data leakage
Preventing data leakage is one of the crucial tasks of the methodology since in the applied data set a single patient with a unique patient id may have more than one X-ray images. The X-ray images of the same patient are present from different days they have visited in the hospital. Thus, while splitting we cannot use the train_test_split command anymore and instead have to come up with a new logic which will split the data at individual patient level. We have performed this by manually assigning 70% (127 patients) of the patients for training purpose and remaining 30% (31 patients) for testing purpose. We had altogether 127 COVID-19 positive patients’ X-ray images for training, on contrary, 31 COVID-19 positive patients’ X-ray images for testing. By doing this, we could be sure that there is no data leakage among training and testing data sets.
3.4. Convolutional Neural Network (CNN)
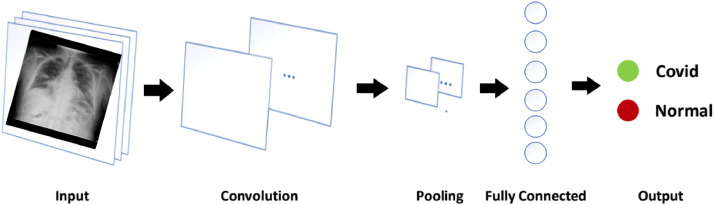
Deep learning techniques are used to reveal those features of the data set such as image and video which are hidden in the original data set. For this, Convolutional Neural Network (CNN) has been significantly applied to extract the features, and this unique characteristic has been immensely applied in medical image analysis that provides a great support in the advancement of health community research [33]. CNN is a type of artificial neural network which has multiple layers, and is expert to process the high volume of data with higher accuracy and less computational cost. The basic structure of CNN comprises convolution, pooling, flattening, and fully connected layers [34]. A basic architecture of CNN is presented in Fig. 3 showing the input X-ray image, networks, pooling and output.
Fig. 3.
Basic CNN architecture for classification and detection of COVID-19.
4. Proposed model and algorithm
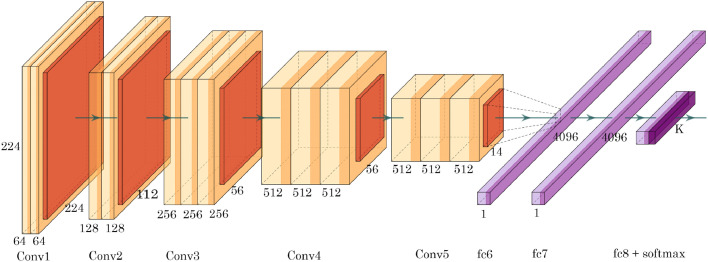
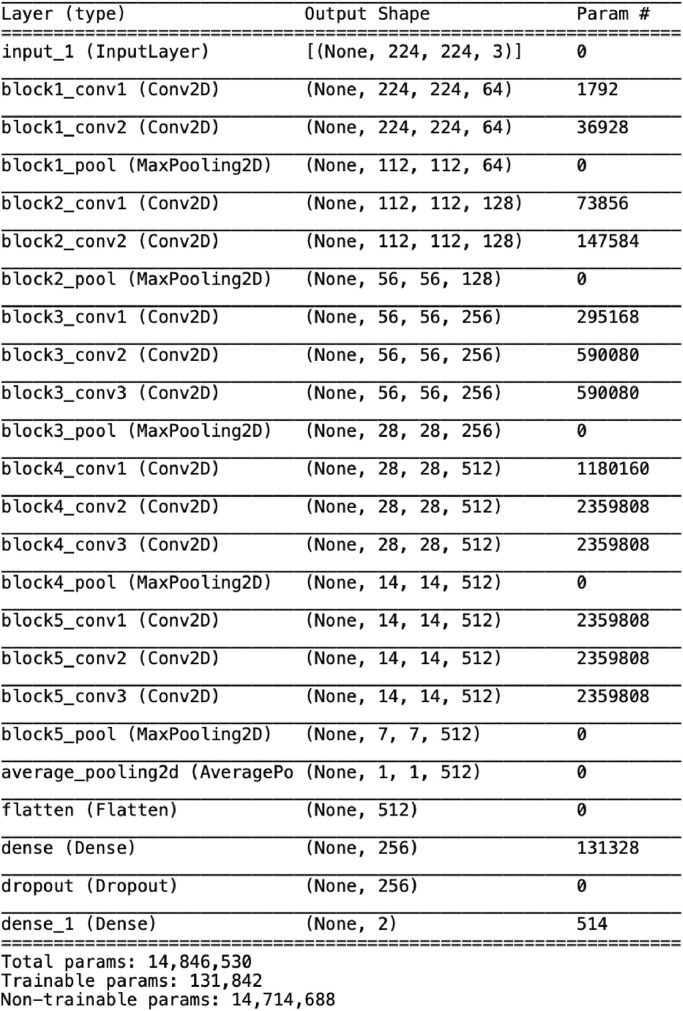
