Abstract
Skeletal muscle tissue regeneration requires quiescent satellite cell activation, proliferation, and differentiation. Regenerative capacity of satellite cells can be studied in vitro by differentiating under low-serum conditions (2% to 5%) to form multinucleated myotubes. Myotubes are fixed and stained, and indices of differentiation are quantified. Jenner and Giemsa stains are typically used for myotube staining; however, this staining process can be variable depending on factors such as stain pH, staining time, and time since stain preparation. This article includes protocols for myoblast isolation, proliferation, and differentiation in vitro; Jenner-Giemsa staining; HEMA 3 staining; and quantification. Representative images using each staining method and quantification are included. The protocols identify critical steps and considerations for cell culture and each staining method and provide an even simpler alternative to Jenner-Giemsa staining.
Keywords: differentiation index, imaging, myoblast isolation, myotube staining
INTRODUCTION
Skeletal muscle tissue is comprised of multinucleated myofibers that can hypertrophy in response to growth stimuli and can be repaired after damage. Growth and repair of skeletal muscle tissue requires satellite cells to be activated, proliferated, differentiated, and fused with existing muscle fibers in a process termed myogenesis (Yin, Price, & Rudnicki, 2013). Some stimuli, such as resistance or high-force exercise and anabolic hormones, promote myogenesis (Bellamy et al., 2014; Goh et al., 2019; Sinha-Hikim, Cornford, Gaytan, Lee, & Bhasin, 2006), whereas stimuli such as alcohol and catabolic hormones Impair myogenesis (Dong, Pan, & Zhang, 2013; McCroskery, Thomas, Maxwell, Sharma, & Kambadur, 2003; Simon et al., 2014; Simon, Ford, et al., 2017). Myogenesis can be studied in vitro using myoblasts isolated from skeletal muscle cultured first in high-serum medium to promote myoblast proliferation and then in low-serum medium to promote differentiation and fusion. Myotube staining allows for quantification of differentiation, including the degree of fusion. Although Jenner-Giemsa staining is traditionally used to stain myotubes, HEMA 3 staining provides comparable results while simplifying the staining protocol and eliminating steps that underlie staining inefficiency.
This article first describes protocols for the isolation of myoblasts from fresh tissue using mechanical and chemical disruption of tissue, passaging and expanding myoblasts in high-serum medium, and differentiating myoblasts into myotubes by switching from high- to low-serum medium. Then, protocols for fixing and staining myotubes with shelf-stable HEMA 3 stains are described, with traditional Jenner-Giemsa staining included as an alternative protocol. Finally, protocols for quantifying common indices of differentiation (i.e., myotube density, fusion index, and myotubes per field) using imaging software are described. Importantly, the use of fresh muscle tissue from human or animal subjects requires approval from an institutional review board or institutional animal care and use committee (IACUC), respectively. Additionally, human subjects must provide written informed consent. The human myoblasts used in this protocol were derived from a primary human myoblast cell line (American Type Culture Collection, PCS-950-010), and myoblasts derived from rhesus macaques were isolated from fresh muscle tissue as part of an IACUC-approved study.
BASIC PROTOCOL 1
PRIMARY MYOBLAST ISOLATION
This protocol provides a method for isolating primary myoblasts from skeletal muscle tissue. We have successfully used this protocol to isolate PAX7+/integrin α7+ myoblasts from rhesus macaque (Duplanty, Siggins, Allerton, Simon, & Molina, 2018; Simon et al., 2014), human, rat, and mice (unpublished) skeletal muscle. After washing muscle tissue, mechanical and enzymatic means are used to disrupt the tissue. The cell–tissue mixture is plated for fibroblast adherence, and the cell–tissue mixture enriched with myoblasts is replated. After a period of incubation, remaining muscle tissue is removed, myoblasts adhere to the plate and form colonies, and cells begin to proliferate and expand.
Materials
Isolation medium (see recipe)
Digest medium (see recipe)
Growth medium (see recipe)
70% ethanol
Freshly collected skeletal muscle in medium
Fetal bovine serum (FBS; e.g., Sigma-Aldrich, cat. no. F0926–500ML)
50-ml beaker
15-ml and 50-ml centrifuge tubes (e.g., Fisher Scientific, cat. nos. 50-809-221 and 50-465-232, respectively)
Fine scissors (e.g., Medline, cat. no. MDS0831525)
Straight forceps (e.g., Medline, cat. no. MDS0421229)
KimWipes (e.g., VWR, cat. no. 21905–026)
35-mm and 100-mm cell culture dishes, uncoated (e.g., Corning, cat. nos. 430165 and 430167, respectively)
Sterile gauze (e.g., Henry Schein, cat. no. 1004380)
Sterile transfer pipettes (e.g., USA Scientific, cat. no. 1020–2510)
Serological pipette and sterile tips, 2 to 25 ml vol. (e.g., Costar Stripettes)
Laboratory rocker, 0.5 to 1.0 Hz (e.g., Reliable Scientific Model 55 Rocking Shaker)
Centrifuge (e.g., Eppendorf Centrifuge 5810 R)
Standard humidified cell culture incubator, 37°C, 5% CO2 (e.g., Sanyo Model MCO-17AIC)
Inverted light microscope (e.g., Nikon DIAPHOT-TMD)
NOTE: All steps should be performed in a sterile biosafety cabinet using sterile technique and appropriate personal protective equipment (PPE).
Tissue preparation
-
1
Prepare isolation medium, digest medium, and growth medium.
-
2Fill 50-ml beaker with 70% ethanol and 50-ml tube with isolation medium. Place scissors and forceps in ethanol. When ready to use, remove ethanol from tools with KimWipe, and dip tools in isolation medium.When removing ethanol with KimWipe, begin at the tip of the tool, and wipe toward the handle to avoid any possible contamination of the tip.
-
3
Collect ~20 to 50 mg fresh muscle tissue, and place immediately in 50-ml tube containing ~15 to 25 ml isolation medium. Place tube on ice until ready to process muscle.
-
4
Pipette ~3 ml isolation medium into three 35-mm cell culture dishes for washing muscle tissue.
-
5
Quickly clean blood and fat from muscle tissue by transferring to sterile gauze using forceps or sterile transfer pipette and blotting.
-
6Immediately transfer muscle sample to 35-mm culture dish, and wash three times by transferring sequentially across the three culture dishes.Forceps, sterile transfer pipette, or both can be used to transfer muscle tissue. It is beneficial to agitate the muscle in each culture dish to ensure complete removal of blood and fat.
Tissue digest
-
7
Fill a fourth 35-mm culture dish with 2 ml digest medium. Transfer muscle sample to digest medium, taking care to avoid transferring isolation medium.
-
8Mince muscle tissue in digest medium using fine scissors.Muscle should be minced as finely as possible, ideally <0.5 mm. Mincing quickly and rotating the culture dish helps to achieve finely minced tissue.
