Short abstract
Objective
Versican (VCAN) has been reported as a potential biomarker in some cancers. However, its role in gastric cancer (GC) is poorly understood.
Methods
Associations between clinical variables and VCAN were assessed. The diagnostic value of VCAN expression in GC patients was determined through receiver operating characteristic (ROC) curve analysis. Cox regression and the Kaplan–Meier method were used to explore clinicopathologic factors related to overall survival in GC patients. The Gene Expression Omnibus and the Human Protein Atlas were used for further validation. Gene set enrichment analysis (GSEA) was performed using The Cancer Genome Atlas dataset.
Results
High expression of VCAN was associated with high stage and T classification in GC. The area under the ROC curve was 0.853. Patients with high VCAN expression had worse prognoses than those with low VCAN expression. Multivariate analysis showed that VCAN was an independent risk factor for overall survival in both cohorts. GSEA identified pathways involved in cancer, ECM-receptor interaction, Wnt signaling, T cell receptor signaling, and chemokine signaling as differentially enriched in GCs with high VCAN expression.
Conclusion
We demonstrated that VCAN is expressed at high levels in GC, and represents a potential independent molecular marker for diagnosis and prognosis of GC.
Keywords: VCAN, gastric cancer, independent predictor, prognosis, biomarker, receiver operating characteristic (ROC) curve
Introduction
Gastric cancer (GC) is the fifth most common cancer and second most important cause of cancer-related death worldwide.1 Of the 677,000 cases diagnosed in developing countries, about half were diagnosed in Eastern Asia, mainly in China.2 It was estimated that approximately 26,370 new cases would be diagnosed in 2016 in the United States.3 Although there has been a gradual decrease in the incidence of GC over the past several decades, the 5-year overall survival rates remain low, as many patients are diagnosed at an advanced stage of GC.4 Low survival rates occur, in part, because GC patients are usually asymptomatic in the early stages of disease. However, when symptoms become obvious, the GC has usually reached an advanced stage and has sometimes metastasized.5 Therefore, biomarkers for early and accurate diagnosis of GC would contribute to improved prognoses for these patients.
VCAN, a large aggregating chondroitin sulfate proteoglycan belonging to the lexicon family, is an important extracellular matrix component, and is closely associated with tumorigenesis.6 Four VCAN isoforms have been identified. The full-length isoform is V0, and the smaller isoforms V1, V2, and V3 result from alternative splicing.7 Previous studies have demonstrated that VCAN improves tumor cell survival, growth, migration, invasion, angiogenesis, and metastasis.8,9 Increased expression of VCAN has been reported in several malignancies, including leukemias as well as brain, colon, liver, prostate, breast, ovarian, oral squamous cell, and lung cancers, and is associated with adverse outcomes.9–15 A recent bioinformatic analysis revealed that VCAN may have an oncogenic role in GC, while another study suggested that VCAN expression predicts favorable outcomes in patients with GC.16,17 However, the clinical significance and prognostic value of VCAN in GC remains largely unclear. Therefore, it is vital to identify reliable biomarkers for GC diagnosis and prognosis. This study aimed to explore the diagnostic and prognostic value of VCAN expression in GC using The Cancer Genome Atlas (TCGA) database. We also sought to further evaluate the biological pathways involved in GC pathogenesis-associated VCAN regulatory networks using Gene Set Enrichment Analysis (GSEA).
Materials and methods
Data collection
The level 3 expression data and mRNA expression profiles (407 cases, including 32 normal samples, Workflow Type: HTSeq-Counts) and clinical information pertaining to survival time for 435 GC patients were downloaded from the TCGA Genomic Data Commons data portal (https://portal.gdc.cancer.gov/repository). We used boxplots to visualize differences in expression for discrete variables. RNA-Seq gene expression HTSeq-Count data for 375 patients were used for further analysis.
Gene set enrichment analysis
GSEA is a computational method that detects whether an a priori defined set of genes show statistically significant differential expression between high and low expression groups.18 Datasets and phenotype label files were generated and uploaded into GSEA software. The phenotype labels were VCAN-high and VCAN-low. Gene set permutations were conducted 1000 times for each analysis. Gene sets with a normal P-value < 0.05 and false discovery rate (FDR) < 0.25 were considered as enriched.
