Abstract
Celiac disease (CD) is a gastrointestinal disorder whose genetic basis is not fully understood. Therefore, we studied a Saudi family with two CD affected siblings to discover the causal genetic defect. Through whole exome sequencing (WES), we identified that both siblings have inherited an extremely rare and deleterious CPED1 genetic variant (c.241 A > G; p.Thr81Ala) segregating as autosomal recessive mutation, suggesting its putative causal role in the CD. Saudi population specific minor allele frequency (MAF) analysis has confirmed its extremely rare prevalence in homozygous condition (MAF is 0.0004). The Sanger sequencing analysis confirmed the absence of this homozygous variant in 100 sporadic Saudi CD cases. Genotype-Tissue Expression (GTEx) data has revealed that CPED1 is abundantly expressed in gastrointestinal mucosa. By using a combination of systems biology approaches like protein 3D modeling, stability analysis and nucleotide sequence conservation analysis, we have further established that this variant is deleterious to the structural and functional aspects of CPED1 protein. To the best of our knowledge, this variant has not been previously reported in CD or any other gastrointestinal disease. The cell culture and animal model studies could provide further insight into the exact role of CPED1 p.Thr81Ala variant in the pathophysiology of CD. In conclusion, by using WES and systems biology analysis, present study for the first-time reports CPED1 as a potential causative gene for CD in a Saudi family with potential implications to both disease diagnosis and genetic counseling.
Keywords: Celiac Disease (CD), Whole exome sequencing, CPED1, Molecular analysis, Autosomal recessive
1. Introduction
Celiac Disease (CD) is a chronic enteropathy elicited by gluten, a storage protein found in rye, barley, and wheat, in genetically susceptible individuals (Fasano and Catassi, 2012). CD has a worldwide prevalence of 0.5%-1%. Recent meta-analysis study has reported the variable prevalence of CD in Asians (0.6%), North Americans (0.5%), South Americans (0.4%), and Europeans (0.8%) (Singh et al., 2018). Interestingly, CD was found to be prevalent in 1.5% of Saudi children (Al-Hussaini et al., 2017). The actual prevalence of CD is seen to be variable among different regions in the kingdom. For example, the highest sero-positivity prevalence was reported in the central region with 3.2%, followed by the southwestern region with 2.1% and the lowest frequency of 1.8% was reported in the western region (Aljebreen et al., 2013). CD primarily damage the small intestinal tissues, although it shows broad clinical manifestations. According to Oslo classification, the clinical presentation of CD, could be either classical, non-classical, silent, latent or refractory among other specific subtypes (Ludvigsson et al., 2013). Therefore, one of the main complications of CD is delayed diagnosis due to its clinical diversity (Fuchs et al., 2018). Nevertheless, duodenal biopsy for small bowel villous atrophy and serological tests of anti-tTG antibodies, anti-endomysium antibodies (EmA), and deamidated gliadin peptide (DGP) antibodies are still considered as gold standard diagnostic tests for confirmation of CD (Volta et al., 2010). Till date, CD treatment involves keeping patients on a strict gluten-free diet to reduce the symptoms, but not the histological manifestations (Wada et al., 2018).
Like other autoimmune diseases, CD is also caused by the complex interactions between genetic, non-genetic factors and immune responses. Gluten consumption along with HLA and non-HLA predisposition alleles contributes to its complexity, thus making CD not just a multifactorial but polygenic disorder. Particularly, HLA-DQ2.5 and HLA-DQ8 haplotypes play a crucial role in stimulating the abnormal reaction in the gastrointestinal tract against gluten. In Saudi population, high risk HLA-DQ alleles were known to be prevalent in 52.7% of healthy children from Riyadh province (Al-Hussaini et al., 2018). Although, HLA genes can increase the risk of the disease, they are not sufficient for developing CD. For example, together HLA DQ2.5 and DQ8 alleles could only explain 25%-40% of the CD genetic risk (Lindfors et al., 2019). This implies that, unknown non-HLA genes also largely contribute to the disease development. The introduction of Genome-wide association studies (GWAS) in CD patients, have helped in discovering the role of 56 non-HLA genetic loci in modifying the disease risk (Hunt et al., 2008, Trynka et al., 2009, Dubois et al., 2010, Trynka et al., 2011). As per the current understanding, non-HLA genetic susceptibility loci only contributes 10% of the current CD risk (Withoff et al., 2016).
Although, GWAS provided a well-recognized knowledge about CD genetics, however most studies were carried out among Caucasians or Asians (Hrdlickova et al., 2018a, Hrdlickova et al., 2018b). This necessitates us to look for genetic markers in other ethnic populations like Saudi Arabians who possess a unique genetic architecture due to the high consanguineous marriage backgrounds (Al-Hussaini et al., 2018, Alnaqeb et al., 2018). Furthermore, majority of the classical GWAS studies were carried out among sporadic cases and could not identify any causal genes. This highlights the need to study CD in rare familial cases to unveil the role of rare variants in genes that cannot be spotted by genome wide association studies conducted in sporadic cases. While common variants in CD are extensively studied through microarray-based genome wide genotyping, studying rare variants has only become feasible with next-generation sequencing approaches like whole exome sequencing (WES), which scans coding regions of the human genome. Therefore, owing to the limited scientific data available on familial aggregation of CD, this study was aimed to find the potential rare variant causal to CD in a Saudi Arabian family using a combination of whole exome sequencing and systems biology approaches.
2. Materials and methods
2.1. Recruitment of CD family and sporadic cases
The Saudi family with two CD affected siblings were referred to our Pediatric Gastroenterology clinic in King Abdulaziz University Hospital. Both these children met the standard diagnostic guidelines of the European Society for Pediatric Gastroenterology, Hepatology and Nutrition (ESPHGAN) for CD (Husby et al., 2012). In brief, CD diagnosis included clinical examinations by pediatric gastroenterologists, intestinal endoscopy and serological test for tissue transglutaminase antibodies (anti-tTG). Multi-generation pedigree was drawn by geneticists after collecting the full family history. In order to validate the genetic findings of CD family, we have additionally recruited 100 sporadic CD cases, who were regularly followed up at our gastroenterology clinic since the time they were diagnosed. This study protocol was approved by the Research Ethics Committee, King Abdulaziz University Hospital, Jeddah (KAUH). The parental written informed consent were obtained after explaining them about our study protocol. Approximately 2.5–3 ml of venous blood samples from all the family members were collected in EDTA tubes and stored at −80 °C until genetic analysis was performed.
2.2. DNA extraction
Genomic DNA was extracted and purified with QIAamp DNA blood Kit, following the manufacturer’s instructions. DNA concentration and purity were measured by Nanodrop (ND-1000 UV–VIS) spectrophotometer.