-
9
Transfer tissue in digest medium to a labeled 15-ml tube.
-
10Rinse dish with 3 ml digest medium, and transfer that medium to the 15-ml tube containing the tissue for a total volume of 5 ml digest medium with tissue.This step allows for the transfer of tissue that might have been left behind during the initial transfer. If a large amount of tissue remains, split this step into two rinses, taking care to transfer as much tissue as possible to the 15-ml tube with digest medium.
-
11
Pipette well using a sterile serological pipette to mechanically agitate the tissue.
-
12Place 15-ml tube containing tissue in digest medium on a rocker at room temperature for 45 to 60 min to agitate.Tissue and medium should move the length of the tube. Set rocker to a speed that is high enough so that tissue does not settle at one end of the tube but low enough to avoid splashing of medium. Speeds of 0.5 to 1.0 Hz work well.
-
13
Remove tube from rocker, and allow tissue to settle at the bottom of the tube for ~1 min at room temperature.
-
14Transfer supernatant to an empty, sterile 15-ml tube, and place on ice.Take care to leave tissue and cells that have settled at the bottom of the tube for a second agitation, described in subsequent steps.
-
15
Add 5 ml fresh digest medium to tube containing tissue, and pipette well to agitate.
-
16Place 15-ml tube containing tissue in digest medium on a rocker at room temperaturefor 45 to 60 min to agitate.Tissue and medium should move the length of the tube. Set rocker to a speed that is high enough so that tissue does not settle at one end of the tube but low enough to avoid splashing of medium. Speeds of 0.5 to 1.0 Hz work well.
Plating for culture
-
17
Remove tube from rocker, and replace supernatant that was stored on ice (step 14) into the tube containing tissue.
-
18Add 2 ml FBS, cap, and gently invert several times to mix.The recommended FBS included in the Materials list is approved by the US Department of Agriculture, sterile filtered, and suitable for cell culture. Other brands may be tested and used. Heat inactivation is not necessary.
-
19
Centrifuge 5 min at 500 × g, room temperature, to obtain pellet containing tissue and cells.
-
20
Remove as much supernatant as possible using a serological pipette without disturbing the cell–tissue pellet. Do not decant.
-
21
Resuspend cell–tissue pellet in 5 ml growth medium.
-
22Transfer medium containing cells–tissue to a 100-mm uncoated culture dish. Add 5 ml growth medium for a total of 10 ml growth medium.If any of the cell–tissue pellet remains in the original centrifuge tube, the additional 5 ml growth medium can be used to rinse the tube and to transfer remaining tissue or cells to the culture dish.
-
23
Place culture dish in a standard humidified cell culture incubator (37°C, 5% CO2) for 5 hr to allow fibroblasts to adhere to the plate.
-
24After 5 hr, tilt plate slightly to collect tissue and medium using a serological pipette, and transfer to a new 100-mm uncoated culture dish, leaving fibroblasts adhered to original culture dish.Do not attempt to rinse plate or take tissue and cells that remain behind, as this could disturb fibroblasts.
-
25
Place culture dish containing tissue and medium in a standard humidified cell culture incubator (37°C, 5% CO2) for 1 week.
-
26After 1 week, change growth medium every other day until 20% to 30% confluent. Monitor growth under a microscope before and after each medium change.For the first and second growth medium changes, small floating tissue pieces will be present. Use a serological pipette to remove growth medium for these changes and to add 10 ml fresh growth medium. After the second medium change, all tissue pieces should either be adhered to the plate or removed, and growth medium can be aspirated using a vacuum aspirator and replaced using a serological pipette.Cells will grow in colonies on the initial plate. Confluence of 20% to 30% in this step refers to overall confluence; however, colonies will appear more confluent. If individual colonies reach 80% to 90% confluent, move on to passage (Basic Protocol 2).
ALTERNATE PROTOCOL 1
PLATING CRYOPRESERVED MYOBLASTS
Although freshly isolated primary myoblasts are preferable, at times a primary myoblast cell line or cryopreserved myoblasts might be used. In such cases, the following protocol replaces the isolation protocol.
Materials
Cryopreserved myoblasts
Growth medium (see recipe)
0.4% trypan blue (e.g., Thermo Fisher Scientific, cat. no. SV3008401) diluted 1:10 (to 0.04%) in sterile phosphate-buffered saline (PBS; e.g., Gibco, cat. no. 10010–023)
37°C water bath (e.g., Fisher Scientific Isotemp Water Bath) with float that fits cryovials
50-ml centrifuge tubes, sterile (e.g., Fisher Scientific, cat. no. 50-465-232)
Centrifuge (e.g., Eppendorf Centrifuge 5810 R)
1.5-ml microcentrifuge tubes (e.g., Thomas Scientific, cat. no. 2591B96)
Hemacytometer with cover slip (e.g., Sigma-Aldrich, cat. no. Z359629)
100-mm cell culture dishes, collagen I coated (e.g., Corning, cat. no. 356450)
Inverted light microscope (e.g., Nikon DIAPHOT-TMD)
Standard humidified cell culture incubator, 37°C, 5% CO2 (e.g., Sanyo Model MCO-17AIC)
NOTE: All steps should be performed in a sterile biosafety cabinet using sterile technique and appropriate PPE.
Place vial of cryopreserved myoblasts in a float in the 37°C water bath for 1 to 2 min to thaw quickly.
Transfer cells to a labeled, sterile 50-ml tube containing 5 ml growth medium for each ml of cells in freeze medium.
Centrifuge 5 min at 500 × g, room temperature, to obtain cell pellet.
- Remove supernatant.Be sure not to disturb cell pellet before removing supernatant, or cells will be lost. When decanting, avoid multiple inversions of the vial.
- Resuspend cells in growth medium.The amount of growth medium can be varied based on desired concentration of cells. We recommend resuspending in 5 to 6 ml growth medium per 1 × 106 expected cells.
- In a separate 1.5-ml microcentrifuge tube, dilute 10 μl cells 1:2 in diluted trypan blue, and count cells using a hemacytometer.If cells are more concentrated, a greater dilution factor can be used.
- Plate desired number of myoblasts per plate on collagen-coated plates.We recommend 2.5–4.0 × 103 cells per cm2. If 100-mm plates are used, we recommend 2.0–3.0 × 105 cells per plate.Other sizes of cell culture dishes can be used; adjust growth medium volume and number of cells accordingly.
- Fill plate with growth medium to desired volume.We recommend 125 to 200 μl growth medium per cm2. For a 100-mm plate, we use 10 ml growth medium.
- Check cells under a microscope, and then place culture dishes in a standard humidified cell culture incubator (37°C, 5% CO2).Cells will appear circular and suspended when freshly plated and will be elongated and adhered after incubation.
