Abstract
Background
A minority of patients with advanced non-small-cell lung cancer (NSCLC) benefit from treatment with immune checkpoint inhibitors (ICIs). Ineffective effector function of activated T and NK cells may lead to reduced tumor cell death, even when these activated effector cells are released from their immune checkpoint brake. Hence, in this study we aimed to assess the association of baseline serum granzyme B, as well as germline variation of the GZMB gene, with clinical outcome to programmed cell death protein 1 (PD-1) blockade.
Methods
A total of 347 patients with stage IV NSCLC who started nivolumab treatment between June 2013 and June 2017 were prospectively included. Baseline serum and whole blood was available, allowing for protein quantification and targeted DNA sequencing. Clinical outcome was based on best overall response (BOR) according to Response Evaluation Criteria in Solid Tumors, V.1.1, progression-free survival (PFS), and overall survival (OS).
Results
Patients with low serum levels of granzyme B had worse PFS (HR: 1.96; 95% CI: 1.12 to 3.43; p=0.018) and worse OS (HR: 2.08; 95% CI: 1.12 to 3.87; p=0.021) than patients with high baseline serum levels. To validate the findings, germline variation of GZMB rs8192917 was assessed. Patients with homozygous and heterozygous variants of GZMB rs8192917 had worse BOR (OR: 1.60; 95% CI: 1.01 to 2.52; p=0.044) and worse PFS (HR: 1.38; 95% CI:1.02 to 1.87; p=0.036) than wild types.
Conclusions
A low baseline serum level of granzyme B and germline variation of GZMB was associated with worse clinical outcome in NSCLC, emphasizing the relevance and additional value of monitoring germline genetic variations which mirror cytotoxic functions of T cells in ICI therapy.
Trail registration number
Dutch Trial Registry (NL6828).
Keywords: immunotherapy, programmed cell death 1 receptor, lung neoplasms, genetic markers, translational medical research
Background
Immune checkpoint inhibitors (ICIs) target immune effector cells, predominantly T cells, which may lead to an effective immune response toward tumor cells.1 2 Although ICIs have shown to significantly improve overall survival (OS) in advanced non-small-cell lung cancer (NSCLC), only a minority of patients benefit from treatment, with an objective response in about one out of five patients.3 4
The molecular mechanisms underlying resistance to ICI therapy are only partly explained. It is postulated that a combination of factors contribute to response, involving tumor biology, tumor microenvironment, the peripheral immune system, as well as germline genetics.5 6 programmed cell death protein 1 (PD-1)-mediated inhibition targets signaling downstream of the T cell receptor via its downstream phosphatase SHP-2, as well as costimulatory molecules.7 8 Importantly, distorted effector function of activated T and NK cells may lead to reduced tumor cell death, even when these activated effector cells are released from their PD-1 brake.
Previously, it has been demonstrated that immunohistochemical staining and in vivo imaging, using positron emission tomography, of intratumoral granzyme B may serve as a predictive biomarker for PD-1 blockade in melanoma.9 Granzyme B is a key serine protease that is secreted to induce apoptosis, primarily by activated T cells and NK cells.10 Distortion of granzyme B activity could be linked to reduced entry, trafficking, and accumulation within the cytoplasm of target cells (eg, tumor cells), and/or linked to reduced enzymatic activity, and hence decreased cleavage of intracellular substrates, such as caspase-3.11–13
Taken together, we hypothesize that low baseline serum levels of granzyme B, and consequently alterations of the GZMB locus, may contribute to resistance mechanisms of PD-1 blockade. The primary objectives of the current analysis were to assess the association of baseline serum granzyme B levels with progression-free survival (PFS) and OS after PD-1 blockade in patients with NSCLC. As a validation, genetic variants of the GZMB gene were related with PFS, OS and best overall response (BOR). Secondary objectives were to assess the relationship between serum granzyme B as well as genetic variations of GZMB and cytotoxic immune cell populations in blood.
Methods
Study design and data collection
Patients with advanced NSCLC who were treated with PD-1 ICIs between June 2013 and June 2017 at the Erasmus University MC (Rotterdam, The Netherlands) and the Amphia Hospital (Breda, The Netherlands) were prospectively included. Patients with NSCLC stage IV who had been treated with nivolumab monotherapy (3 mg/kg intravenous, once every 2 weeks) were included, and patients who were treated with a prior line of immunotherapy were excluded from the study. The study was approved by the independent ethics committee board (MEC 02-1002 and MEC 16-011) and all patients provided written informed consent. Whole blood and baseline serum were available for DNA analysis and protein quantification, respectively. All assays were performed blinded to study endpoints; patients were assigned to a subject number. Patient characteristics included demographic and clinical data. BOR was assessed according to the Response Evaluation Criteria in Solid Tumors V.1.1 (RECIST V.1.1).14 A minimum duration of 90 days for stable disease (SD) was required. Confirmation of partial response (PR) or complete response (CR) was not required. After treatment initiation, radiological evaluation by CT was usually performed every 6 weeks. PFS was defined as the time from the first administration of nivolumab until progressive disease (PD), or death due to any cause, whichever occurred first. OS was defined as the time from the first administration of nivolumab until death due to any cause.