2.3. WES analysis
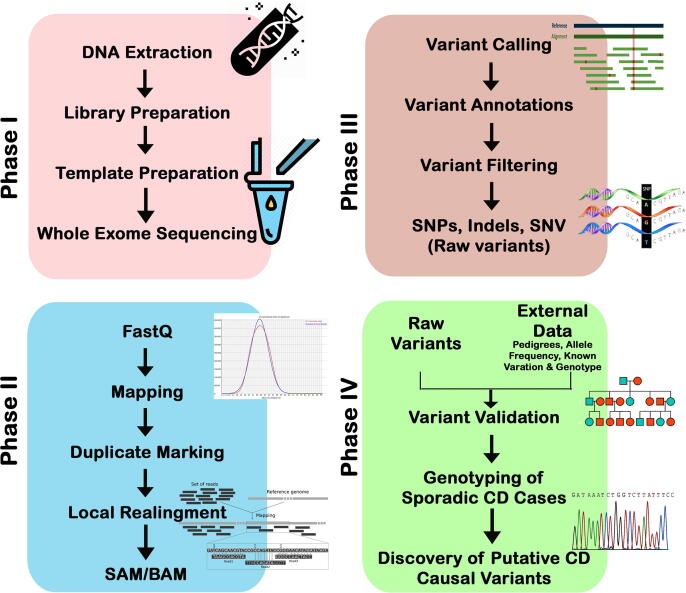
Genomic DNA (100 ng/µl; 260/280 ratio of 1.8–2.0) was used for preparing the library. The whole exome sequencing was performed on HiSeq2000 Next Generation Sequencer (Illumina, San Diego, CA). DNA shearing was done using Agilent Sure Select Target V6 Enrichment capture kit, reaching an optimal size-ranged fragments. The fragmented DNA were hybridized with ultra-long 120-mer biotinylated cRNA library baits. Library concentration and size were examined by capillary electrophoresis. Different adapters were integrated during enrichment, allowing the samples to be amplified for further sequencing. The sequencing reads (in FASTQ format) were aligned against the human genome reference sequence build 38 (GRCH38.p12) supported by BLAST (version 0.6.4d) for variant calling and annotation. The genome analysis toolkit (GATK) was used for downstream high quality variant calling (McKenna et al., 2010). The SAM tools suite was used to differentiate SNPs and short insertion/deletion in each read (Basha et al., 2018). The read coverage was ~ 100×, representing 87% of the targeted region, as (Fig. 1).
Fig. 1.
Workflow whole-exome sequencing; Phase I- Sample preparation; Phase II Sequence read mapping: Phase III Variant calling; Phase IV: Variants validation and Genotyping.
2.4. Variant filtering and candidate gene selection
CD candidate gene list was collected from different databases (NCBI, GWAS catalog, celiac gene panels) to verify their presence in the exome data of our patients. From the WES data, variants were retained if: (1) they passed the sequencing quality control (2) they were found in coding or regulatory sites (3) they had an allele frequency (MAF) of < 1.5% in general populations, African population, East and south Asian population, (4) they had an allele frequency < 1.49 × 10−02 based on the gnomAD database, (5) they are homozygote or compound heterozygotes showing an autosomal recessive inheritance pattern. Moreover, all genes with variants were filtered by their functional relevance to autoimmune diseases in general, and particularly to CD. Genes whose expression was reported in the small intestine or any related parts of the gastrointestinal system were considered potential to CD. The rare frequency of the filtered variants, was further verified in the WES data of 2379 healthy Saudi volunteers, hosted on Saudi Genome Project webserver (SHGP: https://www.saudigenomeproject.org/en/).
2.5. Sanger sequencing of candidate variant
The potential WES-derived candidate variants were further confirmed through Sanger sequencing. For this purpose, we designed oligonucleotide specific primers using open source NCBI Primer Blast webserver (Ye et al., 2012) with an average of 400–600 bp of target amplicon size. (supplementary Table 1). All the sequence reads were aligned and analyzed against reference mRNA (ENST00000310396.10) sequence of CPED1 gene with BioEdit (http://www.mbio.ncsu.edu/) program. Mutation position was determined considering “A” of first ATG codon of the mRNA.
2.6. Computational analysis of candidate protein
The human CPED1 sequences (both nucleotide and amino acid) were aligned against CPED1 sequence of 13 selected primates, with help of comparative genomic orthologue option available in Ensemble browser (www.esembl.org) for examining whether the causative variant is located in a conserved sequence region. We also sought to explore the consequences of candidate rare variant on the protein structure by simulating its 3D structure through either ab-initio or homology modeling approaches, depending on the availability of experimentally solved structure (Rafi et al., 2019). Confidence of each model is quantified by the C-score based on (a) significance of threading template alignments, and (b) convergence parameters of the structure assembly simulations. The best protein selected by highest C-score was subjected to energy minimization by applying the structure to the gromacs-steepest descent energy minimization method using NOMAD-Ref web server (Lindahl et al., 2006). Moreover, the energy minimized native model was entered into DUET software to build the mutated model and to predict variant impact on the protein stability (Abduljaleel, 2019). In addition, backbone atoms variations among superimposed proteins both at whole structure and residue levels were analyzed by calculating their positional root mean square deviation (RMSD) values. RMSD cutoff score considered for complete structure alterations was > 2.0 Å and for residue level it was > 0.2 Å (Ahmed Awan et al., 2020, Masoodi et al., 2019, Kufareva and Abagyan, 2012). Finally, network analysis results of STRING v.9.1 webserver were used to identify the key pathways and gene networks based on known protein–protein interactions between the candidate proteins and other genes (Franceschini et al., 2013, Safaei et al., 2016).
3. Results
3.1. Case presentation
The proband III.1 (3 years) was born to unrelated parents with no family history for celiac disease (Fig. 2). With the introduction of formula milk, she developed gastrointestinal symptoms including severe abdominal pain, bloating and diarrhea. She also demonstrated other symptoms like mild osteoporosis, eczema, severe skin rash, short stature, weight loss and lack of appetite. Besides that, she was previously diagnosed with left congenital renal and ureter disease. Endoscopy test showed fissures on the folds and absence of intestinal villi and her tTG antibody screening confirmed that she is CD positive. Her younger sister III.2 (2 years) suffers from severe diarrhea, vomiting and abdominal bloating is triggered when solid food was introduced at six months of age. She was also put on gluten free diet. She has a normal weight chart and no further complications were reported. All symptoms disappeared after keeping both of them on gluten-free diet.