- Change growth medium every other day until 80% to 90% confluent if expanding or 70% to 80% confluent if differentiating.Remove growth medium using an aspirator, and replace using a sterile serological pipette.
BASIC PROTOCOL 2
MYOBLAST PASSAGE AND EXPANSION
After initial myoblast seeding, myoblast expansion increases cell numbers for downstream applications and, for the purposes of this protocol, for differentiation. Expansion is particularly necessary when isolating myoblasts from fresh tissue which yields fewer cells after initial passage. The desired number of cells needed for downstream applications should be decided ahead of time.
Materials
Confluent myoblasts (see Basic Protocol 1 or Alternate Protocol 1)
Sterile PBS, pH 7.4 (e.g., Gibco, cat. no. 10010–023)
0.25% trypsin-EDTA (e.g., Gibco, cat. no. 25200056)
Growth medium (see recipe)
0.4% trypan blue (e.g., Thermo Fisher Scientific, cat. no. SV3008401) diluted 1:10 (to 0.04%) in sterile PBS (e.g., Gibco, cat. no. 10010–023)
Standard humidified cell culture incubator, 37°C, 5% CO2 (e.g., Sanyo Model MCO-17AIC)
50-ml centrifuge tubes, sterile (e.g., Fisher Scientific, cat. no. 50–465-232)
Centrifuge (e.g., Eppendorf Centrifuge 5810 R)
1.5-ml microcentrifuge tubes (e.g., Thomas Scientific, cat. no. 2591B96)
Hemacytometer with cover slip (e.g., Sigma-Aldrich, cat. no. Z359629)
100-mm cell culture dishes, collagen I coated (e.g., Corning, cat. no. 356450)
Inverted light microscope (e.g., Nikon DIAPHOT-TMD)
NOTE: All steps should be performed in a sterile biosafety cabinet using sterile technique and appropriate PPE.
Cell detachment
-
1
Rinse confluent myoblasts with sterile PBS.
-
2Aspirate PBS and add 2 ml of 0.25% trypsin-EDTA per 100-mm culture dish.Carefully rotate plate to ensure a thin, even trypsin layer.
-
3Incubate in a standard humidified cell culture incubator (37°C, 5% CO2) until myoblasts are just dislodged (about 7 min).Exact time will need to be optimized for each laboratory. When viewed under a microscope, cells will be rounded, and some will float with minimal agitation.
-
4Mechanically dislodge cells by pipetting using the trypsin solution in the plate.Tilt the plate slightly. Hold the pipette so that the tip is ~0.5 cm from the plate, and forcefully expunge liquid directly onto any patches of cells that may still be adhered to the plate. Liquid should not splatter; if it does, reduce force slightly. Groups of adhered cells will appear as translucent patches and will flow with the liquid when dislodged. View under microscope to confirm that cells are dislodged.
-
5Add 2 ml growth medium to trypsinized cells, and gently pipette to mix. Transfer toa labeled, sterile 50-ml tube.Another rinse with 2 ml growth medium can be performed to ensure maximum cell recovery.
Cell count and plating
-
6
Centrifuge 5 min at 500 × g, room temperature, to obtain cell pellet.
-
7Remove supernatant.Be sure not to disturb the cell pellet before removing supernatant or cells will be lost. When decanting, avoid multiple inversions of the vial.
-
8Resuspend cells in growth medium.The amount of growth medium can be varied based on the desired concentration of cells. We recommend resuspending in 5 to 6 ml growth medium per 1 × 106 expected cells.
-
9In a separate 1.5-ml microcentrifuge tube, dilute 10 μl cells 1:2 in diluted trypan blue, and count cells using a hemacytometer.If cells are more concentrated, dilution can be increased.
-
10Plate desired number of myoblasts per plate on collagen-coated plates.We recommend 2.5–4.0 × 103 cells per cm2. If 100-mm plates are used, we recommend 2.0–3.0 × 105 cells per plate. For cells that will be differentiated rather than further expanded, we recommend plating on a smaller plate (e.g., 60-mm plate or 6-well plate) to conserve cells.Other sizes of plates can be used; adjust growth medium volume and number of cells accordingly.
-
11Fill with growth medium to desired volume.We recommend 125 to 200 μl growth medium per cm2. For a 100-mm plate, we use 10 ml growth medium.
-
12View under microscope (4× or 10× magnification recommended) prior to incubation. Place culture dishes in a standard humidified cell culture incubator (37°C, 5% CO2).Cells will appear circular and suspended when freshly plated and will be elongated and spindle-shaped once adhered.
-
13Change growth medium every other day until 80% to 90% confluent or 70% to 80%confluent if moving on to differentiation.Remove growth medium using an aspirator, and replace using a sterile serological pipette.
-
14Repeat steps 1 to 13 for appropriate number of passages to obtain the desired number of cells.Primary cells appear to undergo senescence after passages 5 to 6; therefore, we aim to obtain the desired cell number by passage 4.
BASIC PROTOCOL 3
MYOBLAST DIFFERENTIATION
Myoblast differentiation is a critical step during myogenesis. By changing medium from high- to low-serum conditions in myoblasts that are 70% to 80% confluent, myoblasts are stimulated to differentiate and fuse. After a predetermined number of days of differentiation, cells can be used for downstream applications. This protocol includes a step during which myotubes are fixed in preparation for staining and quantification.
Materials
70% to 80% confluent myoblasts (see Basic Protocols 1 or 2 or Alternate Protocol 1)
Sterile PBS, pH 7.4 (e.g., Gibco, cat. no. 10010–023)
Differentiation medium (see recipe)
100% methanol (e.g., Fisher Chemical, cat. no. A433P-4)
NOTE: All steps should be performed in a sterile biosafety cabinet using sterile technique and appropriate PPE.
Aspirate growth medium from adhered myoblasts that have reached 70% to 80%confluence.
Quickly rinse with sterile PBS, and then aspirate PBS.
- Add differentiation medium.The same volume of medium can be used as in Basic Protocols 1 and 2 and Alternate Protocol 1 (e.g., 125 to 200 μl medium per cm2). We use 4 ml medium for a 60-mm dish.
- Change medium every other day for the desired number of days of differentiation.Optimal differentiation occurs after 5 to 7 days.
When ready to fix cells, remove differentiation medium.
Rinse twice with sterile PBS.
- Incubate with ice-cold methanol (same volume as differentiation medium) for10 min to fix cells.Incubation can be done at room temperature inside a biosafety cabinet as long as methanol is ice-cold when starting.
- Aspirate methanol and leave open in a biosafety cabinet to air dry (~10 min).Once fixed, cells can be immediately stained or stored indefinitely at room temperature.