Statistical analysis
The associations between clinical factors and VCAN expression were evaluated using the Wilcoxon signed-rank test and logistic regression. Clinical factors related to overall survival in GC patients were identified using Cox regression and the Kaplan–Meier method. Multivariate Cox analysis was used to explore the role of VCAN expression in survival along with other clinical features (age, stage, grade, distant metastasis status, lymph node status, and histological subtype). High and low VCAN expression was determined based on the median values. Using the median risk score of VCAN expression as the cutoff value, all patients were divided into low expression or high expression groups. All statistical analyses were performed using R software (V.3.5.1).
Validation using the Gene Expression Omnibus (GEO) and Human Protein Atlas (HPA) databases
To ensure the accuracy of the results from the TCGA cohort, GSE54129 from the GEO database was used to validate VCAN expression levels. The dataset included 111 GC and 21 adjacent non-tumorous tissue samples. Since GSE54129 lacked clinical information, we also used GSE15459 containing 200 primary gastric tumors (192 with complete prognosis information) to validate whether VCAN was an independent predictor of GC prognosis. The HPA is a pathology tool that provides numerous expression profiles of various human proteins. Therefore, we compared the protein expression of VCAN in normal and GC tissues using immunohistochemistry (IHC) data from the HPA database (http://www.proteinatlas.org/).
Results
Associations between VCAN expression and clinical parameters
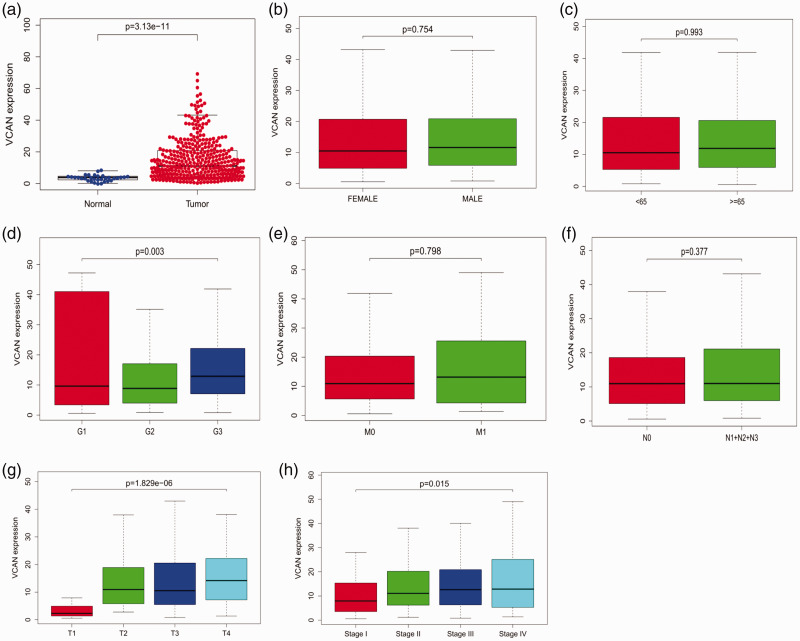
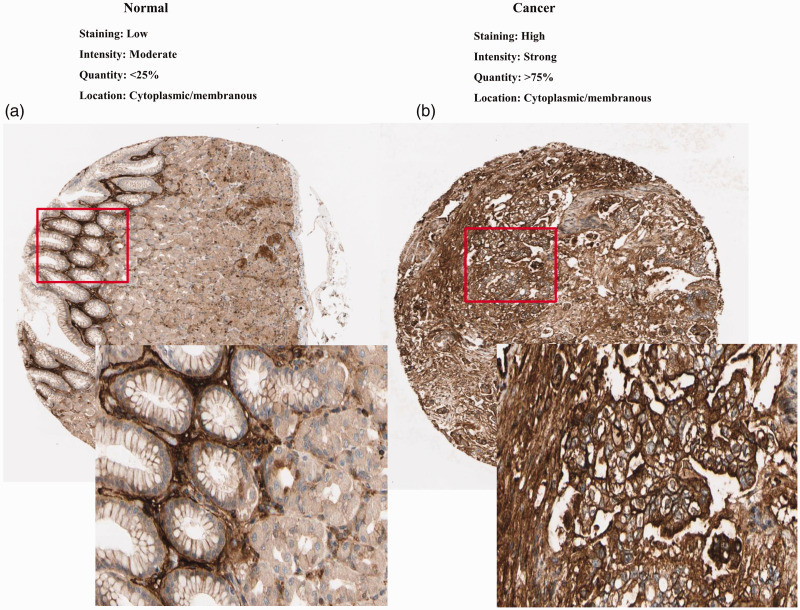
Clinical data pertaining to 375 GC patients from the TCGA were analyzed, and included the patient’s age, gender, clinical stage, histologic grade, tumor, node and metastasis (TNM) classification, survival status, and survival time. As shown in Figure 1 (a–h), comparison of VCAN expression in GC and normal tissues indicated that VCAN expression was elevated in GC (P < 0.001). VCAN expression was also notably associated with histological stage (P = 0.015), clinical stage (P = 0.003), and T classification (P < 0.001). Univariate analysis using logistic regression revealed that VCAN expression