Abstract
The Research Electronic Data Capture (REDCap) data management platform was developed in 2004 to address an institutional need at Vanderbilt University, then shared with a limited number of adopting sites beginning in 2006. Given bi-directional benefit in early sharing experiments, we created a broader consortium sharing and support model for any academic, non-profit, or government partner wishing to adopt the software. Our sharing framework and consortium-based support model have evolved over time along with the size of the consortium (currently more than 3,200 REDCap partners across 128 countries). While the “REDCap Consortium” model represents only one example of how to build and disseminate a software platform, lessons learned from our approach may assist other research institutions seeking to build and disseminate innovative technologies.
Keywords: Medical Informatics, Electronic Data Capture, Clinical Research, Translational Research
Graphical Abstarct
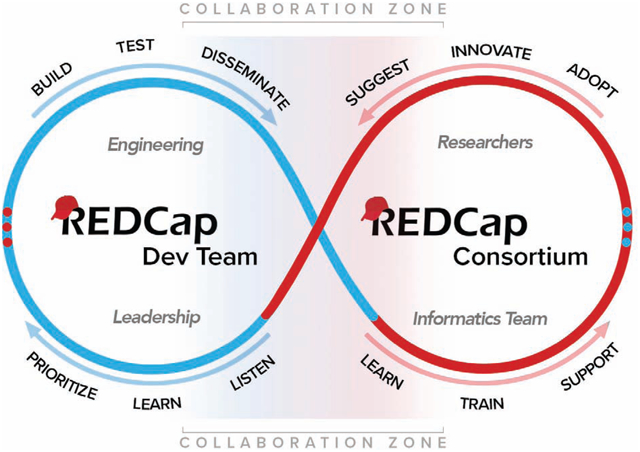
Introduction
Clinical research informatics (CRI) is an evolving field widely recognized as crucial for transforming and reengineering the translational research enterprise [1–3]. Although most institutions recognize the need for growing research informatics support, reductions in grant funding and restricted institutional budgets have diminished the ability of many clinical organizations to hire and retain operations-focused CRI faculty and staff [4,5]. Consequently, limited personnel and resources are stretched to support research needs, forcing sites to restrict programs and services and limit innovation [6–9]. A 2006 initiative of the U.S. National Institutes of Health (NIH) Roadmap for Medical Research introduced the Clinical and Translational Science Awards (CTSAs) to partially address this challenge. The CTSA program incentivizes academic medical centers to evaluate critical needs within and across institutions, propose innovative methods to fill gaps, and ultimately implement collaborative solutions to provide sufficient training and infrastructure enabling scientific teams to design, conduct, and disseminate high-impact research [10–13]. Sharing and leveraging of cross-institutional resources is encouraged by the CTSA program and other cooperative grants [10,14–18].
Even with internal and external expectations to collaborate, there remain significant barriers to sharing clinical research software applications [19] and these challenges are not unique to non-profit research communities [20–23]. In-house software development enables organizations to produce efficient and highly customized applications that accommodate local workflow and IT environments. However, transferring a custom application outside the original environment requires resources and commitment. While many groups may be willing to share source code, the effort required to generalize the code to different IT configurations, supply documentation, test in multiple environments, assist with installations, and provide software training can be prohibitive. The “burden” of sharing is not limited to initial setup, as even well-factored code needs updating when new features are added, bugs are detected, and general security vulnerabilities are exposed and patched. Both sharing and receiving sites need fair and flexible licensing models that respect significant investments needed to develop and share, as well as adopt and contribute to software platforms. Developers need to maximize the value of their intellectual property and limit liability, while adopters need assurance of the long-term sustainability of the platform and development team [24].
Although barriers are significant, sharing can produce bi-directional benefits. Given the right project, sharing model, and collaborative adopters, dissemination can benefit informatics developers, adopting informatics teams, and end users. Systems tend to improve when responsive developers can rely on an active user base for new use cases and feature requests. Research systems with users across multiple institutions also can catalyze scientific collaboration and development across networks [25]. Adopting teams inherit and benefit from a well-developed system, freeing their local developers to innovate in other domains.