BASIC PROTOCOL 4
HEMA 3 STAINING
Fixed cells from Basic Protocol 3 require staining to visualize myonuclei, to distinguish multinucleated myotubes from myoblasts, and to quantify the extent of differentiation. The more commonly used stains require pH adjustment and fresh preparation, where as the HEMA 3 staining method described here uses shelf-stable, ready-to-use stains. HEMA 3 produces robust staining with easy-to-visualize myonuclei.
Materials
Methanol-fixed cells (see Basic Protocol 3)
HEMA 3 Solution I (e.g., Fisher, cat. no. 23–122937)
HEMA 3 Solution II (e.g., Fisher, cat. no. 23–122952)
Orbital shaker (e.g., The Belly Dancer, Stovall Life Science) or rocker (e.g., Reliable Scientific Model 55 Rocking Shaker)
Microscope with camera (e.g., Olympus BX51 microscope with Olympus DP72 camera)
- To methanol-fixed cells, add HEMA 3 Solution I to cover cells.We use 3 ml for a 60-mm culture dish or 1.5 ml for each well of a 6-well plate.
- Incubate with gentle agitation for 10 min.A speed of ~0.5 Hz on an orbital shaker is recommended.
Wash twice with distilled water (1 min each with gentle agitation).
- Remove water and add HEMA 3 Solution II to cover cells.We use 3 ml for a 60-mm culture dish or 1.5 ml for each well of a 6-well plate.
Incubate with gentle agitation for 10 min.
Wash twice with distilled water (1 min each with gentle agitation).
Remove water and leave open in a laminar flow biosafety cabinet to dry.
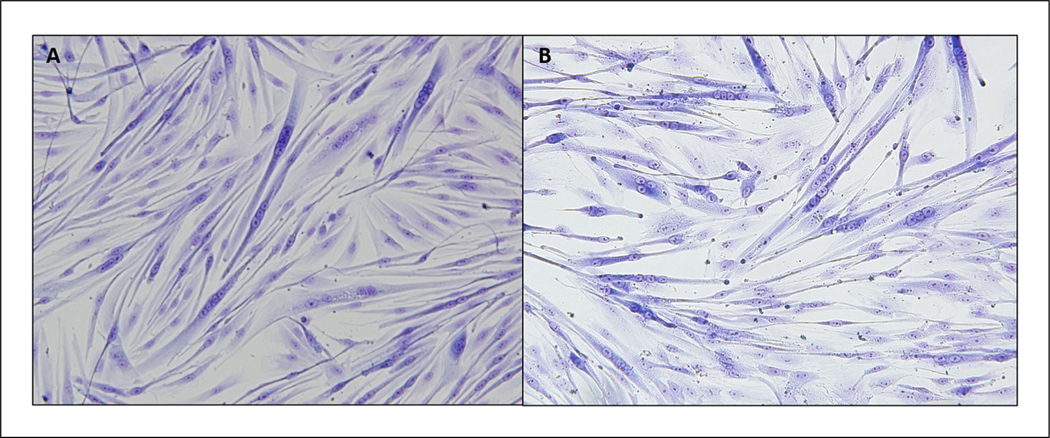
- View under a microscope to ensure staining efficiency. Then place plate upside-down on microscope, and photograph each well.See Figure 1 for an example of efficient staining.We recommend taking 10 to 15 representative images at 10× and 20× magnification.
Figure 1.
Representative images of human (A) and rhesus macaque (B) myoblasts stained with HE 5 days of differentiation.
ALTERNATE PROTOCOL 2
JENNER-GIEMSA STAINING
Jenner-Giemsa staining has traditionally been used for staining of fixed myotubes but requires accurate adjustment of pH and stain preparation. Any differences in stain preparation could result in variations in staining results. Therefore, if this method is used, extra caution should be exercised in pH adjustment and stain preparation. This protocol is slightly modified and expanded from a previous description of Jenner-Giemsa staining (Veliça & Bunce, 2011).
Materials
1 mM sodium phosphate buffer (see recipe)
Jenner staining solution (e.g., Electron Microscopy Sciences, cat. no. 26033–05)
Giemsa staining solution (e.g., Wright-Giemsa Stain; Fisher Healthcare, cat. no. 264–983)
Methanol-fixed cells (see Basic Protocol 3)
Filter paper (e.g., Whatman qualitative filter paper, Grade 1; Millipore Sigma, cat. no. WHA1001325)
Orbital shaker (e.g., The Belly Dancer, Stovall Life Science) or rocker (e.g., Reliable Scientific Model 55 Rocking Shaker)
Microscope with camera (e.g., Olympus BX51 microscope with Olympus DP72 camera)
NOTE: Stains contain methanol and thus should be discarded appropriately.
Prepare 1 mM sodium phosphate buffer.
- Dilute Jenner staining solution 1:3 in 1 mM sodium phosphate buffer. Dilute Giemsa staining solution 1:20 in 1 mM sodium phosphate buffer, and filter with filter paper.Prepare fresh staining solution each day that staining will be performed.
- To methanol-fixed cells, add diluted Jenner staining solution to cover cells.We use 3 ml for a 60-mm culture dish or 1.5 ml for each well of a 6-well plate.
- Incubate with gentle agitation for 10 min.A speed of ~0.5 Hz on an orbital shaker is recommended.
Wash once with distilled water (1 min with gentle agitation).
- Remove water and add diluted Giemsa staining solution to cover cells.We use 3 ml for a 60-mm culture dish or 1.5 ml for each well of a 6-well plate.
Incubate with gentle agitation for 10 min.
Wash twice with distilled water (1 min each with gentle agitation).
Remove water and leave open in a laminar flow biosafety cabinet to dry.
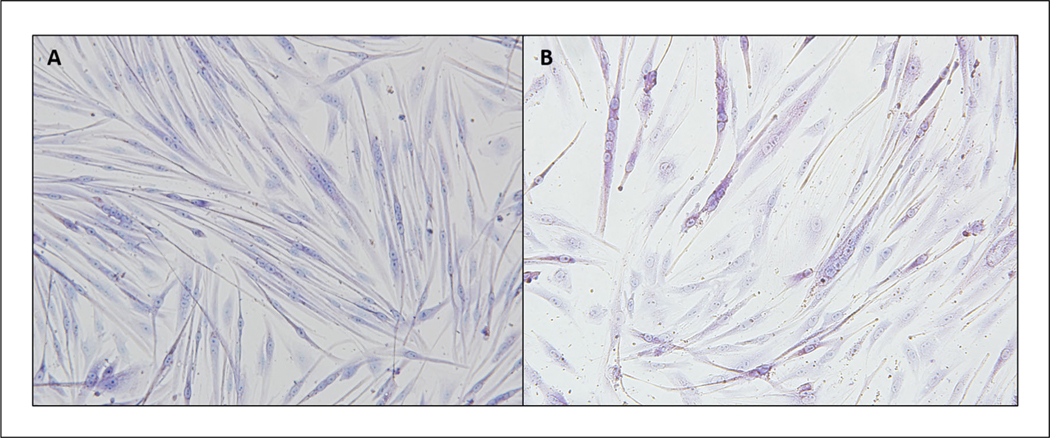
- View under a microscope to ensure staining efficiency. Then place plate upside-down on microscope, and photograph each well.See Figure 2 for an example of efficientWe recommend taking 10 to 15 representative images at 10× and 20× magnification.