was associated with adverse prognostic clinicopathological variables (Table 1). High expression of VCAN in GC was significantly associated with high stage (OR = 2.045 for stage III vs. I) and T classification (OR = 8.5 for T2 vs. T1; OR =7.9 for T3 vs. T1; OR = 12.75 for T4 vs. T1). The high expression of VCAN was also validated in GC tissues in GEO54129 (Figure 2, P < 0.001). These findings revealed that patients with high VCAN expression tended to progress to a more advanced stage than those with low VCAN expression. To further examine VCAN protein expression, we retrieved IHC staining data from the HPA. In normal gastric tissues, the glandular cells usually had low VCAN staining (Figure 3a). However, GC tissues had strong VCAN staining, confirming our findings at the mRNA level (Figure 3b).
Figure 1.
Association of VCAN expression with clinical variables.
a: expression level; b: gender; c: age; d: grade; e: distant metastases; f: lymph node metastasis; g: tumor stage; h: clinical stage; i: relationship between VCAN expression and overall survival in GC patients in TCGA cohort.
Table 1.
Logistic regression of VCAN expression and clinical pathological characteristics.
| Clinical characteristics | Total (N) | Odds ratio for high VCAN expression | 95%CI | P-value |
|---|---|---|---|---|
| Stage classification (II vs I) | 164 | 1.822 | 0.937–3.620 | 0.081 |
| (III vs I) | 203 | 2.045 | 1.081–3.960 | 0.03 |
| (IV vs I) | 91 | 2.211 | 0.951–5.250 | 0.067 |
| T classification (T2 vs T1) | 116 | 8.5 | 2.240–55.855 | 0.0061 |
| (T3 vs T1) | 221 | 7.9 | 2.178–50.905 | 0.0067 |
| (T4 vs T1) | 142 | 12.75 | 3.408–83.230 | 0.001 |
| Gender (male vs female) | 368 | 1.15 | 0.752–1.767 | 0.514 |
| Grade classification (G2 vs G1) | 147 | 0.756 | 0.202–2.834 | 0.67 |
| (G3 vs G1) | 229 | 1.19 | 0.323–4.391 | 0.788 |
| M classification (M1 vs M0) | 350 | 1.09 | 0.480–2.493 | 0.835 |
| N classification (N1 + N2 + N3 vs N0) | 357 | 1.018 | 0.650–1.595 | 0.937 |
| Age (≥65 vs <65) | 368 | 1.25 | 0.826–1.896 | 0.291 |
VCAN: Versican.
CI: confidence interval.
Bold values indicate P < 0.05.
Figure 2.
The expression level of VCAN in gastric cancer and normal tissues.
Figure 3.
Validation of protein expression of VCAN in gastric cancer and normal tissues using the Human Protein Atlas database.