The proposed model depends on the working of deep learning based CNN known as nCOVnet. The applied parameters in this model are tabulated in Table 1 which consist of 24 layers. The first layer indicates the input layer and is fixed with the size of 224 x 224 x 3 pixels which makes it a RGB image. The next 18 layers are the combination of Convolution+ReLU and Max Pooling layers. These layers are part of the pre-trained VGG16 Model proposed in [35] and trained on the ImageNet dataset. ImageNet contains around 15 million annotated images from 22,000 different categories and VGG16 was able to achieve 92.7% accuracy on ImageNet. Therefore, we used the VGG16 model as depicted in Fig. 5 for feature extraction as a base model. Then we have applied a transfer learning model using the proposed 5 different layers and trained the proposed model on the COVID-19 dataset which is shown in Fig. 4 .
Table 1.
Various parameters applied by ‘nCOVnet’ model for detection of COVID-19.
| Layer | Stride | Filter | Pool size | Padding | Depth | No. of Parameters |
|---|---|---|---|---|---|---|
| Input Layer | - | - | - | - | 3 | 0 |
| Conv1 | 1 | 3 x 3 | - | same | 64 | 1792 |
| Conv2 | 1 | 3 x 3 | - | same | 64 | 36,928 |
| Max Pooling | 2 | - | 2 x 2 | - | 64 | 0 |
| Conv1 | 1 | 3 x 3 | - | same | 128 | 73,856 |
| Conv2 | 1 | 3 x 3 | - | same | 128 | 147,584 |
| Max Pooling | 2 | - | 2 x 2 | - | 128 | 0 |
| Conv1 | 1 | 3 x 3 | - | same | 256 | 295,168 |
| Conv2 | 1 | 3 x 3 | - | same | 256 | 590,080 |
| Conv3 | 1 | 3 x 3 | - | same | 256 | 590,080 |
| Max Pooling | 2 | - | 2 x 2 | - | 256 | 0 |
| Conv1 | 1 | 3 x 3 | - | same | 512 | 1,180,160 |
| Conv2 | 1 | 3 x 3 | - | same | 512 | 2,359,808 |
| Conv3 | 1 | 3 x 3 | - | same | 512 | 2,359,808 |
| Max Pooling | 2 | - | 2 x 2 | - | 512 | 0 |
| Conv1 | 1 | 3 x 3 | - | same | 512 | 2,359,808 |
| Conv2 | 1 | 3 x 3 | - | same | 512 | 2,359,808 |
| Conv3 | 1 | 3 x 3 | - | same | 512 | 2,359,808 |
| Max Pooling | 2 | - | 2 x 2 | - | 512 | 0 |
| Average Pooling | None | 4 x 4 | - | valid | 512 | 0 |
| Flatten | - | - | - | - | 512 | 0 |
| Dense | - | - | - | - | 64 | 131,328 |
| Dropout | - | - | - | - | 64 | 0 |
| Dense | - | - | - | - | 2 | 514 |
Fig. 5.
Architecture of the VGG16 Model.
Fig. 4.