Quantitative protein measurement and DNA analysis
Quantitative analysis of granzyme B in serum was performed by a magnetic bead-based assay, using a 1×96-well microplate and magnetic antigranzyme-B-coated beads (Human Magnetic Luminex Assay, R&D Systems Inc, Minneapolis, Minnesota, USA). GZMB c.128T>C (rs8192917) was selected primarily based on its presumed contribution to resistance mechanisms of ICIs.9 A total of 12 single-nucleotide polymorphisms (SNPs) were selected in eight genes based on their contribution to T cell immunity and their correlation with overt T cell responses, such as the autoimmune diseases systemic lupus erythematosus and rheumatoid arthritis.15 16–25 A summary of the correlation of selected SNPs with overt T cell responses is displayed in online supplementary table 1. SNPs with a minor allele frequency (MAF) of <5% were excluded from the analysis. Linkage disequilibrium was tested using the application LDmatrix of LDlink V.5 (https://ldlink.nci.nih.gov/). Distribution of genotypes was tested for Hardy-Weinberg equilibrium (HWE) using the χ2 test. Deviations from HWE may reflect genotyping errors, though, may also be a signal of disease association,26 for example, mutation of tumor suppressor genes. DNA isolation and genotyping for GZMB rs8192917, HLA-A rs60131261, IL10 rs3024493, IL2RA rs3024493, IFNG rs2430561/rs2069705/rs2069718, PDCD1 rs2227981/rs10204525/rs2227982, PTPN11 rs2301756 and ZAP70 rs13420683 has been performed using TaqMan analysis as previously described.27 Whole-blood specimens were used for extraction of DNA. Genotyping was performed using predesigned TaqMan allelic discrimination assays, consisting of two allele-specific minor groove-binding probes. PCRs were performed in a reaction volume containing DNA, primers, and the specific probes were labeled with fluorescent dye.
jitc-2020-000586supp001.pdf (532KB, pdf)
Flow cytometry
Peripheral blood samples were analyzed using multiplex flow cytometry as previously described by Kunert et al.6 In short, absolute cell counts of immune cell populations were determined on whole blood after lysis of red blood cells, and frequencies of particular subsets were determined in more detail on cryopreserved peripheral blood mononuclear cell samples that were stained with a mix of antibodies.
Mutational burden and expression of PD-L1 in tumor tissue
Expression of programmed death-ligand 1 (PD-L1) and tumor mutational burden (TMB) have previously been determined in a subset of patients of cohort 1 (n=26; patients with stage IV NSCLC) using archival formalin-fixed, paraffin-embedded tissue samples.28 Baseline expression of PD-L1 has been determined using antibody clone SP263 (Roche Diagnostics, Tucson, Arizona, USA), whereas TMB has been measured using the Oncomine TML assay (Thermofisher Scientific, Waltham, Massachusetts, USA).
Statistical analysis
To study the relationship between baseline protein levels or SNPs and survival (PFS and OS), Cox regression analysis was used. Patient survival was visualized by the Kaplan-Meier approach. Patients were dichotomized for low or high granzyme B serum levels based on the median value of all patients. Dominant, recessive, and additive models29 were used to test associations between SNPs and clinical outcomes including BOR, PFS, and OS. HRs and 95% CIs were calculated. For the association of SNPs with radiological response, BOR was used as an ordered categorical variable with three levels, namely CR/PR (pooled), SD, and PD, by means of ordinal logistic regression analysis. This method assumes that the relationship between each pair of outcome groups is the same. The proportional odds assumption was checked for all analyses. Univariate ordinal logistic regression was applied for both SNPs and patient factors, and OR and 95% CIs were calculated. Multivariable ordinal logistic analysis was performed including both single SNPs and patient factors, with p<0.1 according to the univariate analyses. All analyses, with a significant relation (p<0.05) between SNP and clinical outcome, were internally validated by bootstrapping,30 where 1000 bootstrap samples were generated (with replacement) and 95% CIs were calculated for either ORs or HRs. Furthermore, bias and bias-corrected 95% CIs were calculated. In general, a p value <0.05 was considered to be statistically significant. Analyses were performed using IBM SPSS Statistics V.24.0.0.1 (Chicago, Illinois, USA) and STATA V.15.1 (StataCorp LP, College Station, Texas, USA).
Results
Patient characteristics
Baseline serum from a total of 78 (cohort 1) and DNA from a total of 322 patients with stage IV NSCLC (cohort 2) was available for analysis (table 1). There was a 16.5% subject overlap between the patient cohorts. All patients were treated with at least one dose of nivolumab at 3 mg/kg once every 2 weeks. Overall, median absolute starting dose was 222 mg (IQR 192–263 mg). The objective response rate was similar in cohort 1 and cohort 2, respectively 19.2% and 16.6%. A minority (7%) of all patients could not be evaluated for response by RECIST V.1.1 due to early death, loss to follow-up or incomplete or absent follow-up tumor assessments. In the univariate analysis of cohort 2, WHO performance status (0 vs ≥1) and sex (male vs female) were significantly associated with PFS (HR: 0.66; 95% CI: 0.46 to 0.95; p=0.027 and HR: 1.38; 95% CI: 1.06 to 1.80; p=0.18, respectively) and OS (HR: 0.52; 95% CI: 0.34 to 0.77; p=0.001 and HR: 1.40; 95% CI: 1.05 to 1.86; p=0.023, respectively). In the multivariable analysis, only WHO performance status and primary tumor type remained significant for their association with, respectively PFS or OS and BOR.
Table 1.