Diagnostic value of VCAN expression in GC
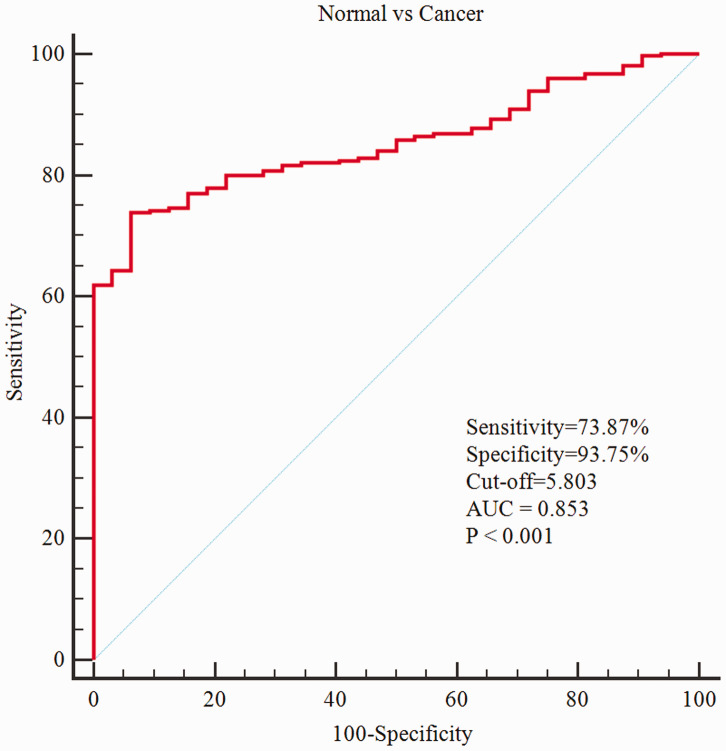
To evaluate the diagnostic value of VCAN, we generated a receiver operating characteristic (ROC) curve using the expression data from 375 GC patients and 32 healthy individuals. The area under the ROC curve was 0.853 [95% confidence interval (CI): 81.5–88.6%], the sensitivity was 73.87% (95%CI: 69.1–78.2%) and the specificity was 93.75% (95%CI: 79.2%–99.2%), which indicated considerable diagnostic value (Figure 4).
Figure 4.
Receiver operating characteristic (ROC) curve for VCAN expression in normal gastric tissue and gastric cancer.
Survival outcomes and multivariate analysis
As demonstrated in Figure 5a, high expression of VCAN was closely associated with poor overall survival (P = 0.001). This relationship was further validated in GSE54129 (Figure 5b, P = 0.002). The univariate analysis indicated that high VCAN expression was significantly associated with poor overall survival [hazard ratio (HR): 1.316; 95%CI: 1.112–1.558; P = 0.0014]. Other clinical factors associated with adverse survival included age, stage, and TNM classification (Table 2). These variables were included in the multivariate analysis. Multivariate Cox analysis revealed that high VCAN expression remained an independent risk factor for overall survival with a HR of 1.305 (95%CI: 1.097–1.551, P = 0.00263), as well as age (HR = 1.037, 95%CI: 1.016–1.058, P < 0.001, Table 2) among GC patients.
Figure 5.
VCAN expression and overall survival in gastric cancer patients in TCGA cohort (a) and the GSE54129 dataset (b).
Table 2.
Univariate and multivariate analysis of the relationship between VCAN expression and overall survival among gastric cancer patients.
|
Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Parameter | HR | 95%CI | P-value | HR | 95%CI | P-value |
| Age | 1.027 | 1.008–1.046 | 0.00556 | 1.037 | 1.016–1.058 | 0.0005 |
| Gender | 1.484 | 0.979–2.247 | 0.06239 | |||
| Grade | 1.368 | 0.947–1.977 | 0.09538 | |||
| Stage | 1.535 | 1.221–1.931 | 0.00024 | 1.318 | 0.860–2.018 | 0.20459 |
| T | 1.298 | 1.023–1.645 | 0.03152 | 1.032 | 0.748–1.425 | 0.8459 |
| M | 2.048 | 1.096–3.827 | 0.02458 | 2.023 | 0.903–4.531 | 0.08671 |
| N | 1.267 | 1.069–1.502 | 0.00639 | 1.126 | 0.887–1.429 | 0.32923 |
| VCAN | 1.316 | 1.112–1.558 | 0.0014 | 1.305 | 1.097–1.551 | 0.00263 |
VCAN: Versican.