Model summary of nCOVnet using VGG16 as a base model and five custom layers as head model.
These 5 layers are an integral part of the head model where the first layer acts as an Average Pooling 2D layer with pool size of (4,4). In average pooling, the average value of all the pixels in the batch is selected unlike max pooling where the maximum value is selected. Next, we use a flatten layer whose role is to apply a flattening transformation on the tensor converting the two-dimensional matrix of features into a vector that can be fed into a fully connected neural network classifier. Once, we have the transformed vector we then input it into a fully dense connected layer with a size of 64 units and a ReLU activation function. Then, we apply dropout with a threshold of 0.5. The dropout layer simply ignores some units (neurons) taking in consideration the threshold value which we provided. Finally, we have an output layer having just two units. Now, the model is optimized using the Adam optimizer [36]. Adaptive moment estimation or Adam is an optimization algorithm which is used in place of vanilla stochastic gradient descent.
4.1. Proposed algorithm
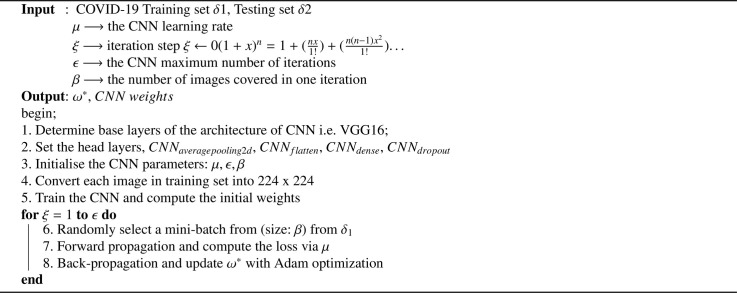
The proposed algorithm ‘nCOVnet’ is based on transfer learning model. The description of notations used in the Algorithm 1 is provided here, δ1 and δ2 refers to the training data set and testing data set respectively. μ, is the learning rate indicating how quickly a model adapts to the problem. Typically, it values nearby zero. ϵ is the total number of iterations the CNN model is trained for also known as epochs. β is another configurable hyper-parameter generally having values in the form of 2n. We can tune these three hyper-parameters (μ, ϵ, and β) based on step 1 to 3 of Algorithm 1 in a sort of hit and trial fashion to achieve the best accuracy with the given data set. The algorithm outputs weights stored in the form of a ‘.h5’ file which are further used for prediction on a completely random X-ray image.
Algorithm 1.
Fast Detection and Classification of COVID-19.
The algorithm proceeds by first using the VGG16 model’s top layers as a base model (refer to step 2 of Algorithm 1) and adding it to the custom 5 layers (refer to step 1 and 2 of Algorithm 1) as the head layers. Then we have applied a for loop ranging from 1 to the total number of iterations (ϵ) that train and update the weights through forward and backward propagation (refer to step 6–10 of Algorithm 1).
5. Experimental evaluation
For evaluation of proposed model, we have calculated the confusion matrix, Area Under Curve (AUC) of Receiver Operating Characteristics (ROC), and plotted the training accuracy.
5.1. Confusion matrix
Confusion matrix is based on four primary parameters known as True Positive (TP), False Positive (FP), False Negative (FN), and True Negative (TN) as shown in Table 2 . Here, rows represent the ‘Actual class values’, and the attributes are the ‘Predicted/Detected class values’. This has been prominently used for evaluating the performance of model.
Table 2.
Confusion matrix used for detection of Covid-19.
| Predicted Class |
|||
|---|---|---|---|
| Covid | other | ||
| Actual Class | Covid | ||
| other | |||
From confusion matrix on the test data set, we have achieved the sensitivity of 97.62% and specificity of 78.57%. Where Sensitivity refers to the measure of True Positives (TP) or COVID-19 positive patients in this case. Therefore, out of the 42 positive patients in used training data set we were able to correctly predict COVID-19 in 41 of them providing 97.62% probability. This implies that we can detect COVID-19 in infected patients, X-Ray with only 2.38% error. Specificity refers the measure of True Negatives (TN) or COVID-19 negative patients in this case. So, out of the 42 negative patients in used testing data set, we were able to correctly predict that a person is not infected by COVID-19 in 33 patients providing 78.57% probability. This implies that we can detect the COVID-19 in infected patients’ X-ray with an error rate of 21.43%. By calculating the accuracy of proposed model through the confusion matrix we have achieved an overall accuracy of 88.10% considering the limited amount of data used for training.