Patient characteristics
| Cohort 1 (n=78) | Cohort 2 (n=322) | |
| Age at start | ||
| Mean (range in years) | 63.6 (35–78) | 64.7 (29–85) |
| n | % | n | % | |
| Sex | ||||
| Male | 48 | 61.5 | 202 | 62.7 |
| Female | 30 | 38.5 | 120 | 37.3 |
| Primary tumor | ||||
| Adenocarcinoma | 52 | 66.7 | 200 | 62.1 |
| Squamous cell carcinoma | 17 | 21.8 | 96 | 29.8 |
| Large cell carcinoma | – | – | 22 | 6.8 |
| Lung cancer with unknown pathology | 9 | 11.5 | 4 | 1.2 |
| WHO PS | ||||
| 0 | 18 | 23.1 | 54 | 16.8 |
| 1 | 39 | 50.0 | 188 | 58.4 |
| 2 | 1 | 1.3 | 7 | 2.2 |
| 3 | – | – | 1 | 0.3 |
| Unknown | 20 | 25.6 | 72 | 22.4 |
| Prior lines | ||||
| 0 | 2 | 2.6 | 1 | 0.3 |
| 1 | 60 | 76.9 | 217 | 67.4 |
| 2 | 13 | 16.7 | 82 | 25.5 |
| 3 | 2 | 2.6 | 15 | 4.7 |
| 4 | 1 | 1.3 | 6 | 1.9 |
| 5 | – | – | 1 | 0.3 |
| Smoking status | ||||
| History or current smoker | 56 | 71.8 | 238 | 73.9 |
| Never smoker | 8 | 10.3 | 20 | 6.2 |
| Unknown | 14 | 17.9 | 64 | 19.9 |
| Ethnicity | ||||
| Caucasian | 74 | 94.9 | 308 | 95 |
| Other | 4 | 5.1 | 5 | 2 |
| Unknown | 0 | – | 9 | 3 |
Baseline characteristics of NSCLC cohort 1 and cohort 2. 16.5% of the patients (n=53) from cohort 1 were overlapping with cohort 2.
NSCLC, non-small-cell lung cancer; WHO PS, WHO performance status.
Serum levels of granzyme B are associated with clinical outcome
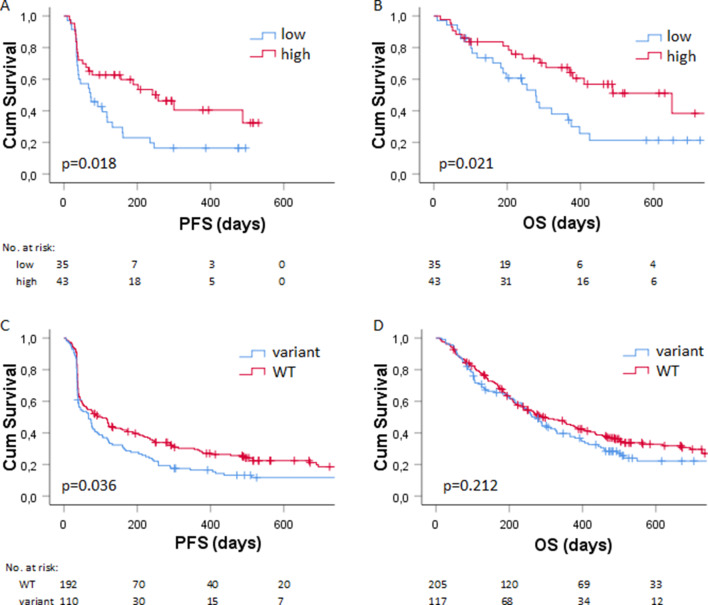
Serum levels of granzyme B were determined in cohort 1 (online supplementary figure 1), having a median concentration of 1.27 pg/mL, which was used as the cut-off to dichotomize patients with low and high levels of serum granzyme B (as described in the Method section). Patients with low baseline serum levels of granzyme B had a significant worse PFS (HR: 1.96; 95% CI: 1.12 to 3.43; p=0.018, figure 1A) than patients with high levels of baseline serum granzyme B. Likewise, low levels of granzyme B were significantly associated with worse OS (HR: 2.08; 95% CI: 1.12 to 3.87; p=0.021, figure 1B) compared with high levels of granzyme B.
Figure 1.
Association of serum granzyme B and GZMB genotype with survival (A) Kaplan-Meier plots showing the (A) PFS (in days) and (B) OS (in days) of patients in cohort 2 (n=78) with low (in blue) versus high (in red) baseline serum levels of granzyme B. The (C) PFS and (D) OS of patients in cohort 2 (n=322) with wild type (in red) versus variant germline GZMB (in blue). Numbers at risk are shown below the graph. OS, overall survival; PFS, progression-free survival.