HR: hazard ratio.
CI: confidence interval.
Bold values indicate P < 0.05.
Validation using GEO databases
Univariate Cox analysis using GSE15459 revealed that high expression of VCAN was associated with lower overall survival in GC patients, with a HR of 1.534 (95%CI: 1.265–1.859, P < 0.001). A multivariate adjustment for other factors indicated that VCAN expression, as well as stage, were independent prognostic factors with a HR of 1.350 (95%CI: 1.109–1.644, P = 0.0027) in the GSE15459 dataset (Table 3).
Table 3.
Univariate and multivariate analysis of the relationship between VCAN expression and overall survival among gastric cancer patients validated using the GSE15459 dataset.
|
Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Parameter | HR | 95%CI | P-value | HR | 95%CI | P-value |
| Age | 0.999 | 0.983–1.015 | 0.96787 | |||
| Gender | 1.402 | 0.908–0.127 | 0.127097 | |||
| Stage | 2.789 | 2.140–3.635 | <0.001 | 2.701 | 2.058–3.547 | <0.001 |
| VCAN | 1.534 | 1.265–1.859 | <0.001 | 1.35 | 1.109–1.644 | 0.002703 |
VCAN: Versican; HR: hazard ratio; CI: confidence interval; Bold values indicate P < 0.05.
GSEA identifies VCAN-associated signaling pathways
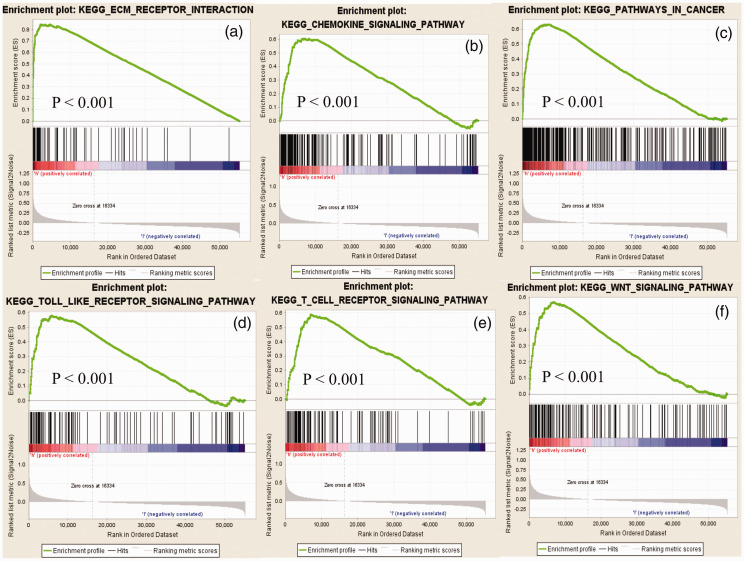
To screen for potential signaling pathways that were differentially activated in GC, we performed GSEA comparing the high and low VCAN expression datasets. Gene sets with nominal P-value < 0.05 and FDR q-value < 0.25 were considered as significantly enriched. GSEA revealed significant differences in the enrichment of the MSigDB collection (h.all.v6.2.symbols.gmt). The most significantly enriched signaling pathways based on their normalized enrichment scores were identified. As shown in Figure 6, gene sets related to extracellular matrix receptor interaction, cancer, chemokine signaling, Toll-like receptor signaling, T cell receptor signaling, and Wnt signaling were differentially associated with the VCAN high expression phenotype.
Figure 6.
Enrichment plots from gene set enrichment analysis (GSEA). The GSEA results revealed that genes involved in extracellular matrix-receptor interaction (a), chemokine signaling (b), cancer (c), Toll-like receptor signaling (d), T cell receptor signaling (e), and WNT signaling (f) were differentially enriched in VCAN-associated gastric cancer.