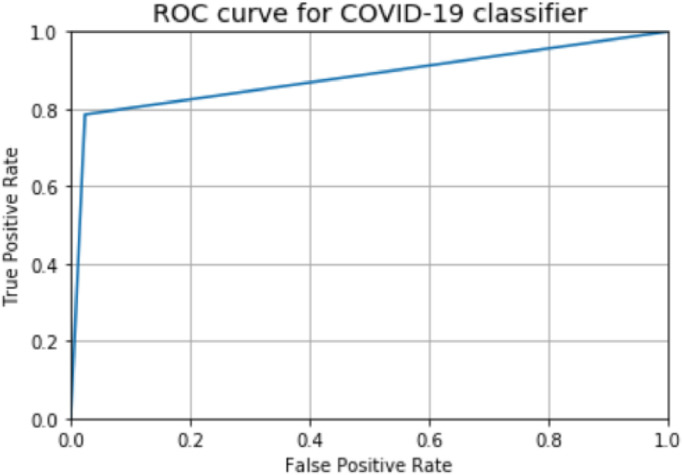
5.2. Receiver operating characteristics (ROC)
ROC is a 2-dimensional graph, plots between the true positive rate (TPR) and false negative rate (FPR), in other words, it can be defined as the trade-off between sensitivity and specificity [37]. We have plotted the Receiver Operating Characteristic (ROC) curve with True Positive Rate on the y-axis and False Positive Rate on the x-axis. In medical diagnosis higher area under curve (AUCs) are required. As a result, in clinical data analysis research, its computation allows the data scientist to simplify the data analysis with predictive knowledge on medical research [38].
The calculation of AUC is done by the well known trapezoidal method. This method calculates AUC by dividing the area into a number of sections with equal width. Here, trapezoid (T) refers to the integration of points - (a, b) from a functional form which is divided into n equal pieces. The summation of the area of each section by the area of the trapezium formed when the upper end is replaced by a chord and the sum of these approximations provides the final numerical result of the AUC. The trapezoidal rule is indicated as a definite function integral and the points of domain subdivision of the integration (a, b) are labelled as {x 0, x 1, ⋅⋅⋅xn}; where . Further, the function T(a, b, n) is defined by using Eq. (1) [39].
| (1) |
Hence, this is an effective way to calculate the accuracy of ROC produced by model. The plotted AUC of ROC as shown in Fig. 6 of proposed model is above the threshold level is calculated as 0.88095 which is a good indication of model and comes under the good rank of classification, and also considered to be ‘excellent’ in the field of medical diagnosis [38], [40].
Fig. 6.
ROC curve for nCOVnet.
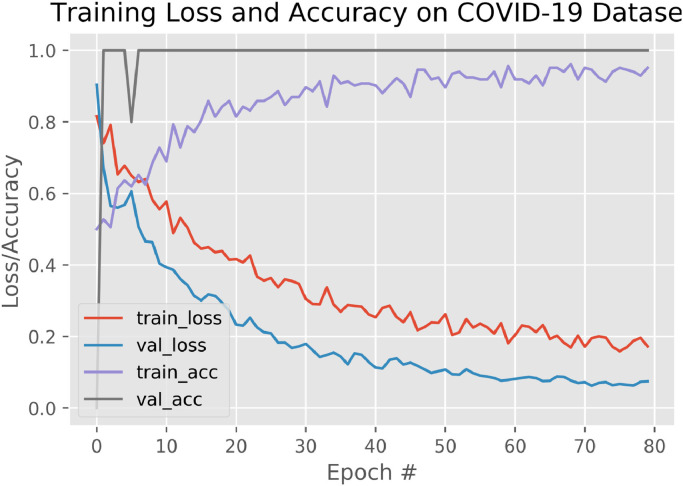
5.3. Training accuracy
We have trained the proposed model for 80 epochs and set the learning rate to be 0.0001. The training accuracy as seen in the graph lies around . Whereas, the training loss goes as low as 0.2% as highlighted in Fig. 7 . Although obtained results represents that the training accuracy is up to 97% which could be considered as one of the good measure for assessing classification models especially in the field of medical diagnosis. It is because during calculating training accuracy we have assigned equal cost to both false negatives and false positives. Therefore, we have considered the confusion matrix as a better evaluation parameter as it provides the accuracy separately for each metrics i.e. true positives, true negative, false positives, and false negatives.