Germline variation of GZMB is associated with clinical outcome
To validate the findings in a second cohort by a different approach,31 genetic alterations of GZMB c.128T>C (rs8192917), together with a mechanism-of-action-based panel of other SNPs, were related to PFS, OS and BOR in cohort 2. Investigated SNPs with the corresponding haplotype frequencies, MAF and HWE statistics are shown in online supplementary table 2. After correction for WHO performance status and sex in multivariable analysis, patients with homozygous and heterozygous variants of GZMB rs8192917 had worse PFS (HR: 1.38; 95% CI: 1.02 to 1.87; p=0.036; table 2, figure 1C). This finding could be internally validated (HR 95% CI: 1.02 to 1.87; bias: 0.003; bias-corrected 95% CI: 1.03 to 1.88; p=0.036; online supplementary table 3). Remaining SNPs were not associated with PFS after correction for patient factors (table 3). No significant association of SNPs with OS was observed after correction of patient factors (table 3). Of note, homozygous and heterozygous variants of GZMB rs8192917 were not significantly associated with OS (HR: 1.19; 95% CI: 0.91 to 1.57; p=0.212; table 2, figure 1D). Homozygous and heterozygous variants of GZMB rs8192917 were significantly associated with worse BOR (OR: 1.60; 95% CI: 1.01 to 2.52; p=0.044; table 2) and could be internally validated for the association with BOR (OR 95% CI: 1.05 to 2.55, bias: 0.007; bias-corrected 95% CI: 1.09 to 2.57; p=0.030, online supplementary table 3). All the results from both univariate (p<0.1) and multivariate analyses of the association with PFS, OS or BOR are shown in table 3. Results from the internal validation of factors that were significantly associated after correction for patient factors (p value <0.05) are shown in online supplementary table 3. For a subset of patients, both serum levels of granzyme B and germline variation of GZMB c.128T>C (rs8192917) were available for analysis. Patients with homozygous and heterozygous variants of GZMB rs8192917 (n=19) had significant lower serum levels of granzyme B compared with patients with wild-type GZMB (n=34; mean difference 8.6 pg/mL, 95% CI 1.77 to 15.38, p=0.015).
Table 2.
Association between GZMB genotype and clinical outcome
| Parameter | Test variables | Univariate analysis | Multivariate analysis | ||
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| PFS | |||||
| GZMB c.128T>C | CC+CT vs TT | 1.39 (1.07 to 1.80) | 0.013* | 1.38 (1.02 to 1.87) | 0.036* |
| WHO | 0 vs ≥1 | 0.66 (0.46 to 0.95) | 0.027* | 0.61 (0.42 to 0.89) | 0.01* |
| Sex | Male vs female | 1.38 (1.06 to 1.80) | 0.018* | 1.29 (0.95 to 1.76) | 0.103 |
| OS | |||||
| GZMB c.128T>C | CC+CT vs TT | 1.19 (0.91 to 1.57) | 0.212 | ||
| WHO | 0 vs ≥1 | 0.52 (0.35 to 0.77) | 0.001* | ||
| Sex | Male vs female | 1.40 (1.05 to 1.86) | 0.023* | ||
| Parameter | Test variables | Univariate analysis | Multivariate analysis | ||
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| BOR | |||||
| GZMB c.128T>C | CC +CT vs TT | 1.63 (1.04 to 2.57) | 0.033* | 1.60 (1.01 to 2.52) | 0.044* |
| Primary tumor | Other vs adeno | 0.63 (0.40 to 0.98) | 0.041* | 0.63 (0.41 to 0.99) | 0.045* |
Univariate and multivariate analysis of the association of germline variation of GZMB and PFS, OS, or BOR. Patient factors associated with OS/PFS (p value<0.1) were included in the multivariate analysis. Significance is marked by *.
BOR, best overall response; OS, overall survival; PFS, progression-free survival; WHO, WHO performance status.
Table 3.
Association between investigated SNPs and clinical outcome
| Parameter | Test variables | HR (95% CI) | P value |
| Univariate analysis (PFS) | |||
| IFNG c.874T>A | AA+AT vs TT | 0.76 (0.58 to 0.99) | 0.045 |
| IL2RA c.64+5006T>C | CC+CT vs TT | 0.80 (0.62 to 1.04) | 0.098 |
| WHO performance status | 0 vs ≥1 | 0.66 (0.46 to 0.95) | 0.027 |
| Sex | Male vs female | 1.38 (1.06 to 1.80) | 0.018 |
| Multivariate analysis (PFS) | |||
| IFNG c.874T>A | AA+AT vs TT | 0.79 (0.58 to 1.07) | 0.131 |
| WHO performance status | 0 vs ≥1 | 0.66 (0.45 to 0.95) | 0.024 |
| Sex | Male vs female | 1.26 (0.92 to 1.71) | 0.148 |
| | | | |
| IL2RA c.64+5006T>C | CC+CT vs TT | 0.81 (0.60 to 1.10) | 0.177 |
| WHO performance status | 0 vs ≥1 | 0.65 (0.45 to 0.93) | 0.019 |
| | | | |
| Sex | Male vs female | 1.29 (0.95 to 1.76) | 0.101 |
| IFNG c.874T>A | AA+AT vs TT | 0.76 (0.55 to 1.05) | 0.093 |
| WHO performance status | 0 vs ≥1 | 0.52 (0.34 to 0.77) | 0.001 |
| Sex | Male vs female | 1.45 (1.03 to 2.02) | 0.32 |
| Univariate analysis (OS) | |||
| IFNG c.874T>A | AA+AT vs TT | 0.72 (0.55 to 0.95) | 0.021 |
| WHO PS | 0 vs ≥1 | 0.52 (0.35 to 0.77) | 0.001 |
| Sex | Male vs female | 1.40 (1.05 to 1.86) | 0.023 |
| Multivariate analysis (OS) | |||
| IFNG c.874T>A | AA+AT vs TT | 0.76 (0.55 to 1.05) | 0.093 |
| WHO performance status | 0 vs ≥1 | 0.52 (0.34 to 0.77) | 0.001 |
| Sex | Male vs female | 1.45 (1.03 to 2.02) | 0.32 |
| Parameter | Test variables | OR (95% CI) | P value |
| Univariate analysis (BOR) | |||
| IL10 c.387+284C>A | AA vs AA +AC | 0.25 (0.07 to 0.89) | 0.033 |
| ZAP70 −21–4127C>A | AA vs CC+AC | 0.50 (0.25 to 0.98) | 0.045 |
| Primary tumor | Other vs adeno | 0.63 (0.40 to 0.98) | 0.041 |
| Multivariate analysis (BOR) | |||
| IL10 c.387+284C>A | AA vs AA +AC | 0.24 (0.07 to 0.84) | 0.026 |
| Primary tumor | Other vs adeno | 0.61 (0.39 to 0.96) | 0.031 |
| | | | |
| ZAP70 −21–4127C>A | AA vs CC+AC | 0.49 (0.24 to 0.97) | 0.042 |
| Primary tumor | Other vs adeno | 0.61 (0.39 to 0.96) | 0.033 |
Univariate and multivariate analysis of the association between germline SNPs (except those related to GZMB variation; those are shown in table 2) and PFS, OS, or BOR. Only SNPs that were associated with OS/PFS/BOR (p value <0.1) are shown and included in the multivariate analysis. Significance is marked by *.