Figure 2.
Representative images of human (A) and rhesus macaque (B) myoblasts stained with Jenner and Giemsa stains after 5 days of differentiation.
BASIC PROTOCOL 5
QUANTIFICATION OF MYOTUBE DENSITY
Myotube density measurement, one of several measures of differentiation, provides one measure of insight into the extent of myoblast differentiation into myotubes. Protein-rich myotubes stain more darkly than myoblasts, so this protocol uses staining intensity to quantify myotube density. This protocol was originally described by Velica and Bunce (2011).
Materials
10× myotube images (see Basic Protocol 4 or Alternate Protocol 2)
Computer running:
NIS-Elements software
Excel or other spreadsheet analysis software
- Open 10× image in NIS-Elements software.The instructions provided in this protocol are for NIS-Elements version 4.51.Other imaging processing software is also acceptable, but menus and options may be set up differently. NIS-Elements software is for Nikon microscopes, but image files obtained using other microscopes and camera systems can be uploaded and analyzed using this software. Alternately, software available to the user can be used to measure intensity based on the settings and requirements of that particular software.
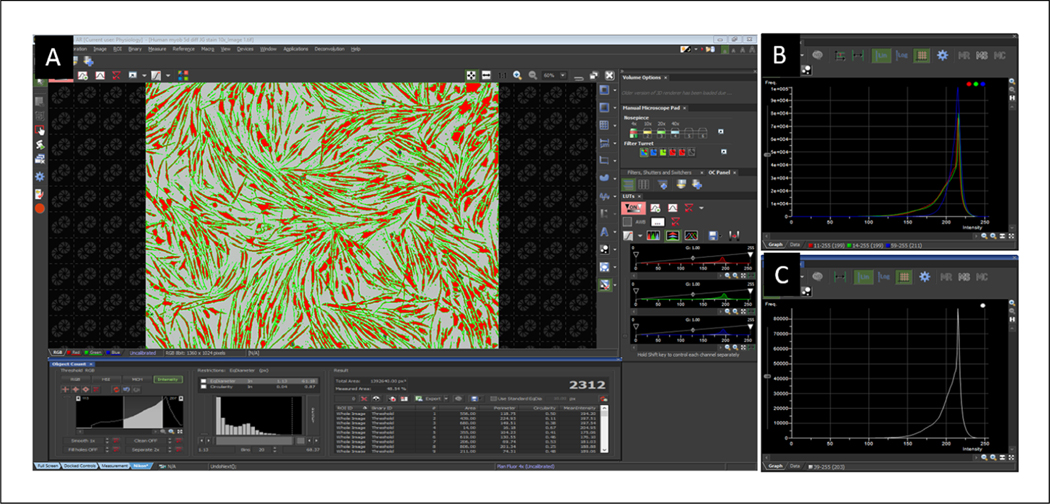
- Open “Object Count” toolbar by clicking View, Analysis controls, and Object count and “Histogram” toolbar by clicking View, Visualization controls, and Histogram. Using the Object Count toolbar, adjust intensity threshold to highlight portions of the image considered to be positive staining (Fig. 3A).The toolbar will produce data including mean intensity readings.
Export data to Excel, and determine median in “MeanIntensity” column; name spreadsheet and Excel file appropriately. Record median in a summary Excel spreadsheet.
Convert image to grayscale by clicking Image, Convert, and Convert to gray(Fig. 3B,C).
- Export histogram toolbar data to Excel; name appropriately.This gives a pixel count for all intensity readings.
Repeat steps 1 to 5 for all images of a single sample.
- Calculate average median intensity for all images of a single sample; record in summary spreadsheet in Excel.This number will be the positive intensity threshold applied to all images and used for quantification.
Sum pixel numbers (frequency column in grayscale) for each image of a single sample from 0 to threshold from step 7. Record in summary spreadsheet.
Average pixel sums for all images of a single sample to calculate myotube density.
Repeat steps 1 to 9 for images from all samples.
Figure 3.
Representative images of myotubes with positive staining highlighted in NIS-Element (A) and histograms before (B) and after (C) converting the image to grayscale.
BASIC PROTOCOL 6
QUANTIFICATION OF FUSION INDEX
Counting the number of myonuclei that are contained within multinucleated myotubes versus nonfused myoblasts provides a second index of the extent to which myoblasts have differentiated in the predefined time period (e.g., 5 days). The ratio of myonuclei in myotubes to total myonuclei is the fusion index. This protocol provides a method to quantify the fusion index using ImageJ and spreadsheet software.
Materials
20× myotube images (see Basic Protocol 4 or Alternate Protocol 2)
Computer running:
ImageJ (available at http://imagej.nih.gov/ij/download.html)
Excel or other spreadsheet analysis software
NOTE: Before beginning, determine the threshold number of myonuclei within a cell to consider the cell a myotube. We typically require three myonuclei for a myotube, but two myonuclei can also be used as the threshold provided the definition is consistent throughout quantification.
Open 20× image in ImageJ.
- Sharpen image by clicking Process, Sharpen.This makes borders of nuclei and myotubes more visible.
Select “Multi-point” icon to bring up the multi-point tool.