Fig. 7.
Training curve of loss and Accuracy for nCOVnet models.
The problem of treating false positives and false negatives with equal costs is that we cannot afford to diagnose a COVID-19 positive patient as negative, since in such a case the patient may go back to the society thinking She/he is no longer a COVID-19 patient and this may result in community spread of the disease.
5.4. Prediction
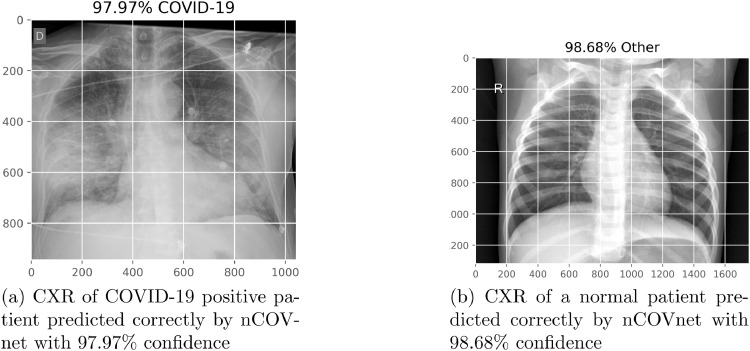
When we have applied prediction on the proposed model, it is able to classify the COVID-19 patient correctly with 97.97% confidence as shown in Fig. 8 . And applying prediction on COVID-19 negative patients the model was able to provide the correct result with 98.68% confidence. The measure of confidence can help us to only select those results which the nCOVnet is confident about. We have predicted a COVID-19 patient correctly with only 2.03% error in under 5 seconds, and we know that in the case of COVID-19 positive patients we have achieved an accuracy of 97.62%. From the obtained results this can be seen that the proposed nCOVnet model can be used as a substitute to RT-PCR which takes around hours for detecting COVID-19 patient. Since nCOVnet predicts with a confidence measure we can use the RT-PCR testing in the few cases where nCOVnet is not confident about to decrease the chances of errors.
Fig. 8.
Prediction results of Covid-19.
6. Discussion and conclusions
This is a proven fact that rigorous testing and social distancing are some of the most important measures to be taken by the governments in different parts of the world to control the COVID-19 pandemic. There are mainly two types of tests that are being conducted throughout the world to detect COVID-19, the antibody test and the RT-PCR test. The antibody test can find whether or not the immune system has encountered the virus and is an indirect way of testing. Since anti-bodies can take up to 9–28 days after the infection has set in, it is a very slow process and by that time the infected person can spread the disease if not properly isolated. On the other hand, RT-PCR testing is rather fast and can detect COVID-19 in around 4–6 hours. Looking at the magnitude of the pandemic this is also not too fast and RT-PCR testing has other limitations as well. One such limitation is the high cost of importing the chemicals and other elements used in the kits. One test can cost up to 60$, and the rate may vary in different parts of the world depending on the availability. Which brings us to the next limitation, availability, not every part of the world has the same requirements and resources. Some countries have a greater population and less availability of kits whereas some countries have more kits than required.