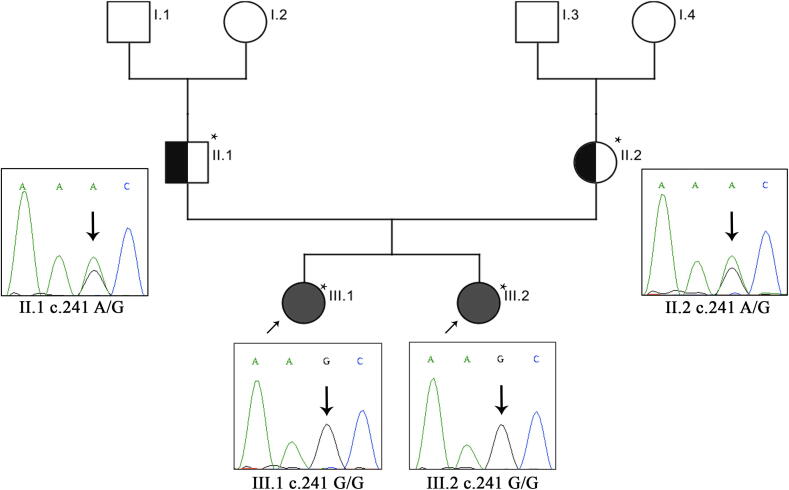
Fig. 2.
Sanger sequencing analysis of CD Family. Probands are indicated by the (arrow). Exome sequenced individuals are indicated with an (*) mark showing the electrophoretic trace for mutations of the CPED1 gene. The probands are homozygous to the mutation in exons 2 (c.241 G/G). Both parents are heterozygous for the mutation (c.241 A/G).
3.2. Exome sequencing
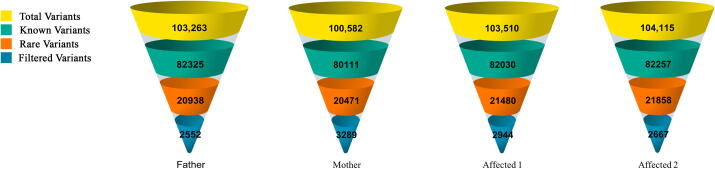
The terms ‘variant’ or ‘mutation’ represents each other in this manuscript. Exome data had an output of approximately 100 K of variants for each individual. Non-coding variants (downstream, upstream, intergenic and intronic variants) were excluded, except the regulatory region variants (3′-UTR and 5′-UTR). All data were filtered to exclude shared variants with MAF of > 0.015 based on 1000 Genomes Project data. The exome data were screened for both homozygous and heterozygous mutations. In these patients, about 2600 unique and rare variants were detected in the form of missense, frameshift and regulatory region mutations (Fig. 3).
Fig. 3.
The genetic variants yield of CD family. Yellow section indicates the whole variants generated from whole exome sequencing. Green section indicates known variants with MAF > 1.5%. Orange section indicates variants with MAF < 1.5%. Blue section indicates final filtered variants. Note: Filtering criteria for variants: exonic, unknown or extremely rare (MAF = <0.015%).
All the filtered variants were further segregated based on their functional annotations to CD, or other gastrointestinal disease with known autoimmune background. The final list consisted of 16 pathogenic variants (8 missense, 2 frameshift and 6 UTR), spanning 9 genes located on different chromosomes: 7,12,14,18,19 and 22. All these genes were known to have a putative functional relationship with inflammatory responses or autoimmune diseases (Table 1, Table 2). Among these 16 variants, a homozygous missense mutation in cadherin-like and PC-esterase domain containing 1 (CPED1) gene is the only one which survived our stringent variant filtering criteria, as detailed in our methods. Interim release of UK Biobank data (UKBB) on gene-phenotype association reported a number of CPED1 variants are significantly associated with gastrointestinal clinical phenotypes (Staley et al., 2016, Kamat et al., 2019). The putative CD causal variant of CPED1 gene is found in homozygous condition in both siblings, whereas it was seen in heterozygous condition in parents.
Table 1.
List of homozygous nucleotide variants which showed autosomal recessive inheritance pattern in CD family.
| No. | Gene name | Chromosomal position | Ref/Alt Bases | Effect | HGVS.c | HGVS.p | dbSNP ID | 1000G | ExAC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | HEBP1 | 12: 13,000,192 | C/CGGCGGCAGGGCGGCAG | 5′ UTR variant | c.-94_-79dupCTGCCGCCCTGCCGCC | . | rs71064363 | . | . |
| 2 | NFATC1 | 18: 79,400,367 | C/CGCCCCG | 5′ UTR variant | c.-14_-9dupCGGCCC | . | rs749710747 | . | . |
| 3 | CPED1 | 7: 120,989,862 | A/G | Missense variant | c.241A > G | p.T81A | rs117047013 | 0.0011 | 0.0011 |
Table 2.
List of compound heterozygous nucleotide variants following autosomal recessive inheritance pattern in CD family.
| No. | Gene name | Chromosomal position | Ref/Alt Bases | Effect | HGVS.c | HGVS.p | dbSNP ID | 1000G | ExAC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CYP27B1 | 12: 57,766,052 | G/C | Missense variant | c.341C > G | p.T114R | rs1247839240 | . | . |
| CYP27B1 | 12: 57,766,119 | C/T | Missense variant | c.274G > A | p.A92T | rs958027919 | . | . | |
| 2 | HIF1A | 14–61,697,832 | A/AT | 5′ UTR variant | c.-9dupT | . | rs769491528 | . | . |
| HIF1A | 14–61,738,090 | C/T | Missense variant | c.1325C > T | p.T442I | rs41508050 | 0.0039 | 0.0036 | |
| 3 | SMAD4 | 18: 51,082,732 | G/GT | 3′ UTR variant | c.*4277dupT | . | rs559471969 | . | . |
| SMAD4 | 18:51,084,012 | G/GCACA | 3′ UTR variant | c.*5574_*5577dupCACA | . | rs1491420266 | . | . | |
| 4 | MUC16 | 19: 8,888,826 | T/TCCGA | Frameshift variant | c.40672_40673insTCGG | p.K13558fs | rs769228524 | . | . |
| MUC16 | 19: 8,974,159 | GA/G | Frameshift variant | c.6979delT | p.S2327fs | rs759867982;rs208917 | . | . | |
| MUC16 | 19: 8,974,184 | T/C | Missense variant | c.6955A > G | p.I2319V | rs200019597;rs208918 | . | 0.0003 | |
| 5 | FCGBP | 19: 39,886,005 | T/G | Missense variant | c.8174A > C | p.N2725T | rs2916067 | . | 0.00005 |
| FCGBP | 19: 39,902,423 | C/T | Missense variant | c.4405G > A | p.G1469R | rs143680639 | . | 0.0738 | |
| 6 | SLC7A4 | 22: 21,029,052 | C/T | 3′ UTR variant | c.*3G > A | . | rs140809496 | 0.0023 | . |
| SLC7A4 | 22: 21,031,031 | G/T | Missense variant | c.782C > A | p.A261D | . | . | . |
This homozygous nucleotide variant c. 241A > G (rs117047013), results in the substitution of novel Threonine amino acid at 81st residue position to Alanine. This mutation appeared to have an allele frequency of 0.02985 (~2%) among 2379 in Saudi Arabia people, whereas only one individual had a homozygous genotype (1/2379 = 0.0004). In the 1000 genome project, allele frequency was reported to be 0.1% with only one homozygous individual. Moreover, in gnomAD, which hosts the genetic data of ~ 251 K individuals from across the world, the frequency of this variant is 0.1%. Hence, the rare frequency of this variant highly suggests it to be a pathogenic and has a potential role in pathophysiology of CD.