BOR, best overall response; OS, overall survival; PFS, progression-free survival; SNPs, single-nucleotide polymorphisms; WHO PS, WHO performance status.
Serum levels of granzyme B are accompanied by presence of T cells with terminal differentiation and exhausted phenotype
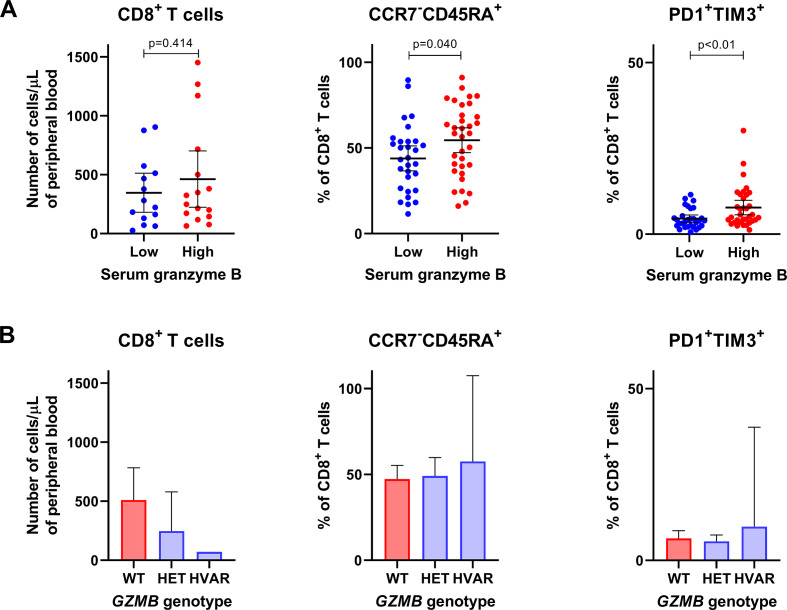
Next, we assessed the relationship between serum granzyme B as well as genetic variations of GZMB and cytotoxic immune cell populations in blood. Overall, there was a tendency that patients with lower serum granzyme B levels had lower absolute numbers of peripheral CD56+ NK cells (mean 141 vs 205 cells/µL, p=0.16; online supplementary figure 2) and CD8+ T cells (346 vs 463 cells/µL, p=0.41; figure 2A) when compared with patients with higher serum granzyme B levels. In fact, patients with homozygous or heterozygous variants of GZMB rs8192917 had a higher number of CD8+ T cells but not CD56+ NK cells than patients with wild-type GZMB (figure 2B and online supplementary figure 2B). Further, we studied whether granzyme B levels or GZMB genotypes correlate with frequencies of those immune cell phenotypes that correspond to either terminal differentiation (CCR7-CD45RA+) or exhaustion (PD1+ and TIM3+) of CD8+ T cells, as signs of T cell cytotoxicity. Interestingly, patients with lower serum granzyme B levels had a lower proportion of CCR7−CD45RA+CD8+ T cells (mean 44 vs 54 %, p=0.040) as well as PD1+TIM3+CD8+ T cells (mean 4.5 vs 7.8 %, p=0.001; figure 2A). No significant association nor trend could be observed between GZMB genotype and frequencies of T cells with these phenotypes (figure 2B).
Figure 2.
Association between serum granzyme B and T cell populations with phenotypes that correspond to functional responses. Associations are shown between (A) serum granzyme B levels or (B) GZMB genotype and T cell populations in blood. CD8+ T cells is considered as a subset of T lymphocytes with cytotoxic functions. CCR7−CD45RA+ cytotoxic T cells correspond with terminal differentiation, and PD1+TIM3+ cytotoxic T cells correspond with exhaustion, both signs of T cell activation or cytotoxicity. HET, heterozygous variant; HVAR, homozygous variant; WT, wild type.