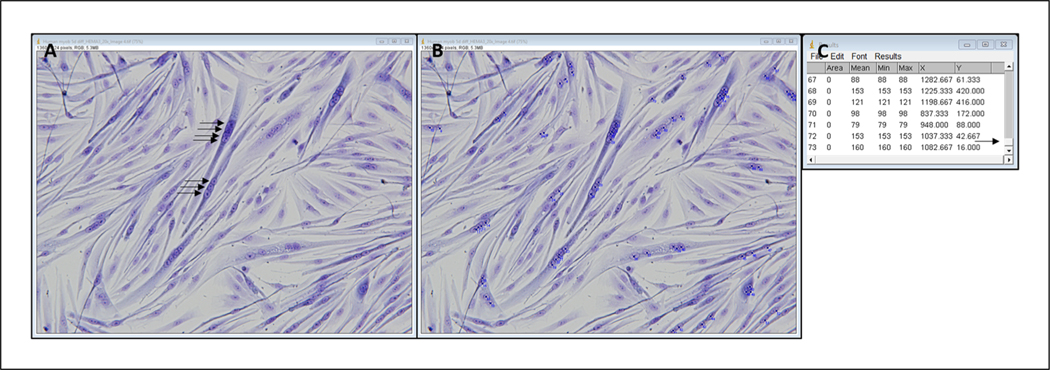
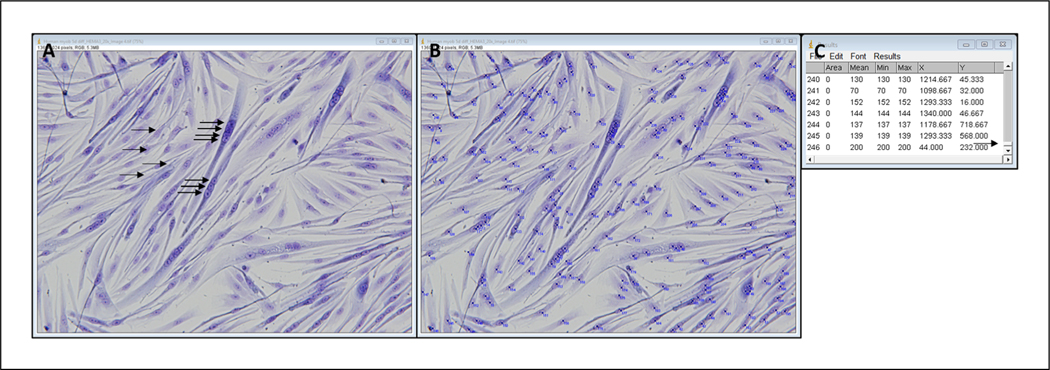
Click on each myonucleus that is within a myotube (Fig. 4A,B).
- When all myonuclei in myotubes have been selected, obtain a final count by clicking Analyze, Measure; record final number from the far-left column in an Excel spreadsheet.This number indicates the number of fused myonuclei. See example results box in Figure 4C.
Divide fused myonuclei by total myonuclei to calculate fusion index for a single image.
Repeat steps 1 to 7 for all images of a single sample.
Average fusion indices for each image to obtain a fusion index for the sample.
Repeat for each sample.
Figure 4.
Representative image of HEMA 3–stained myotubes before (A) and after (B) selecting fused nuclei and example results diagram (C). Arrows in A represent a subset of nuclei that will be selected. Arrow in C shows that the results table is scrolled to the bottom. Left column contains a count of each point selected, so the last number in that column is the number of fused nuclei counted.
Figure 5.
Representative image of HEMA 3–stained myotubes before (A) and after (B) selecting total nuclei and example results diagram (C). Arrows in A represent a subset of nuclei that will be selected. Arrow in C shows that the results table is scrolled to the bottom. Left column contains a count of each point selected, so the last number in that column is the number of total nuclei counted.
BASIC PROTOCOL 7
QUANTIFICATION OF MYOTUBES PER FIELD
Quantifying the number of myotubes in each field of view offers a complementary assessment of myotube density. This technique is performed by selecting all myotubes within a given image using ImageJ and averaging results from all images of a given sample.
Materials
20× myotube images (see Basic Protocol 4 or Alternate Protocol 2)
Computer running:
ImageJ (available at http://imagej.nih.gov/ij/download.html)
Excel or other spreadsheet analysis software
NOTE: The threshold number of myonuclei for a cell to be considered a myotube should be the same in this protocol as in Basic Protocol 6.
Open 20× image in ImageJ.
- Sharpen image by clicking Process, Sharpen.This makes borders of nuclei and myotubes more visible.
Select “Multi-point” icon to bring up the multi-point tool.
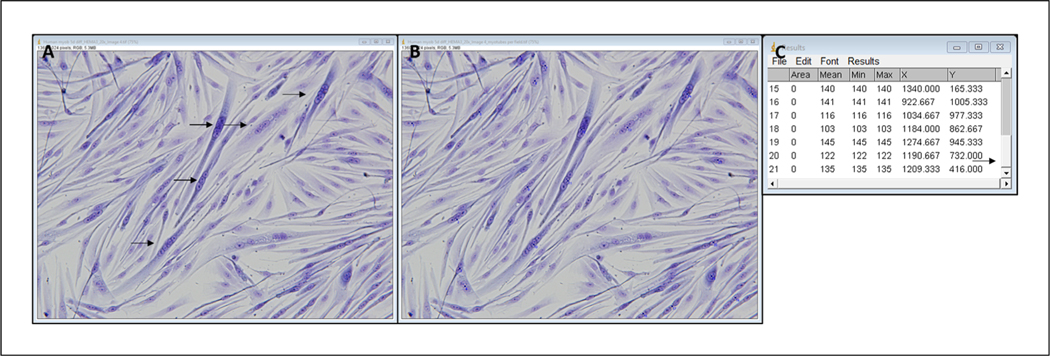
- When all myotubes have been selected, click Analyze, Measure; record final number from the far-left column in an Excel spreadsheet.This number indicates the number of myotubes in the image. See example results box in Figure 6C.
Repeat steps 1 to 5 for all images of a single sample.
Average myotubes per field for each image to obtain the average number of myotubes per field for the sample.
Repeat for each sample.
Figure 6.
Representative image of HEMA 3–stained myotubes before (A) and after (B) selecting myotubes and example results diagram (C). Arrows in A represent a subset of myotubes that will be selected. Arrow in C shows that the results table is scrolled to the bottom. Left column contains a count of each point selected, so the last number in that column is the number of myotubes counted.
REAGENTS AND SOLUTIONS
Differentiation medium
474 ml Ham’s F12 Nutrient Mixture with 1 mM L-glutamine (e.g., GE Healthcare Life Sciences, cat. no. SH30026.01)
10 ml horse serum, heat inactivated (e.g., Thermo Fisher, cat. no. 26050088)
5 ml penicillin-streptomycin (e.g., Gibco, cat no. 15140–122)
0.5 ml fungizone (i.e., 250 μg/ml amphotericin B; e.g., Gibco, cat. no. 15290–018)
10 ml of 200 mM L-glutamine (e.g., Gibco, cat. no. 25030–081)
Store at 4°C for up to 1 month
A total of 474 ml Ham’s F12 Nutrient Mixture can be obtained by removing 26 ml from a 500-ml bottle.
Antibiotic-antimycotic (5 ml of 100×; e.g., Gibco, cat. no. 15240–062) can be used in place of penicillin-streptomycin and fungizone.
Digest medium
To a sterile 50-ml tube, add:
15 ml isolation medium (see recipe)
15 ml of 0.25% trypsin-EDTA (e.g., Gibco, cat. No. 25200056)
Cap and invert several times to mix
Prepare freshly before use
If not immediately needed, store at 4°C, but allow medium to reach room temperature before use.