These limitations can be overcome with the proposed nCOVnet model. Proposed model can detect a COVID-19 positive patient in under 5 seconds. With the limited amount of data we had, we were able to achieve 97.62% true positive rate. By adding more samples of chest X-ray to the training data set, we can get higher accuracy with the same model architecture that we have already used. nCOVnet also addresses the issue of unavailability of RT-PCR kits, as it only needs an X-Ray machine which is already present in most of the hospitals throughout the world. So countries will no longer have to wait for large shipments of RT-PCR kits. With the fast detection of COVID-19, we will be able to contact and isolate the COVID-19 patients and reduce community transmission. The pandemic is still in the Stage-2 in many nations across the globe and they will not be able to afford the highly expensive kits and COVID-19 pandemic is not going away any time soon unless proper testing measures are taken by every country. Many of the early studies which promised accuracy up to did not take consider the possible data leakage which we addressed while training nCOVnet and thus the results we got are unbiased. This model could help to hospital administration and medical experts in order to make necessary steps to manage the COVID-19 patients after getting its fast detection.
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
No funding was received for this work.
We further confirm that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
References
- 1.Coronaviruses. https://www.niaid.nih.gov/diseases-conditions/coronaviruses, Last accessed on May 2020; 2020.
- 2.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11(3):210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iqbal H.M., Romero-Castillo K.D., Bilal M., Parra-Saldivar R. The emergence of novel-coronavirus and its replication cycle-an overview. J Pure Appl Microbiol. 2020;14(1) [Google Scholar]
- 4.Siddiqui M.K., Morales-Menendez R., Gupta P.K., Iqbal H.M., Hussain F., Khatoon K. Correlation between temperature and covid-19 (suspected, confirmed and death) cases based on machine learning analysis. J Pure Appl Microbiol. 2020;14:1017–1024. doi: 10.22207/JPAM.14.SPL1.40. 6201. [DOI] [Google Scholar]
- 5.Bilal M., Nazir M., Parra-Saldivar R., Iqbal H.M. 2019-ncov/covid-19 - approaches to viral vaccine development and preventive measures. J Pure Appl Microbiol. 2020;14(1) [Google Scholar]
- 6.WHO. Coronavirus disease (covid-2019) r&d. https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus/en/Last accessed on Mar 2020; 2019.
- 7.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., et al. Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv preprint arXiv:2002093342020. [DOI] [PMC free article] [PubMed]
- 8.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., et al. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv preprint arXiv:2003050372020.
- 9.Fanelli D., Piazza F. Analysis and forecast of covid-19 spreading in china, italy and france. Chaos Solitons Fractals. 2020;134:109761. doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M.. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004020602020.
- 11.Alimadadi A., Aryal S., Manandhar I., Munroe P.B., Joe B., Cheng X.. Artificial intelligence and machine learning to fight covid-19. 2020. [DOI] [PMC free article] [PubMed]
- 12.worldometers. Coronavirus cases. https://www.worldometers.info/coronavirus/ Last accessed on May 2020.
- 13.Xie Y., Wang X., Yang P., Zhang S. Covid-19 complicated by acute pulmonary embolism. Radiology. 2020;2(2):e200067. doi: 10.1148/ryct.2020200067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandemic A.E.. Coronavirus disease (covid-2019) r&d. https://foreignpolicy.com/2020/03/09/coronavirus-economic-pandemic-impact-recession/Last accessed on Mar 2020; 2020.
- 15.Mahmud M., Kaiser M.S., Hussain A.. Deep learning in mining biological data. arXiv preprint arXiv:2003001082020. [DOI] [PMC free article] [PubMed]
- 16.Mahmud M., Kaiser M.S., Hussain A., Vassanelli S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans Neural Netw Learn Syst. 2018;29(6):2063–2079. doi: 10.1109/TNNLS.2018.2790388. [DOI] [PubMed] [Google Scholar]
- 17.Harsono I.W., Liawatimena S., Cenggoro T.W. Lung nodule detection and classification from thorax ct-scan using retinanet with transfer learning. J King Saud Univ-ComputInf Sci. 2020 [Google Scholar]
- 18.Singh G.A.P., Gupta P. Performance analysis of various machine learning-based approaches for detection and classification of lung cancer in humans. Neural Comput Appl. 2019;31(10):6863–6877. [Google Scholar]
- 19.Klang E. Deep learning and medical imaging. J Thorac Dis. 2018;10(3):1325. doi: 10.21037/jtd.2018.02.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan+ F., Gao+ Y., Wang J., Shi W., Shi N., Han M., et al. Lung infection quantification of covid-19 in ct images with deep learning. arXiv preprint arXiv:2003046552020.