Relationship between serum granzyme B, expression of PD-L1 and TMB
Interaction analysis for PFS and OS was performed between serum granzyme B, and PD-L1 and TMB in a subset of evaluable patients. In the univariate analysis, TMB (HR: 7.31; 95% CI: 1.5 to 34.7; p=0.01) was significantly correlated with PFS, while expression of PD-L1 (HR: 4.30; 95% CI: 0.9 to 19.8; p=0.06) or serum granzyme B (HR: 2.34; 95% CI: 0.9 to 5.9; p=0.07) did not meet the threshold for significance (online supplementary table 4). Moreover, only serum granzyme B was significantly correlated with OS (HR: 2.95; 95% CI: 1.09 to 7.93; p=0.03; online supplementary table 5). Multivariate analysis of these biomarkers (namely, serum granzyme B, PD-L1 and TMB) for PFS and OS did not meet significance. Of note, the HRs were in favor of higher levels of PD-L1 (≥50%), TMB (≥10 mut/Mb) and serum granzyme B (≥1.27 pg/mL), yet expression of PD-L1 (%), TMB (mut/Mb) and serum granzyme B levels (pg/mL) were not significantly correlated with each other (online supplementary table 6).
Discussion
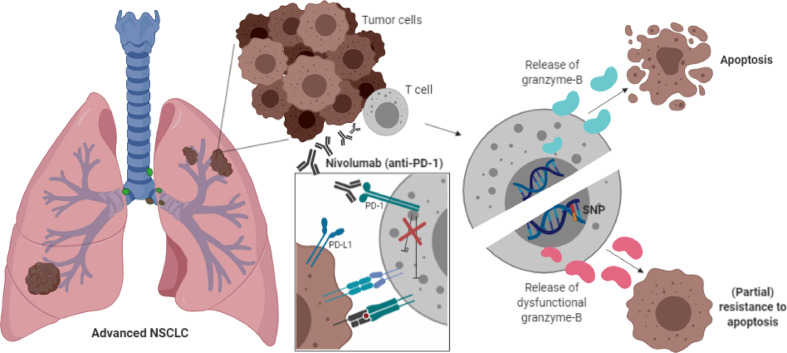
The present study demonstrates that serum levels of granzyme B and alterations of the GZMB locus may contribute to resistance mechanisms of PD-1 blockade. This concept (figure 3) is based on our findings that (1) low serum levels of granzyme B and (2) a common germline polymorphism of the GZMB gene were associated with worse clinical outcome after PD-1 blockade in patients with metastatic NSCLC. Moreover, serum levels of granzyme B were positively correlated with peripheral abundance of T cell populations that were highly differentiated or were expressing the immune checkpoints PD-1 and TIM3. The results are inline with our hypothesis that cytotoxic effector functions of lymphocytes (ie, through granzymes) are critical for the anti-PD-1 antibody-mediated immune effects.
Figure 3.
Mechanism of action. Schematic overview of the proposed mechanism of action of the GZMB SNP. Presence of the GZMB c.128T>C SNP indicates the expression of variant granzyme B isotypes (indicated as pink granzyme B molecules) that might lead to impaired cytotoxic effector functions of T cells, resulting in impaired apoptosis of tumor cells and partial resistance to anti-PD-1 blockade. The molecular mechanism of the PD-1 antibody nivolumab is shown in the white box. This figure was created by DPH in BioRender.com. PD-1, programmed cell death 1 receptor; SNP, single-nucleotide polymorphism.
Granzyme B levels were determined at baseline in patients with stage IV NSCLC prior to start with PD-1 ICIs. The median granzyme B level in this cohort was 1.27 pg/mL (range 0–88 pg/mL), which was lower than reported levels in healthy controls, for example, a median plasma granzyme B of 5.7 pg/mL (IQR 3.7–7.1 pg/mL; n=38)32 or 11.5 pg/mL (range 1–130 pg/mL; n=54).33 Measurements of granzyme B in serum were consistent with measurements in plasma.33 Our findings confirm the hypothesis that lower granzyme B levels in patients with metastatic cancer mirror the immune microenvironment of tumors favoring tumor growth by halting the antitumor response of cytotoxic immune cells. This may be a result of those germline variations that impact cytotoxic functions of lymphocytes.
Besides germline polymorphisms in the GZMB gene, polymorphisms in other genes have been investigated as well for their association with clinical outcome. Interestingly, these germline variations were not consistently found to correlate with PFS, OS, or BOR. Interpretations of these findings are challenging, and do not necessarily implicate an absence of effects of non-GZMB SNPs. Although SNPs were rationally selected by their correlations with autoimmunity (online supplementary table 2), we cannot exclude that SNPs in the investigated genes contribute to therapy resistance, as well as epigenetic or post-transcriptional regulation of gene expression or function. For example, PD-1 ICIs are suggested to only minimally target responses mediated by T cell receptors (involving ZAP70) and their primary method of action is most likely explained by relieving SHP-2 inhibition of the CD28 costimulatory pathway.8 Genomic alterations in tumors may involve loss of tumor antigens that are recognized by T cells, loss of antigen-presenting machinery components, tumor-induced inactivation of T cell signaling and insensitivity to effector functions of lymphocytes, death receptors or interferons.34
The nonsynonymous substitution of GZMB c.128T>C (rs8192917) was found to be in strong linkage disequilibrium with other common (MAF >15%) exonic variants, representing a haplotype block (data publicly available from The Single Nucleotide Polymorphism Database (https://www.ncbi.nlm.nih.gov/snp/; online supplementary figure 3). This indicates that two predominant isoforms of granzyme B are expressed in the European population, defining two predominant isoforms, and is consistent with previous reports that genetic variation of GZMB leads to change of three amino acids of the mature protein (Q48P88Y245 to R48A88H245).15 35 Interestingly, the two isoforms have a similar expression, stability and proteolytic activity, however, the variant granzyme B isotype was observed to be incapable of inducing apoptosis in tumor cell lines in contrast to the functional effects of the more common isotype.36
Interaction analysis between granzyme B, expression of PD-L1 in tumor and TMB showed that these markers were not intercorrelated, which suggests their value as independent biomarkers for clinical outcome after PD-1 ICIs. Although our findings in regard to serum granzyme B and GZMB germline variation are confirmed in a large patient cohort and by internal validation, further investigations need to be performed to determine the clinical value as a predictive or prognostic biomarker of response to PD-1 blockade. These investigations include comparative analysis with these currently used biomarkers in an extended patient cohort, and evaluation of the diagnostic accuracy and mapping of the sensitivity of serum granzyme B in a prospective clinical trial. The results may not be directly applicable to other ethnic groups, as the study was performed in a European population.