Growth medium
434 ml Ham’s F12 Nutrient Mixture with 1 mM L-glutamine (e.g., GE Healthcare Life Sciences, cat. no. SH30026.01)
50 ml FBS (e.g., Sigma-Aldrich, cat. no. F0926–500ML)
0.5 ml of 2.5 μg/ml recombinant human fibroblast growth factor (e.g., R&D systems, cat. no. 233-FB) in sterile PBS (e.g., Gibco, cat. no. 10010–023)
5 ml penicillin-streptomycin (e.g., Gibco, cat. no. 15140–122)
0.5 ml fungizone (i.e., 250 μg/ml amphotericin B; e.g., Gibco, cat. no. 15290–018)
10 ml of 200 mM L-glutamine (e.g., Gibco, cat. no. 25030–081)
Store at 4°C for up to 1 month
A total of 434 ml Ham’s F12 Nutrient Mixture can be obtained by removing 66 ml from a 500-ml bottle.
Heat inactivation of FBS is not necessary.
Antibiotic-antimycotic (5 ml of 100×; e.g., Gibco, cat. no. 15240–062) can be used in place of penicillin-streptomycin and fungizone.
Isolation medium
48.5 ml F12 Ham’s Nutrient Mixture with 1 mM L-glutamine (e.g., GE Healthcare Life Sciences, cat. no. SH30026.01)
0.5 ml penicillin-streptomycin (e.g., Gibco, cat. no. 15140–122)
0.05 ml fungizone (i.e., 250 μg/ml amphotericin B; e.g., Gibco, cat. no. 15290–018)
1 ml of 200 mM L-glutamine (e.g., Gibco, cat. no. 25030–081)
Store at 4°C for up to 1 month
Antibiotic-antimycotic (5 ml of 100×; e.g., Gibco, cat. no. 15240–062) can be used in place of penicillin-streptomycin and fungizone.
Sodium phosphate buffer, 1 mM
Prepare stock A by combining 27.6g sodium phosphate monobasic monohydratein 1000 ml deionized water.
Prepare stock B by combining 28.4 g sodium phosphate dibasic in 1000 mldeionized water.
Combine 110 ml stock A and 6 ml stock B.
Dilute to 232 ml with deionized water.
- Adjust pH to 5.6 with HCl or NaOH, if necessary. Store at room temperature for up to 1 yearVisually inspect buffer before use to ensure absence of fungal growth.
COMMENTARY
Background Information
After muscle injury or atrophy, satellite cells undergo the process of myogenesis for repair and regeneration of muscle tissue. Myogenesis is characterized by sequential phases including satellite cell activation, myoblast proliferation, differentiation, and fusion with existing myofibers (Yin et al., 2013). Stimuli that affect myogenesis can be studied in vitro using isolated myoblasts. Differentiating myoblasts can be fixed, stained, and visualized, and indices of differentiation can be quantified. Using the protocols described herein, we have previously found that myoblasts isolated from simian immunodeficiency virus–infected male rhesus macaques subjected to chronic binge alcohol administration have reduced myoblast differentiation potential compared to those who were not administered alcohol (Simon et al., 2014; Simon, Ford, et al., 2017). The use of these protocols to assess differentiation provide support for hypothesized mechanisms underlying alcoholic myopathy (Molina, Simon, Amedee, Welsh, & Ferguson, 2018; Simon, Jolley, & Molina, 2017).
To accurately visualize differentiating myoblasts and quantify indices of differentiation, cells must be fixed and stained. The more commonly used Jenner and Giemsa stains were originally developed as variations of the Romanowsky stain traditionally used in hematology (Barcia, 2007). The stains contain basic (e.g., methylene blue) and acidic (e.g., eosin Y) components to differentially stain the nuclear membrane and cytosolic proteins. Several iterations of these stains have been developed and are used for similar purposes (Woronzoff-Dashkoff, 1993; Wright, 1902). Application of these stains to myotubes and quantification have been previously described (Veliça & Bunce, 2011) and widely used. HEMA 3 solutions are a further development of these stains that contain the necessary basic (methylene blue) and acidic (eosin Y) components premixed with potassium and sodium phosphate buffers, but they are shelf-stable and offer several advantages over Jenner and Giemsa stains. Namely, HEMA 3 stains require no preparation or pH adjustment yet produce comparable, robust, and reliable staining results. Therefore, we propose the use of these stains as a preferable alternative to the Jenner-Giemsa staining method.
Critical Parameters and Troubleshooting
For all cell culture experiments, a sterile working environment and clean cell culture incubator are essential. Failing to maintain a sterile working environment or contaminating any media during the myoblast isolation, expansion, or differentiation phases may introduce bacteria or fungi into the cultures. We suggest disinfecting the biosafety cabinet and all pipettes before use, turning on the laminar air flow at least 15 min before using the hood, and disinfecting the biosafety cabinet and all pipettes after using the hood. We also suggest autoclaving nonfilter pipette tips for vacuum aspiration and flushing the vacuum aspirator with sterile water followed by 70% ethanol after use. Growth of spores in culture indicates fungal contamination, and an opaque nonadherent layer indicates bacterial bloom. If cultures become contaminated, they should be bleached for 30 min and discarded, and the cell culture incubator should be disinfected. Media can be ruled out as a contaminant if other cultures incubated with the same media are contamination free or if a culture dish with medium alone incubated for several days is contamination free.
During expansion, cells can grow to 80% to 90% confluence before passaging, but for differentiation it is critical that growth medium is switched to differentiation medium when cells are ~70% to 80% confluent. If myoblasts become too dense prior to differentiation, it reduces the ability of cells to fuse and form myotubes.
Common problems with staining include light staining and localized points of excessive staining. Light staining for both staining methods described herein (HEMA 3 and Jenner-Giemsa) can be due to insufficient length of staining. If staining is too light, try increasing the time of incubation. For Jenner-Giemsa staining, light staining may also be due to incorrect pH of buffer. Verify that sodium phosphate buffer pH is 5.6. Finally, light staining for Jenner-Giemsa can be due to old stains. We recommend making fresh Jenner and Giemsa stains each time this staining method is performed. The use of shelf-stable HEMA 3 for staining eliminates the potential problems of incorrect pH and frequent preparation of fresh stains. Localized points of excessive staining maybe due to either debris in the fixed cells or insufficient washing. The latter is a greater problem with Jenner-Giemsa staining since Giemsa stain contains small granules. Proper filtration of Giemsa stain is important for granule removal. If debris is suspected, ensure that lids are kept on fixed cells at all times except when pipetting solutions into dishes and during imaging. During drying steps, culture dishes should be kept in a laminar flow biosafety cabinet to prevent debris from entering dishes. If insufficient washing is suspected, be sure that distilled water covers the surface of the culture dish during wash steps and that no stain is left in the dish after washing. If necessary, an extra wash step can be performed at the end of the protocol to remove excess stain.