- 21.Liu J., Pan Y., Li M., Chen Z., Tang L., Lu C. Applications of deep learning to mri images: asurvey. Big Data Min Anal. 2018;1(1):1–18. [Google Scholar]
- 22.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., et al. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711052252017.
- 23.Alqudah A., Qazan S., Alqudah A.. Automated systems for detection of covid-19 using chest x-ray images and lightweight convolutional neural networks2020; 10.21203/rs.3.rs-26500/v1. [DOI]
- 24.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Cao Y., Alcantara M., Liu B., Brunette M., Peinado J. 2017 IEEE International Conference on Image Processing (ICIP) IEEE; 2017. Tx-cnn: detecting tuberculosis in chest x-ray images using convolutional neural network; pp. 2314–2318. [Google Scholar]
- 26.Butt C., Gill J., Chun D., Babu B.A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020:1. doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanelli D., Piazza F. Analysis and forecast of covid-19 spreading in China, Italy and France. Chaos Solitons Fractals. 2020;134:109761. doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chimmula V.K.R., Zhang L. Time series forecasting of covid-19 transmission in canada using lstm networks. Chaos Solitons Fractals. 2020:109864. doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salman F.M., Abu-Naser S.S., Alajrami E., Abu-Nasser B.S., Alashqar B.A. Covid-19 detection using artificial intelligence. Int J Acad EngRes (IJAER) 2020;4(3):18–25. [Google Scholar]
- 32.Cohen J.P., Morrison P., Dao L.. Covid-19 image data collection. arXiv preprint arXiv:2003115972020.
- 33.Choe J., Lee S.M., Do K.-H., Lee G., Lee J.-G., Lee S.M. Deep learning–based image conversion of ct reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292(2):365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 34.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep learning. [Google Scholar]
- 35.Simonyan K., Zisserman A.. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:140915562014.
- 36.Kingma D.P., Ba J.. Adam: a method for stochastic optimization. arXiv preprint arXiv:141269802014.
- 37.Streiner D.L., Cairney J. What’s under the roc? an introduction to receiver operating characteristics curves. Can J Psychiatry. 2007;52(2):121–128. doi: 10.1177/070674370705200210. [DOI] [PubMed] [Google Scholar]
- 38.Siddiqui M.K., Morales-Menendez R., Ahmad S. Application of receiver operating characteristics (roc) on the prediction of obesity. Braz Arch Biol Technol. 2020;63(Human and Animal Health) [Google Scholar]
- 39.Yeh S.-T. Proceedings of the 27th Annual SAS® User Group International (SUGI’02) 2002. Using trapezoidal rule for the area under a curve calculation. [Google Scholar]
- 40.Korsten M. 7 Reflections on the future of gastroenterology–unmet needs. Vol. 52. 2007. Application of summary receiver operating characteristics (sroc) analysis to diagnostic clinical testing; p. 76. [PubMed] [Google Scholar]
- 41.Siddiqui M.K, Islam M.Z., Kabir M.A. A novel quick seizure detection and localization through brain data mining on ECoG dataset. Neural Comput & Applic. 2019;29(4):5595–5608. doi: 10.1007/s00521-018-3381-9. [DOI] [Google Scholar]
- 42.Siddiqui M.K., Morales-Menendez R., Huang X. A review of epileptic seizure detection using machine learning classifiers. Brain Inf. 2020;7(1):1–18. doi: 10.1186/s40708-020-00105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]