3.3. Validation of family specific variant using Sanger sequencing
Sanger sequencing has confirmed that both affected sisters (III.1 and III.2) were homozygous for the mutation c.241A > G (GG genotype), while both parents (II.1 and II.2) were heterozygous carriers (AG genotype), and that this variant was inherited in the family in an autosomal recessive fashion. This variant was confirmed to be absent in 98% of Saudi CD sporadic cases whereas the remaining 2% were found to be carriers for the heterozygous mutation, confirming the unique familial segregation of this variant, showing a strong association of CPED1 gene to be the causative in celiac disease.
3.4. Computational analysis of CPED1 mutant and wild type gene products
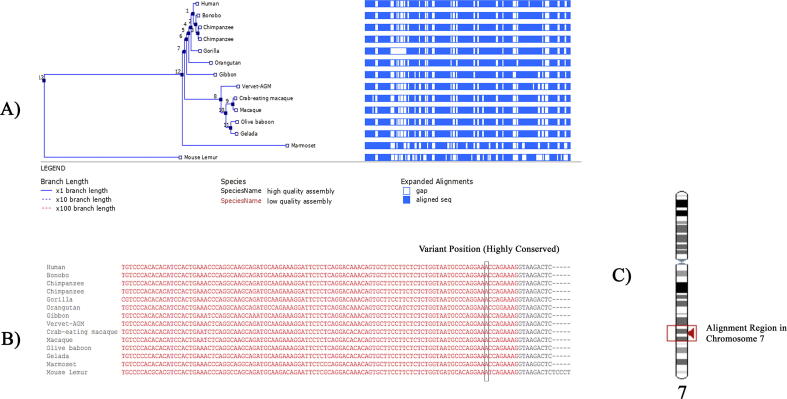
Cross-species alignment of CPED1 nucleotide sequences showed that this variant site falls in a highly conserved region (Fig. 4), further confirming our initial suspicion that this variant is pathogenic. Additionally, amino acid sequence analysis too confirmed the high-level conservation of the residue and its location across nine primates, indicating its critical role to the function of CPED1 protein.
Fig. 4.
(A) Phylogenetic tree of the human Cadherin-like and PC-esterase domain containing 1 (CPED1) aligned with 13 primates showing the aligned sequence in dark blue sections and gaps in white sections. (B) Nucleotide sequence Alignment of human and primates CPED1 gene showing the highly conservative site of CPED1 variant. (C) Alignment region containing CPED1 variant in chromosome 7.
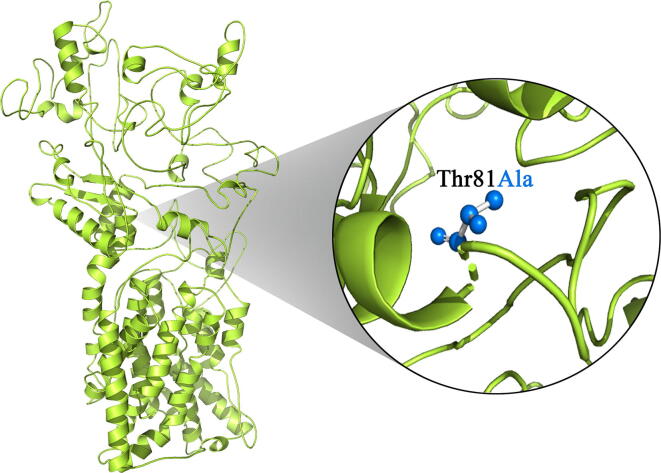
While an open reading frame for CPED1 gene has been defined, but its protein structure at three-dimensional level has not been solved through conventional x-ray crystallography methods. Therefore, we have adopted an ab-initio modeling approach using I-TASSER that uses only the highest significance in the string alignments to obtain CPED1 protein model. The predicted CPED1 protein model had the C-score of −1.16, TM-score of 0.57 ± 0.15 and RMSD score of 11.8 ± 4.5 Å. Overall, these scores indicate the high quality of the predicted structure and accurate folding of CPED1 protein chains. Furthermore, superimposition at the protein residue level revealed significant divergence of CPED1 protein in mutant condition (RMSD score is 1.8735 Å), (Fig. 5).
Fig. 5.
Molecular view of protein superimposition of native (green in color) and mutant (blue in color) version of CPED1 proteins.
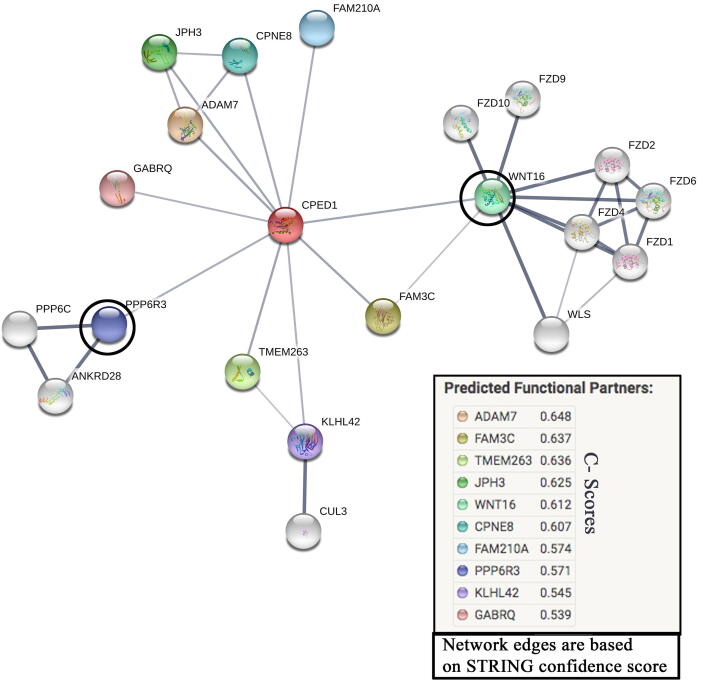
DUET-Integrated computational approach (ΔΔG) of the two methods (mCSM and SDM) that uses SVM regression with a Radial Basis Function kernel has predicted that the p.Thr81Ala substitution destabilizes the CPED1 protein (ΔΔG is −0.562 Kcal/mol). Furthermore, protein–protein interaction network analysis showed the functional association of CPED1 with ten protein partners, of which Wnt family member 16 (WNT16) and Protein Phosphatase 6 Regulatory subunit 3 (PPP6R3) proteins had the highest confidence scores of 0.61 and 0.57, respectively (Fig. 6).