Discussion
VCAN is a large extracellular matrix proteoglycan with an apparent mass of more than 1000 kDa. VCAN overexpression is a prognostic biomarker for poor disease survival in several tumor types, including endometrial cancer, ovarian cancer, oral squamous cell carcinoma, and gastric and gastrointestinal stromal tumors.14,19–23 However, the clinical implications of VCAN expression as a biomarker in GC have not been well studied. In the present study, bioinformatic analysis of high throughput RNA-sequencing data from TCGA revealed significantly increased VCAN expression in gastric carcinomas compared with the adjacent normal gastric mucosa, in agreement with the results of previous studies.19,20,24,25 VCAN was expressed at markedly higher levels in serous ovarian cancer samples compared with normal tissues.14 Together with our study in GC, these data suggested that VCAN mRNA expression levels may be associated with tumor development. Further, we found that VCAN expression was also strongly associated with histological stage, clinical stage, and T classification. A previous study reported that relative expression of VCAN in different hepatocellular carcinoma grades and stages showed progressive upregulation in more aggressive cancers, consistent with our results in GC patients.15 Kaplan–Meier curves for overall survival revealed that high expression of VCAN was associated with poor outcomes in GC patients. Univariate and multivariate Cox analyses of both TCGA and GEO databases indicated the VCAN expression was a potential independent marker for poor prognosis in GC, and ROC analysis confirmed the diagnostic value of VCAN expression in gastric cancer. As revealed in the multivariate analysis, age was an independent risk factor for overall survival in GC patients. This finding was consistent with a previous study of patients with gastrointestinal stromal tumors.26,27
It has been reported that abnormal expression of VCAN is associated with changes in cell proliferation, differentiation, adhesion, and the homeostasis and integrity of the extracellular matrix.28 VCAN regulates the organization of the extracellular matrix and contributes to tumor growth, and increased VCAN expression may be necessary for angiogenesis and metastasis in tumors.29–31 VCAN, along with CD44 and hyaluronan, can form a macromolecular complex in the extracellular matrix, which is believed to result in invasion and metastasis by promoting tumor cell motility.32,33 To further evaluate the roles of VCAN in GC, we performed GSEA using TCGA data. GSEA showed that genes involved in extracellular matrix receptor interaction, cancer, chemokine signaling, Toll-like receptor signaling, T cell receptor signaling, and Wnt signaling were differentially associated with the VCNA high expression phenotype. This indicated that VCAN may serve as a potential prognostic marker for diagnosis and prognosis in GC. Extracellular matrix receptor interactions have been shown to be involved in cell proliferation and invasion of many cancer cell types, including GC.34,35 Chronic inflammation promotes cancer development, and inflammation has been identified as a hallmark of cancer.36,37 Inflammatory effectors and cytokines within the tumor microenvironment can either improve antitumor immune responses or support tumor pathogenesis,37,38 in agreement with our results. Activation of the Wnt signaling pathway has been reported to play an important role in GC development.39 Downregulation of FEZF1-AS1 was shown to suppress activation of the Wnt/β-catenin signaling pathway in GC, and could therefore be used as a new biomarker for the treatment of GC.40 Together with our results, these data support the view that the Wnt signaling pathway is closely related to GC. Although we have demonstrated that VCAN expression is a potential independent molecular marker for the diagnosis and prognosis in GC, the major limitations of our study should be noted. Our results were based on bioinformatic analysis. Further experimental validation needs to be carried out in the future to verify the biological significance of VCAN expression in GC.
Conclusion
Based on analysis of the TCGA and GEO databases, we provide evidence that high VCAN expression may be a potential diagnostic and prognostic molecular marker in GC. Furthermore, the Wnt and chemokine signaling pathways may be key regulators of VCAN expression in GC.