For quantification, consistency in using imaging software is key. This is particularly important when setting intensity thresholds for myotube density. Ensure that intensity thresholds are set such that areas of localized excessive staining and blank areas with no myotubes are excluded and that these settings are applied consistently with each image. For fusion index and myotubes per field, it is essential to predetermine how many myonuclei a cell must contain to be considered a myotube (typically two or three), and this threshold must be applied consistently throughout quantification. Focusing the microscope during imaging and sharpening the image in ImageJ aid in discerning edges of myonuclei for quantification. If myonuclei are difficult to discern after sharpening, it is possible that stains were applied for too short or too long a time period (i.e., staining is too light or too dark) or that the microscope was not appropriately focused during imaging. If the latter is suspected, cells can be reimaged.
Understanding Results
Myoblast isolation and expansion produces a large number of myoblasts that can be used for downstream applications, including differentiation, staining, and quantification as described in Basic Protocols 3 through 7. During expansion, a confluent 100-mm culture dish can yield 3.0 × 106 cells. After differentiation, HEMA 3 staining should result in myotubes stained light purple with dark purple staining of chromatin and nuclear membranes. Myotubes can be classified as containing a minimum of two or three nuclei per cell, but the threshold should be used consistently for all images to be analyzed. Subsequent analysis will depend on experimental design. Myoblasts or differentiated myotubes can also be used for other downstream applications including assessing mitochondrial function (Duplantyetal.,2018)and anabolic functions such as insulin-responsive glucose uptake and protein synthesis.
Time Considerations
The initial day of myoblast isolation from fresh tissue takes 4 hr, including incubation steps. For initial plating, myoblasts isolated from fresh tissue should be 20% to 30% confluent in ~2 to 4 weeks, whereas the initial passage beginning with cryopreserved myoblasts should be 80% to 90% confluent within ~1 week. Each subsequent myoblast passage should be 80% to 90% confluent within ~1 week. When myoblasts are designated for differentiation, medium is changed from growth to differentiation medium at 70% to 80% confluency, and differentiation should be carried out for a predetermined number of days. The processes of cell fixation and HEMA 3 staining take <1 hr each. Extra time should be set aside for Jenner-Giemsa stain preparation if that staining method is selected. Quantification (Basic Protocols 5, 6, and 7) is time consuming, but total time depends on the number of images.
Acknowledgments
The authors would like to thank Dr. Patricia Molina for her scientific expertise and guidance and Curtis Vande Stouwe and Naveena Chalapati for their valuable technical assistance. The research underlying the development of the protocols was supported by grants from the National Institute on Alcohol Abuse and Alcoholism (1F32AA027982-01A1, 5K01AA024494-03, 5P60AA009803-25, and 5T32AA007577-20).
Literature Cited
- Barcia JJ (2007). The Giemsa stain: Its history and applications. International Journal of Surgical Pathology, 15, 292–296. doi: 10.1177/1066896907302239. [DOI] [PubMed] [Google Scholar]
- Bellamy LM, Joanisse S, Grubb A, Mitchell CJ, McKay BR, Phillips SM, . . . Parise G. (2014). The acute satellite cell response and skeletal muscle hypertrophy following resistance training. PLoS One, 9, e109739. doi: 10.1371/journal.pone.0109739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Pan JS, & Zhang L. (2013). Myostatin suppression of Akirin 1 mediates glucocorticoid induced satellite cell dysfunction. PLoS One, 8, e58554. doi: 10.1371/journal.pone.0058554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplanty AA, Siggins RW, Allerton T, Simon L, & Molina PE (2018). Myoblast mitochondrial respiration is decreased in chronic binge alcohol administered simian immunodeficiency virus-infected antiretroviral treated rhesus macaques. Physiological Reports, 6, e13625. doi: 10.14814/phy2.13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh Q, Song T, Petrany MJ, Cramer AA, Sun C, Sadayappan S, . . . Millay DP (2019). Myonuclear accretion is a determinant of exercise-induced remodeling in skeletal muscle. eLife, 8, E44876. doi: 10.7554/elife.44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCroskery S,Thomas M,Maxwell L,Sharma M, & Kambadur R. (2003). Myostatin negatively regulates satellite cell activation and self-renewal. The Journal of Cell Biology, 162, 1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina PE, Simon L, Amedee AM, Welsh DA, & Ferguson TF (2018). Impact of alcohol on HIV disease pathogenesis, comorbidities and aging: Integrating preclinical and clinical findings. Alcohol and Alcoholism, 53, 439–447. doi: 10.1093/alcalc/agy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Ford SM, Song K, Berner P, Vande Stouwe C, Nelson S, . . . Molina PE (2017). Decreased myoblast differentiation in chronic binge alcohol-administered simian immunodeficiency virus-infected male macaques: Role of decreased miR-206. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 313, R240–R250. doi: 10.1152/ajpregu.00146.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Jolley SE, & Molina PE (2017). Alcoholic myopathy: Pathophysiologic mechanisms and clinical implications. Alcohol Research: Current Reviews, 38, 207–217. [PMC free article] [PubMed] [Google Scholar]
- Simon L, LeCapitaine N, Berner P, Vande Stouwe C, Mussell JC, Allerton T, . . . Molina PE (2014). Chronic binge alcohol consumption alters myogenic gene expression and reduces in vitro myogenic differentiation potential of myoblasts from rhesus macaques. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 306, R837–844. doi: 10.1152/ajpregu.00502.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha-Hikim I, Cornford M, Gaytan H, Lee ML, & Bhasin S. (2006). Effects of testosterone supplementation on skeletal muscle fiber hypertrophy and satellite cells in community dwelling older men. The Journal of Clinical Endocrinology and Metabolism, 91, 3024–3033. doi: 10.1210/jc.2006-0357. [DOI] [PubMed] [Google Scholar]
- Veliça P, & Bunce CM (2011). A quick, simple and unbiased method to quantify C2C12 myogenic differentiation. Muscle & Nerve, 44, 366–370. doi: 10.1002/mus.22056. [DOI] [PubMed] [Google Scholar]
- Woronzoff-Dashkoff KP (1993). The EhrlichChenzinsky-Plehn-Malachowski-Romanowsky-Nocht-Jenner-May-Grunw ald-Leishman-¨ Reuter-Wright-Giemsa-Lillie-Roe-Wilcox stain. The mystery unfolds. Clinics in Laboratory Medicine, 13, 759–771. doi: 10.1016/s0272-2712(18)30406-2. [DOI] [PubMed] [Google Scholar]
- Wright JH (1902). A rapid method for the differential staining of blood films and malarial parasites. The Journal of Medical Research, 7, 138–144. [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, & Rudnicki MA (2013). Satellite cells and the muscle stem cell niche. Physiological Reviews, 93, 23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]