Fig. 6.
Predicted interactions in protein network analysis for CPED1 (red colored node) were exported from STRING online database (http://string-db.org). Circled protein nodes indicate the strongest protein partners for CPED1 protein.
4. Discussion
HLA loci, spanning several genes and variants by and large is the major known genetic risk factor contributing to the development of CD (Abadie et al., 2020). In quest of solving the non-HLA genetic basis of CD, several GWAS studies with case-control study design were conducted in several ethnic populations. However, most studies have reported the association of genetic variants which could explain disease risk only for the specific ethnic population. First GWAS was conducted with 778CD cases and 1422 controls revealed a genetic variant (rs13119723) in a linkage disequilibrium block encompassing the KIAA1109/Tenr/IL2/IL21 genes, as a novel susceptibility factor for CD among British population (van Heel et al., 2007). Later on, the list of potential CD risk variants was further expanded with CCR3, IL12A, IL18RAP, RGS1, SH2B3 and TAGAP genes which regulates immune reactions in humans (Hunt et al., 2008). GWAS has so far identified 57 risk loci spanning multiple genes like PTPN2, IL18RAP, CCR4, ICOSLG, TAGAP, DUSP10, PUS10, etc. (Dubois et al., 2010, Festen et al., 2011, Garner et al., 2014, Hrdlickova et al., 2018a, Hrdlickova et al., 2018b, Ostensson et al., 2013, Trynka et al., 2011, Zhernakova et al., 2011). However, these GWAS studies have been conducted on patients of European and American ethnicity, leaving a gap of knowledge on CD from Arab population, which has high inbreeding rate (Al-Hussaini et al., 2018, Alnaqeb et al., 2018). Single case-control study reported from Saudi Arabia had described that a 518 Thr/Thr polymorphism in MMEL1 had an independent association with susceptibility to CD in Saudi patients (Saadah et al., 2015). However, all the above studies only highlighted statistically associated common variants but not the specific causal factors of CD etiology.
On the contrary, WES has created a paradigm shift in genetic diagnosis of complex diseases, which are otherwise very difficult to be exclusively studied by conventional Sanger sequencing (Li et al., 2018, Maffucci et al., 2016). Extending the power of WES and deep resequencing, a large cohort of large families with many affected members of Caucasian origin was unable to identify any new CD mutations (Mistry et al., 2015). Moreover, a recent study on a Saudi family found the putative protective role of a rare homozygous insertion (c.1683_1684insATT) in AK5 gene which modifies the risk of Saudi population against CD development (Al-Aama et al., 2017). However, with the recent availability of Saudi Human Genome Project (SHGP) data, it is now feasible to find the true positive disease causative genes (Al-Hamed et al., 2019, Yemni et al., 2019).
Here, by using WES, a hypothesis free survey to scan potential coding rare variants, we identified a rare CPED1 genetic mutation (c. 241A > G). Remarkably, this novel variant has not been previously associated with any disease. This study is the first to propose CPED1 as a causative gene of CD in a Saudi family. This gene is located on the long arm of chromosome 7 at the position of 31.31. It is composed of 3081 nucleotides, 23 exons and encodes a 1,026 amino acid long protein (Zhu et al., 2012). This protein comprises an N-terminal signal peptide that functions in the guidance of the protein product to the secretory pathway. It consist of two domains mainly involved in acyl-transferase and acyl-esterase activities for the modification of glycoproteins, a cadherin-like domain, and a PC esterase domain with an N-terminal fused ATP-grasp domain (Anantharaman and Aravind, 2010). Our in-silico analysis showed a decreased stability of the assembly of these two domains which may affect the protein integrity and might also contribute to CD.
Moreover, genetic background of CD has been reported to share a common susceptibility risk-loci with other immune diseases like inflammatory bowel disease and rheumatoid arthritis. Meta-analysis of GWAS data has reported four shared risk loci in PUS10, PTPN2, TAGAP, and IL18RAP between CD and Crohn’s disease due to their common genetic pathway (Festen et al., 2011), and seven shared loci with rheumatoid arthritis (Alexandra Zhernakova et al., 2011) counting on the shared mechanism between the two diseases in antigen presentation and T-cell activation. Furthermore, a cross‐disease study based on gene expression showed the upregulation of FASLG, PLEK, CCR4, and TAGAP genes in both CD and ulcerative colitis (Medrano et al., 2019). Overall, our results show a similar correlation that links CPED1 as a CD causal gene based on the genome wide association studies in UKBB with inflammatory bowel disease, rheumatoid arthritis and multiple intestinal diseases (Supplementary table 2). Moreover, GWAS among 3,475 Finnish ancestry individuals, found the association of intronic CPED1 variant (rs77670845) with alterations in plasma interleukin 2 levels (IL-2) (Ahola-Olli et al., 2017). Interleukin 2 has an important role in immune system especially in the autoimmune disease prevention, where IL-2 promotes the differentiation of particular immature T-cells into regulatory T-cells, which suppress other T-cells that are otherwise primed to attack normal healthy cells in the body (Zhang et al., 2019). The role of IL-2 in immunity reportedly to act against gluten consumption in CD patients (Tye-Din et al., 2019) suggesting the use of IL-2 high serum level as a reliable diagnostic approach in celiac patients (Goel et al., 2019).