Availability of data and materials
The datasets used during the present study are available from TCGA (https://portal.gdc.cancer.gov/repository) database, or from the corresponding author upon reasonable request.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Ethics statement
Not applicable.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD
Wenfei Li https://orcid.org/0000-0001-5913-2305
References
- 1.Fitzmaurice C, Dicker D, Pain A, et al. The Global Burden of Cancer 2013. JAMA Oncol 2015; 1: 505–527. DOI: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. DOI: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. DOI: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol 2014; 20: 1635–1649. DOI: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung WK, Wu MS, Kakugawa Y, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol 2008; 9: 279–287. DOI: 10.1016/s1470-2045(08)70072-x. [DOI] [PubMed] [Google Scholar]
- 6.Edwards IJ. Proteoglycans in prostate cancer. Nat Rev Urol 2012; 9: 196–206. DOI: 10.1038/nrurol.2012.19. [DOI] [PubMed] [Google Scholar]
- 7.Ricciardelli C, Rodgers RJ. Extracellular matrix of ovarian tumors. Semin Reprod Med 2006; 24: 270–282. DOI: 10.1055/s-2006-948556. [DOI] [PubMed] [Google Scholar]
- 8.Du WW, Yang W, Yee AJ. Roles of versican in cancer biology–tumorigenesis, progression and metastasis. Histol Histopathol 2013; 28: 701–713. DOI: 10.14670/hh-28.701. [DOI] [PubMed] [Google Scholar]
- 9.Fujii K, Karpova MB, Asagoe K, et al. Versican upregulation in Sezary cells alters growth, motility and resistance to chemotherapy. Leukemia 2015; 29: 2024–2032. DOI: 10.1038/leu.2015.103. [DOI] [PubMed] [Google Scholar]
- 10.Yen CY, Huang CY, Hou MF, et al. Evaluating the performance of fibronectin 1 (FN1), integrin alpha4beta1 (ITGA4), syndecan-2 (SDC2), and glycoprotein CD44 as the potential biomarkers of oral squamous cell carcinoma (OSCC). Biomarkers 2013; 18: 63–72. DOI: 10.3109/1354750x.2012.737025. [DOI] [PubMed] [Google Scholar]
- 11.Suwiwat S, Ricciardelli C, Tammi R, et al. Expression of extracellular matrix components versican, chondroitin sulfate, tenascin, and hyaluronan, and their association with disease outcome in node-negative breast cancer. Clin Cancer Res 2004; 10: 2491–2498. [DOI] [PubMed] [Google Scholar]
- 12.Ricciardelli C, Mayne K, Sykes PJ, et al. Elevated levels of versican but not decorin predict disease progression in early-stage prostate cancer. Clin Cancer Res 1998; 4: 963–971. [PubMed] [Google Scholar]
- 13.Suhovskih AV, Aidagulova SV, Kashuba VI, et al. Proteoglycans as potential microenvironmental biomarkers for colon cancer. Cell Tissue Res 2015; 361: 833–844. DOI: 10.1007/s00441-015-2141-8. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, Albitar L, LeBaron R, et al. Up-regulation of stromal versican expression in advanced stage serous ovarian cancer. Gynecol Oncol 2010; 119: 114–120. DOI: 10.1016/j.ygyno.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naboulsi W, Megger DA, Bracht T, et al. Quantitative Tissue Proteomics Analysis Reveals Versican as Potential Biomarker for Early-Stage Hepatocellular Carcinoma. J Proteome Res 2016; 15: 38–47. DOI: 10.1021/acs.jproteome.5b00420. [DOI] [PubMed] [Google Scholar]
- 16.Kim NS, Lee HH, Jung CK, et al. Versican expression in tumor epithelial cells is correlated with a good prognosis in gastric cancer. Anticancer Res 2014; 34: 5613–5619. [PubMed] [Google Scholar]
- 17.Jiang K, Liu H, Xie D, et al. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol Lett 2019; 17: 3191–3202. DOI: 10.3892/ol.2019.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. DOI: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen XH, Lin WR, Xu MD, et al. Prognostic significance of Versican expression in gastric adenocarcinoma. Oncogenesis 2015; 4: e178. DOI: 10.1038/oncsis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Setoguchi T, Kikuchi H, Yamamoto M, et al. Microarray analysis identifies versican and CD9 as potent prognostic markers in gastric gastrointestinal stromal tumors. Cancer Sci 2011; 102: 883–889. DOI: 10.1111/j.1349-7006.2011.01872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pukkila M, Kosunen A, Ropponen K, et al. High stromal versican expression predicts unfavourable outcome in oral squamous cell carcinoma. J Clin Pathol 2007; 60: 267–272. DOI: 10.1136/jcp.2005.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodama J, Hasengaowa Y, Kusumoto T, et al. Prognostic significance of stromal versican expression in human endometrial cancer. Ann Oncol 2007; 18: 269–274. DOI: 10.1093/annonc/mdl370. [DOI] [PubMed] [Google Scholar]