An important aspect of our results is that germline genetics related to cytotoxic effector functions of lymphocytes contribute to mechanisms of resistance to ICI therapy. Previously, it has been shown that germline variations in regard to the antigen presentation machinery are related to response to ICIs, that is HLA-1 heterozygosity.37 Considering the urgent need to understand how genetic variation affects response to ICIs, as reviewed by Havel et al,5 further work is warranted to identify novel genetic loci. We believe that apart from genes that are involved in antigen presentation, other immune-modulating genes may also contribute to response and resistance mechanisms of ICIs, of which granzyme B is an example. Determination of germline variations of patients with cancer contributing to response to ICIs may well be performed by future genome-wide association studies.
Conclusions
While the immune-oncology field rapidly advances, our understanding of ICI resistance mechanisms is limited and effective patient selection is lacking. Our data demonstrate that granzyme B is an important player in the antitumor immunity, as a consequence of the finding that low serum levels of granzyme B and germline variation of the GZMB gene contributes to resistance to PD-1 blockade in advanced NSCLC. As recent clinical trial results underline effective biomarker-based patient selection, future trials that incorporate granzyme B into diagnostics are considered as a promising step in this direction.
Acknowledgments
We thank Kazem Nasserinejad (Erasmus MC, Rotterdam, The Netherlands) for his assistance with the statistics, and Wim Dik (Erasmus MC, Rotterdam, The Netherlands) for performing the LUMINEX assay.
Footnotes
Contributors: DPH, SB, AAMvdV, RHNvS, JA and RHJM contributed to the conception and design of the study. DPH drafted the manuscript. EO-DH contributed to the acquisition, analysis of data, and contributed statistical review of data. DPH, SLWK, AAMvdV, RD, RHNvS, JA and RHJM took part in interpretation of data. DPH, NS, AAMvdV and JA performed radiological evaluation. DPH and RD performed additional analysis of peripheral immune cell phenotypes. EB, NS and SEB contributed logistic, technical and material support. All authors contributed to critical revision of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding: This research was funded by the Departments of Medical Oncology, Pulmonology, and Clinical Chemistry, Erasmus MC, Rotterdam, The Netherlands.
Competing interests: CHvdL reports non-financial support from BMS, MSD, Roche, Boehringer Ingelheim and AstraZeneca outside the submitted work. AAMvdV reports support from BMS, MSD, Sanofi, Roche, Novartis, Pfizer, Pierre Fabre, Ipsen, Eisai and Bayer outside the submitted work. JA reports personal fees from MSD, BMS, Amphera, Eli-Lilly, Takeda, Bayer, Roche, Boehringer Ingelheim, AstraZeneca outside the submitted work, and has a patent allogenic tumor cell lysate licensed to Amphera, a patent combination immunotherapy in cancer and a patent biomarker for immunotherapy pending. RHJM reports grants and non-financial support from Astellas, Bayer and Boehringer Ingelheim, grants from Cristal Therapeutics and Pamgene, grants and personal fees from Novartis, Servier, grants and non-financial support from Pfizer, grants from Roche, Sanofi, outside the submitted work.
Patient consent for publication: Not required.