Because CPED1 is a novel candidate gene with largely unknown functions, CPED1 mutations are few and limited to 20 citations in PubMed. Data from Human Protein Atlas (https://www.proteinatlas.org/) indicate that CPED1 is expressed in several tissues and most abundant in the gastrointestinal mucosa. Likewise, CPED1 expressed in myeloid dendritic cells (DC), plasmacytoid DC, intermediate monocyte, classical monocyte and non-classical monocyte. Protein network predictions of CPED1 has linked it to the protein PPP6R3, which is known for its important role in maintaining immune self-tolerance (Coulibaly et al., 2019). Another predicted protein–protein association was on CPED1 with WNT16 protein. WNT signaling has a fundamental role in cellular development, proliferation and cell fate regulation (Loh et al., 2016). WNT signaling is important for the intestinal cells’ proliferation, gut development (Gregorieff et al., 2005) and alteration in such signaling pathways can affect the intestinal epithelial proliferation as it is expressed early in human infants (Dudhwala et al., 2019). This suggests a therapeutic potential as it activates the WNT signaling to compromise the intestinal epithelia in IBD and CD patients (Kuo, 2005). WNT16 activates the β-catenin/T-cell factor (TCF) pathway in dendritic cells. Recent studies have reported that the activation of Wnt/β-catenin pathway in DCs plays to solve a crucial puzzle in the mucosal tolerance and the suppression of chronic autoimmune pathologies (Suryawanshi et al., 2016). Emerging evidence support our hypothesis that mutations in CPED1 gene may alter the WNT signaling in DCs of the intestinal epithelium, hence it most likely contributes to the celiac pathogenesis.
Human GWAS have repeatedly mapped the uncharacterized CPED1 gene to the bone mineral density (BMD) phenotypes and with osteoporosis development (Chesi et al., 2017, Estrada et al., 2012, Medina-Gomez et al., 2012, Medina-Gomez et al., 2018). These findings may also explain the short stature phenotype in our patients and the growth failure in Saudi CD patients (Saadah, 2020). Shifting from genetic causes to environmental factors, one study showed how cesarean delivery can play a crucial role on developing CD by influencing the mucosal immune system through the infant’s microbiome (Decker et al., 2011). Although more recent studies have reported otherwise (Dydensborg Sander et al., 2018, Koletzko et al., 2018), we found both affected individuals in this family were born by cesarean section.
There are few limitations to which this study would sincerely like to admit. First, the affected sisters have no other sibling to compare the inheritance pattern of variant and also the negative family history to CD. Secondly, CPED1 has unknown function, making the in-silico analysis more challenging to support our claim.
In conclusion, we showed how WES method can be used as a powerful genetic strategy in resolving the molecular basis of complex or polygenic diseases like CD and also presents an opportunity to discover new potential candidate genes and therapeutic targets helpful in disease diagnosis and treatment. Our WES analysis has identified a genetic missense variant in CPED1 gene c.(241A > G) as a novel causal gene for CD in a Saudi family. This study expands the spectrum of celiac mutations that could be useful to the genetic diagnosis and counseling of families with CD. It is important to point out the importance of cell line based functional studies from gastrointestinal system to confirm the specific molecular mechanisms through which CPED1 mutations contributes to the biological pathways connected to CD.
Acknowledgments
Acknowledgement
This project was funded by the National Plan for Science, Technology, and Innovation (MAARIFAH) – King Abdulaziz City for Science and Technology – the Kingdom of Saudi Arabia – award number (13-MED2225-03). The authors also acknowledge with thanks the Science and Technology Unit, King Abdulaziz University for technical support.
Author contribution statement
RE, OS and NS conceptualization and design of the study; HB and BB formal analysis; HB, BB, NS and RE investigation; HB,OR and HA methodology; NS and OS resources; RE, NS and BB supervision; HB, RE validation, HB, NS, HA, AS, OA, JS, AA, KN, NA, BB, JA, RE and OS and writing original draft, review and editing; HB and BB data curation, software and visualization; NS funding acquisition and project administration.
Declaration of Competing Interest
Authors declares that they have no conflict of interest.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2020.04.011.
Contributor Information
Ramu Elango, Email: relango@kau.edu.sa.
Omar Ibrahim Saadah, Email: osaadah@kau.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abadie, V., Kim, S.M., Lejeune, T., Palanski, B.A., Ernest, J.D., Tastet, O., Jabri, B., et al., 2020. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. doi:10.1038/s41586-020-2003-8 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Abduljaleel Z. Comprehensive structure-function analysis of causative variants in retinal pigment epithelium specific 65 kDa protein associated Leber Congenital Amaurosis. Non-coding RNA Res. 2019;4(4):121–127. doi: 10.1016/j.ncrna.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Awan Z., Bima A., Rashidi O.M., Jamil K., Khan I.A., Almukadi H.S., Banaganapalli B. Low resolution protein mapping and KB-R7943 drug-protein molecular interaction analysis of long-QT syndrome linked KCNH2 mutations. All Life. 2020;13(1):183–193. doi: 10.1080/26895293.2020.1737249. [DOI] [Google Scholar]
- Ahola-Olli A.V., Wurtz P., Havulinna A.S., Aalto K., Pitkanen N., Lehtimaki T., Raitakari O.T. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am. J. Hum. Genet. 2017;100(1):40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Aama J.Y., Shaik N.A., Banaganapalli B., Salama M.A., Rashidi O., Sahly A.N., Saadah O.I. Whole exome sequencing of a consanguineous family identifies the possible modifying effect of a globally rare AK5 allelic variant in celiac disease development among Saudi patients. PLoS One. 2017;12(5):e0176664. doi: 10.1371/journal.pone.0176664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hamed M.H., Alsahan N., Rice S.J., Edwards N., Nooreddeen E., Alotaibi M., Imtiaz F. Bialleleic PKD1 mutations underlie early-onset autosomal dominant polycystic kidney disease in Saudi Arabian families. Pediatr. Nephrol. 2019;34(9):1615–1623. doi: 10.1007/s00467-019-04267-x. [DOI] [PubMed] [Google Scholar]