- 23.Cheon DJ, Tong Y, Sim MS, et al. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res 2014; 20: 711–723. DOI: 10.1158/1078-0432.ccr-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theocharis AD, Vynios DH, Papageorgakopoulou N, et al. Altered content composition and structure of glycosaminoglycans and proteoglycans in gastric carcinoma. Int J Biochem Cell Biol 2003; 35: 376–390. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Wang L, Yang J, et al. Expression of Versican and its clinical significance in gastric carcinoma. Chin J Pathol 2014; 43: 473–477. [PubMed] [Google Scholar]
- 26.Song W, Lv CG, Miao DL, et al. Development and validation of a nomogram for predicting survival in patients with gastrointestinal stromal tumours. Eur J Surg Oncol 2018; 44: 1657–1665. DOI: 10.1016/j.ejso.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Sun F, Chen L, et al. Prognostic value of visceral pleural invasion in non-small cell lung cancer: a propensity score matching study based on the SEER registry. J Surg Oncol 2017; 116: 398–406. DOI: 10.1002/jso.24677. [DOI] [PubMed] [Google Scholar]
- 28.Wight TN. Versican: a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol 2002; 14: 617–623. [DOI] [PubMed] [Google Scholar]
- 29.Wu YJ, La Pierre DP, Wu J, et al. The interaction of versican with its binding partners. Cell Res 2005; 15: 483–494. DOI: 10.1038/sj.cr.7290318. [DOI] [PubMed] [Google Scholar]
- 30.Norian JM, Malik M, Parker CY, et al. Transforming growth factor beta3 regulates the versican variants in the extracellular matrix-rich uterine leiomyomas. Reprod Sci 2009; 16: 1153–1164. DOI: 10.1177/1933719109343310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang BL, Zhang Y, Cao L, et al. Cell adhesion and proliferation mediated through the G1 domain of versican. J Cell Biochem 1999; 72: 210–220. [DOI] [PubMed] [Google Scholar]
- 32.Ricciardelli C, Russell DL, Ween MP, et al. Formation of hyaluronan- and versican-rich pericellular matrix by prostate cancer cells promotes cell motility. J Biol Chem 2007; 282: 10814–10825. DOI: 10.1074/jbc.M606991200. [DOI] [PubMed] [Google Scholar]
- 33.Ween MP, Oehler MK, Ricciardelli C. Role of versican, hyaluronan and CD44 in ovarian cancer metastasis. Int J Mol Sci 2011; 12: 1009–1029. DOI: 10.3390/ijms12021009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Q, Zhang B, Wang Z, et al. Integrated bioinformatics analysis reveals novel key biomarkers and potential candidate small molecule drugs in gastric cancer. Pathol Res Pract 2019; 215: 1038–1048. DOI: 10.1016/j.prp.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Cao L, Chen Y, Zhang M, et al. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ 2018; 6: e5180. DOI: 10.7717/peerj.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani A. Molecular pathways linking inflammation and cancer. Curr Mol Med 2010; 10: 369–373. [DOI] [PubMed] [Google Scholar]
- 37.Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer 2013; 13: 525–541. DOI: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 38.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest 2015; 125: 3347–3355. DOI: 10.1172/jci80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Wang D, Sun X, et al. ADAM17 promotes lymph node metastasis in gastric cancer via activation of the Notch and Wnt signaling pathways. Int J Mol Med 2019; 43: 914–926. DOI: 10.3892/ijmm.2018.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Zhang P, Zhu H, et al. Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathway. Biomed Pharmacother 2017; 96: 1103–1108. DOI: 10.1016/j.biopha. 2017.11.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from TCGA (https://portal.gdc.cancer.gov/repository) database, or from the corresponding author upon reasonable request.