Ethics approval: Ethics approval of Multomab study (Medical Ethical Board Erasmus MC ref. 16-011), patient signed written informed consent provided before study entry.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 2018;18:153–67. 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced Nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunert A, Basak EA, Hurkmans DP, et al. CD45RA+CCR7- CD8 T cells lacking co-stimulatory receptors demonstrate enhanced frequency in peripheral blood of NSCLC patients responding to nivolumab. J Immunother Cancer 2019;7:149. 10.1186/s40425-019-0608-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui E, Cheung J, Zhu J, et al. T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science 2017;355:1428–33. 10.1126/science.aaf1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamphorst AO, Wieland A, Nasti T, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 2017;355:1423–7. 10.1126/science.aaf0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larimer BM, Wehrenberg-Klee E, Dubois F, et al. Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res 2017;77:2318–27. 10.1158/0008-5472.CAN-16-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol 2015;15:388–400. 10.1038/nri3839 [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Cai SF, Fehniger TA, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 2007;27:635–46. 10.1016/j.immuni.2007.08.014 [DOI] [PubMed] [Google Scholar]
- 12.Nagata S. Apoptosis by death factor. Cell 1997;88:355–65. 10.1016/S0092-8674(00)81874-7 [DOI] [PubMed] [Google Scholar]
- 13.Pinkoski MJ, Hobman M, Heibein JA, et al. Entry and trafficking of granzyme B in target cells during granzyme B-perforin-mediated apoptosis. Blood 1998;92:1044–54. 10.1182/blood.V92.3.1044 [DOI] [PubMed] [Google Scholar]
- 14.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Andersen G, Yorgov D, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet 2016;48:1418–24. 10.1038/ng.3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M, Liu Y, Liu Y, et al. Genetic polymorphisms of GZMB and vitiligo: a genetic association study based on Chinese Han population. Sci Rep 2018;8:13001. 10.1038/s41598-018-31233-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos PS, Criswell LA, Moser KL, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet 2011;7:e1002406. 10.1371/journal.pgen.1002406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurreeman FAS, Daha NA, Chang M, et al. Association of IL2RA and IL2RB with rheumatoid arthritis: a replication study in a Dutch population. Ann Rheum Dis 2009;68:1789–90. 10.1136/ard.2008.106393 [DOI] [PubMed] [Google Scholar]
- 19.Wang X-X, Chen T. Meta-analysis of the association of IL2RA polymorphisms rs2104286 and rs12722489 with multiple sclerosis risk. Immunol Invest 2018;47:431–42. 10.1080/08820139.2018.1425699 [DOI] [PubMed] [Google Scholar]
- 20.Lee YH, Bae S-C. Association between interferon-γ +874 T/A polymorphism and susceptibility to autoimmune diseases: a meta-analysis. Lupus 2016;25:710–8. 10.1177/0961203315624557 [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Cho S-K, Sestak A, et al. Interferon-gamma gene polymorphisms associated with susceptibility to systemic lupus erythematosus. Ann Rheum Dis 2010;69:1247–50. 10.1136/ard.2009.117572 [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Li Y, Deng C, et al. The associations between PD-1, CTLA-4 gene polymorphisms and susceptibility to ankylosing spondylitis: a meta-analysis and systemic review. Rheumatol Int 2016;36:33–44. 10.1007/s00296-015-3327-9 [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Hu L-H, Li Y-R, et al. Programmed cell death 1 gene polymorphisms is associated with ankylosing spondylitis in Chinese Han population. Rheumatol Int 2011;31:209–13. 10.1007/s00296-009-1264-1 [DOI] [PubMed] [Google Scholar]
- 24.Narumi Y, Isomoto H, Shiota M, et al. Polymorphisms of PTPN11 coding SHP-2 as biomarkers for ulcerative colitis susceptibility in the Japanese population. J Clin Immunol 2009;29:303–10. 10.1007/s10875-008-9272-6 [DOI] [PubMed] [Google Scholar]
- 25.Bouzid D, Fourati H, Amouri A, et al. Association of ZAP70 and PTPN6, but not BANK1 or CLEC2D, with inflammatory bowel disease in the Tunisian population. Genet Test Mol Biomarkers 2013;17:321–6. 10.1089/gtmb.2012.0372 [DOI] [PubMed] [Google Scholar]
- 26.Namipashaki A, Razaghi-Moghadam Z, Ansari-Pour N. The essentiality of reporting Hardy-Weinberg equilibrium calculations in population-based genetic association studies. Cell J 2015;17:187–92. 10.22074/cellj.2016.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bins S, Basak EA, El Bouazzaoui S, et al. Association between single-nucleotide polymorphisms and adverse events in nivolumab-treated non-small cell lung cancer patients. Br J Cancer 2018;118:1296–301. 10.1038/s41416-018-0074-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurkmans DP, Kuipers ME, Smit J, et al. Tumor mutational load, CD8+ T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients. Cancer Immunol Immunother 2020. 10.1007/s00262-020-02506-x. [Epub ahead of print: 12 Feb 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis CM. Genetic association studies: design, analysis and interpretation. Brief Bioinform 2002;3:146–53. 10.1093/bib/3.2.146 [DOI] [PubMed] [Google Scholar]
- 30.Efron B. Bootstrap methods: another look at the jackknife. Ann Stat 1979;7:1–26. 10.1214/aos/1176344552 [DOI] [Google Scholar]
- 31.It's time to talk about ditching statistical significance. Nature 2019;567:283. 10.1038/d41586-019-00874-8 [DOI] [PubMed] [Google Scholar]
- 32.de Jong HK, Garcia-Laorden MI, Hoogendijk AJ, et al. Expression of intra- and extracellular granzymes in patients with typhoid fever. PLoS Negl Trop Dis 2017;11:e0005823. 10.1371/journal.pntd.0005823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spaeny-Dekking EH, Hanna WL, Wolbink AM, et al. Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. J Immunol 1998;160:3610–6. [PubMed] [Google Scholar]
- 34.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 2016;375:819–29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrara TM, Jin Y, Gowan K, et al. Risk of generalized vitiligo is associated with the common 55R-94A-247H variant haplotype of GZMB (encoding granzyme B). J Invest Dermatol 2013;133:1677–9. 10.1038/jid.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIlroy D, Cartron P-F, Tuffery P, et al. A triple-mutated allele of granzyme B incapable of inducing apoptosis. Proc Natl Acad Sci U S A 2003;100:2562–7. 10.1073/pnas.0437935100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. 10.1126/science.aao4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-000586supp001.pdf (532KB, pdf)