- Al-Hussaini A., Alharthi H., Osman A., Eltayeb-Elsheikh N., Chentoufi A. Genetic susceptibility for celiac disease is highly prevalent in the Saudi population. Saudi J. Gastroenterol. 2018;24(5):268–273. doi: 10.4103/sjg.SJG_551_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hussaini A., Troncone R., Khormi M., AlTuraiki M., Alkhamis W., Alrajhi M., Chentoufi A.A. Mass screening for celiac disease among school-aged children: toward exploring celiac Iceberg in Saudi Arabia. J. Pediatr. Gastroenterol. Nutr. 2017;65(6):646–651. doi: 10.1097/MPG.0000000000001681. [DOI] [PubMed] [Google Scholar]
- Aljebreen A.M., Almadi M.A., Alhammad A., Al Faleh F.Z. Seroprevalence of celiac disease among healthy adolescents in Saudi Arabia. World J. Gastroenterol. 2013;19(15):2374–2378. doi: 10.3748/wjg.v19.i15.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnaqeb D., Hamamy H., Youssef A.M., Al-Rubeaan K. Assessment of Knowledge, Attitude and Practice Towards Consanguineous Marriages Among a Cohort of Multiethnic Health Care Providers in Saudi Arabia. J. Biosoc. Sci. 2018;50(1):1–18. doi: 10.1017/S0021932016000675. [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. Novel eukaryotic enzymes modifying cell-surface biopolymers. Biol. Direct. 2010;5:1. doi: 10.1186/1745-6150-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha M., Demeer B., Revencu N., Helaers R., Theys S., Bou Saba S., Vikkula M. Whole exome sequencing identifies mutations in 10% of patients with familial non-syndromic cleft lip and/or palate in genes mutated in well-known syndromes. J. Med. Genet. 2018;55(7):449–458. doi: 10.1136/jmedgenet-2017-105110. [DOI] [PubMed] [Google Scholar]
- Chesi A., Mitchell J.A., Kalkwarf H.J., Bradfield J.P., Lappe J.M., Cousminer D.L., Grant S.F. A Genomewide Association Study Identifies Two Sex-Specific Loci, at SPTB and IZUMO3, Influencing Pediatric Bone Mineral Density at Multiple Skeletal Sites. J. Bone Miner. Res. 2017;32(6):1274–1281. doi: 10.1002/jbmr.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly A., Velásquez S.Y., Sticht C., Figueiredo A.S., Himmelhan B.S., Schulte J., Thiel M. AKIRIN1: a potential new reference gene in human natural killer cells and granulocytes in sepsis. Int. J. Mol. Sci. 2019;20(9):2290. doi: 10.3390/ijms20092290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker E., Hornef M., Stockinger S. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Gut Microbes. 2011;2(2):91–98. doi: 10.4161/gmic.2.2.15414. [DOI] [PubMed] [Google Scholar]
- Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., van Heel D.A. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudhwala Z.M., Drew P.A., Howarth G.S., Moore D., Cummins A.G. Active β-Catenin Signaling in the Small Intestine of Humans During Infancy. Digest. Diseases Sci. 2019;64(1):76–83. doi: 10.1007/s10620-018-5286-y. [DOI] [PubMed] [Google Scholar]
- Dydensborg Sander S., Hansen A.V., Stordal K., Andersen A.N., Murray J.A., Husby S. Mode of delivery is not associated with celiac disease. Clin. Epidemiol. 2018;10:323–332. doi: 10.2147/CLEP.S152168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano A., Catassi C. Clinical practice. Celiac disease. N. Engl. J. Med. 2012;367(25):2419–2426. doi: 10.1056/NEJMcp1113994. [DOI] [PubMed] [Google Scholar]
- Festen E.A., Goyette P., Green T., Boucher G., Beauchamp C., Trynka G., Rioux J.D. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet. 2011;7(1):e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., Jensen, L.J., et al., 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucl. Acids Res., 41(Database issue), D808-815. doi:10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed]
- Fuchs V., Kurppa K., Huhtala H., Maki M., Kekkonen L., Kaukinen K. Delayed celiac disease diagnosis predisposes to reduced quality of life and incremental use of health care services and medicines: a prospective nationwide study. United Eur. Gastroenterol. J. 2018;6(4):567–575. doi: 10.1177/2050640617751253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C., Ahn R., Ding Y.C., Steele L., Stoven S., Green P.H., Neuhausen S.L. Genome-wide association study of celiac disease in North America confirms FRMD4B as new celiac locus. PLOS ONE. 2014;9(7):e101428. doi: 10.1371/journal.pone.0101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel G., Tye-Din J., Qiao S.-W., Russell A., Mayassi T., Ciszewski C., Anderson R. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci. Adv. 2019;5:eaaw7756. doi: 10.1126/sciadv.aaw7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129(2):626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hrdlickova B., Mulder C.J., Malamut G., Meresse B., Platteel M., Kamatani Y., Bonder M.J. A locus at 7p14. 3 predisposes to refractory celiac disease progression from celiac disease. Eur. J. Gastroenterol. Hepatol. 2018;30(8):828. doi: 10.1097/MEG.0000000000001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova B., Mulder C.J., Malamut G., Meresse B., Platteel M., Kamatani Y., Kumar V. A locus at 7p14.3 predisposes to refractory celiac disease progression from celiac disease. Eur. J. Gastroenterol. Hepatol. 2018;30(8):828–837. doi: 10.1097/MEG.0000000000001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., van Heel D.A. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husby, S., Koletzko, S., Korponay-Szabo, I.R., Mearin, M.L., Phillips, A., Shamir, R., Nutrition., 2012. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr., vol. 54, 1, pp. 136–160. doi:10.1097/MPG.0b013e31821a23d0. [DOI] [PubMed]
- Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J., Staley J.R. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koletzko, S., Lee, H.S., Beyerlein, A., Aronsson, C. A., Hummel, M., Liu, E., Group, T.S., 2018. Cesarean Section on the Risk of Celiac Disease in the Offspring: The Teddy Study. J. Pediatr. Gastroenterol. Nutr., vol. 66, 3, pp. 417–424. doi:10.1097/MPG.0000000000001682. [DOI] [PMC free article] [PubMed]
- Kufareva I., Abagyan R. Methods of protein structure comparison. Methods Mol. Biol. 2012;857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, C., 2005. Modulation of gastrointestinal epithelium proliferation through the Wnt signaling pathway. In: Google Patents.
- Li R., Zheng Y., Li Y., Zhang R., Wang F., Yang D., Gao Z. Common variable immunodeficiency with genetic defects identified by whole exome sequencing. BioMed Res. Int. 2018;3724630:2018. doi: 10.1155/2018/3724630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E., Azuara C., Koehl P., Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucl. Acids Res. 2006;34(Web Server issue):W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindfors K., Ciacci C., Kurppa K., Lundin K.E., Makharia G.K., Mearin M.L., Kaukinen K. Coeliac disease. Nat. Rev. Disease Primers. 2019;5(1):3. doi: 10.1038/s41572-018-0054-z. [DOI] [PubMed] [Google Scholar]
- Loh Kyle M., van Amerongen R., Nusse R. Generating Cellular Diversity and Spatial Form: Wnt Signaling and the Evolution of Multicellular Animals. Develop. Cell. 2016;38(6):643–655. doi: 10.1016/j.devcel.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Ludvigsson J.F., Leffler D.A., Bai J.C., Biagi F., Fasano A., Green P.H., Ciacci C. The Oslo definitions for coeliac disease and related terms. Gut. 2013;62(1):43–52. doi: 10.1136/gutjnl-2011-301346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci P., Filion C.A., Boisson B., Itan Y., Shang L., Casanova J.-L., Cunningham-Rundles C. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front. Immunol. 2016;7:220. doi: 10.3389/fimmu.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi T.A., Shaik N.A., Burhan S., Hasan Q., Shafi G., Talluri V.R. Structural prediction, whole exome sequencing and molecular dynamics simulation confirms p. G118D somatic mutation of PIK3CA as functionally important in breast cancer patients. Comput. Biol. Chem. 2019;80:472–479. doi: 10.1016/j.compbiolchem.2019.05.012. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez C., Kemp J.P., Estrada K., Eriksson J., Liu J., Reppe S., Rivadeneira F. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8(7):e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez C., Kemp J.P., Trajanoska K., Luan J., Chesi A., Ahluwalia T.S., Rivadeneira F. Life-Course Genome-wide Association Study Meta-analysis of Total Body BMD and Assessment of Age-Specific Effects. Am. J. Hum. Genet. 2018;102(1):88–102. doi: 10.1016/j.ajhg.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano L.M., Pascual V., Bodas A., López-Palacios N., Salazar I., Espino-Paisán L., Núñez C. Expression patterns common and unique to ulcerative colitis and celiac disease. Annals Human Genet. 2019;83(2):86–94. doi: 10.1111/ahg.12293. [DOI] [PubMed] [Google Scholar]
- Mistry V., Bockett N.A., Levine A.P., Mirza M.M., Hunt K.A., Ciclitira P.J., van Heel D.A. Exome sequencing of 75 individuals from multiply affected coeliac families and large scale resequencing follow up. PLOS ONE. 2015;10(1):e0116845. doi: 10.1371/journal.pone.0116845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostensson M., Monten C., Bacelis J., Gudjonsdottir A.H., Adamovic S., Ek J., Torinsson-Naluai A. A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease. PLOS ONE. 2013;8(8):e70174. doi: 10.1371/journal.pone.0070174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafi S.K., Fernandez-Jaen A., Alvarez S., Nadeau O.W., Butler M.G. High Functioning Autism with Missense Mutations in Synaptotagmin-Like Protein 4 (SYTL4) and Transmembrane Protein 187 (TMEM187) Genes: SYTL4- Protein Modeling, Protein-Protein Interaction, Expression Profiling and MicroRNA Studies. Int. J. Mol. Sci. 2019;20(13) doi: 10.3390/ijms20133358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadah, O.I., NM, A. L. (2020). Celiac disease in Saudi children with isolated short stature: is it rare or are we not screening rigorously enough? J. Pediatr. Endocrinol. Metab., vol. 33, 1, pp. 89–93. doi:10.1515/jpem-2019-0348. [DOI] [PubMed]
- Saadah O.I., Shaik N.A., Banaganapalli B., Salama M.A., Al-Harthi S.E., Wang J., Al-Aama J.Y. Replication of GWAS Coding SNPs Implicates MMEL1 as a Potential Susceptibility Locus among Saudi Arabian Celiac Disease Patients. Dis Mark. 2015;2015:351673. doi: 10.1155/2015/351673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaei A., Rezaei Tavirani M., Arefi Oskouei A., Zamanian Azodi M., Mohebbi S.R., Nikzamir A.R. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol. Hepatol. Bed Bench. 2016;9(2):114–123. [PMC free article] [PubMed] [Google Scholar]
- Singh P., Arora A., Strand T.A., Leffler D.A., Catassi C., Green P.H., Makharia G.K. Global prevalence of celiac disease: systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2018;16(6) doi: 10.1016/j.cgh.2017.06.037. 823–836 e822. [DOI] [PubMed] [Google Scholar]
- Staley J.R., Blackshaw J., Kamat M.A., Ellis S., Surendran P., Sun B.B., Butterworth A.S. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi A., Tadagavadi R.K., Swafford D., Manicassamy S. Modulation of Inflammatory Responses by Wnt/beta-Catenin Signaling in Dendritic Cells: A Novel Immunotherapy Target for Autoimmunity and Cancer. Front. Immunol. 2016;7:460. doi: 10.3389/fimmu.2016.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., van Heel D.A. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43(12):1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Zhernakova A., Romanos J., Franke L., Hunt K.A., Turner G., Wijmenga C. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58(8):1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- Tye-Din J.A., Skodje G.I., Sarna V.K., Dzuris J.L., Russell A.K., Goel G., Anderson R.P. Cytokine release after gluten ingestion differentiates coeliac disease from self-reported gluten sensitivity. United Eur. Gastroenterol. J. 2019;2050640619874173 doi: 10.1177/2050640619874173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39(7):827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta U., Fabbri A., Parisi C., Piscaglia M., Caio G., Tovoli F., Fiorini E. Old and new serological tests for celiac disease screening. Exp. Rev. Gastroenterol. Hepatol. 2010;4(1):31–35. doi: 10.1586/egh.09.66. [DOI] [PubMed] [Google Scholar]
- Wada H., Hayashida M., Sato T., Minowa S., Ikezaki O., Mitsui T., Hisamatsu T. A Caucasian American patient with celiac disease diagnosed in Japan and successfully treated with a gluten-free diet. Clin. J. Gastroenterol. 2018;11(1):23–28. doi: 10.1007/s12328-017-0794-4. [DOI] [PubMed] [Google Scholar]
- Withoff S., Li Y., Jonkers I., Wijmenga C. Understanding Celiac Disease by Genomics. Trends Genet. 2016;32(5):295–308. doi: 10.1016/j.tig.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemni E.A., Monies D., Alkhairallah T., Bohlega S., Abouelhoda M., Magrashi A., Al-Tassan N. Integrated analysis of whole exome sequencing and copy number evaluation in Parkinson's Disease. Sci. Rep. 2019;9(1):3344. doi: 10.1038/s41598-019-40102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Gothe F., Pennamen P., James J.R., McDonald D., Mata C.P., Lenardo M.J. Human interleukin-2 receptor β mutations associated with defects in immunity and peripheral tolerance. J. Exp. Med. 2019;216(6):1311–1327. doi: 10.1084/jem.20182304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Stahl E.A., Trynka G., Raychaudhuri S., Festen E.A., Franke L., Plenge R.M. Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci. PLOS Genet. 2011;7(2):e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Qiu J., Magrane G., Abedalthagafi M., Zanko A., Golabi M., Chehab F.F. Duplication of C7orf58, WNT16 and FAM3C in an obese female with a t(7;22)(q32.1;q11.2) chromosomal translocation and clinical features resembling Coffin-Siris Syndrome. PLOS ONE. 2012;7(12):e52353. doi: 10.1371/journal.pone.0052353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.