The Research Electronic Data Capture (REDCap) data management platform is an example of a CTSA-funded clinical research software application that has grown beyond its single institutional use case to be shared with institutions across the world [26]. Since its initial development at Vanderbilt University, REDCap has been adopted by more than 3,207 partners in 128 countries as of mid-December 2018 (Figures 1 and 2, Table 1). REDCap is a secure web application for building and managing online surveys and databases, designed to support data capture for research studies [26]. The diverse applications of REDCap (Figure 3) include support of basic science research studies [27,28], data collection for clinical trials [29,30], registries and cohort studies [31–35], quality reviews for clinical practice [36–39], comparative effectiveness trials [40–42], patient questionnaires [43–45], clinical decision support applications [46], and operational support [47]. REDCap has limited personnel requirements (one support person can easily manage hundreds of projects [48]) but does require typical web infrastructure including one or more secure web servers running PHP, MySQL/MariaDB, and SMTP email services.
Figure 1:
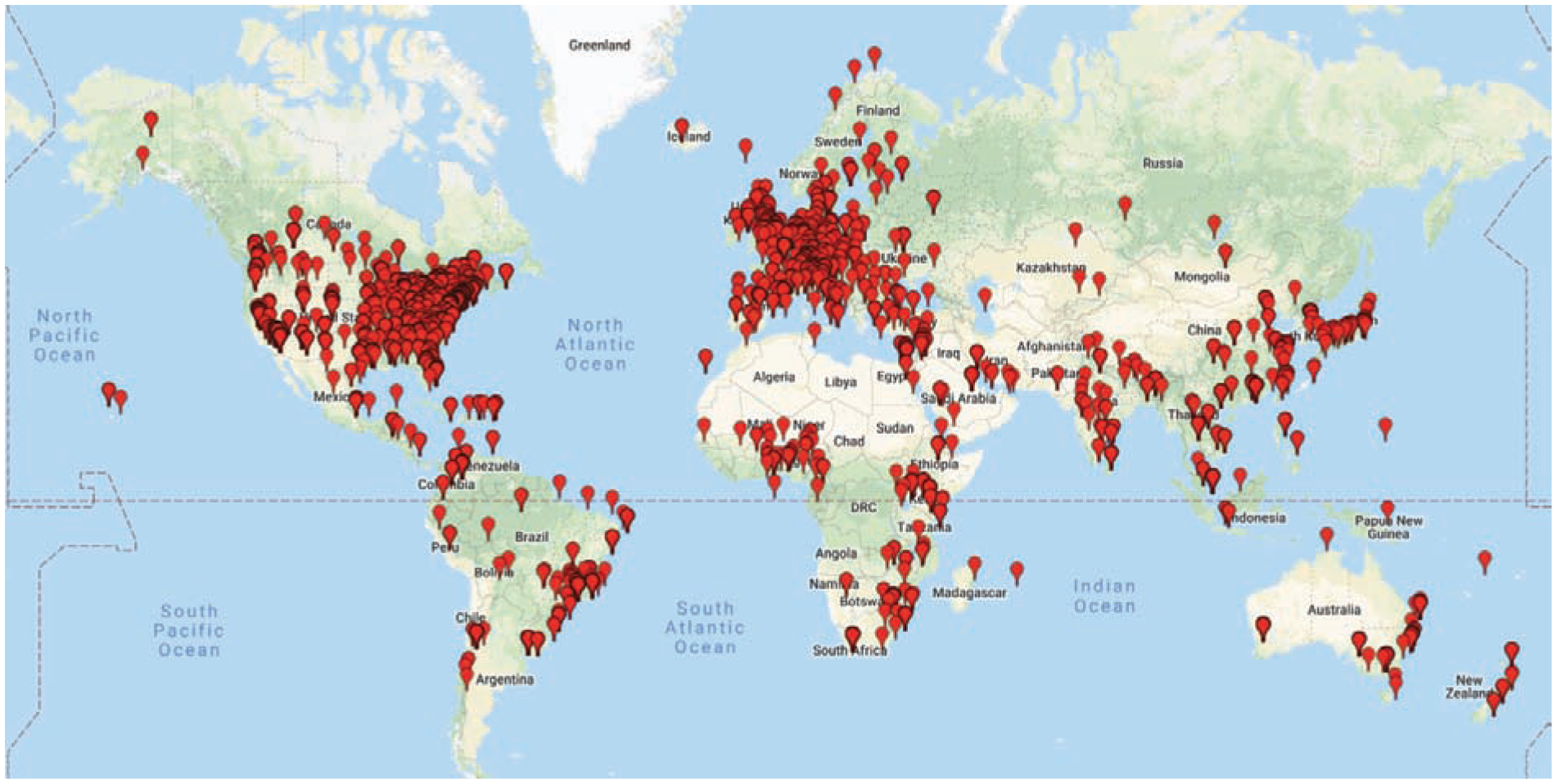
Map of Consortium (December 19, 2018) – 3,207 partner institutions in 128 countries
Figure 2:
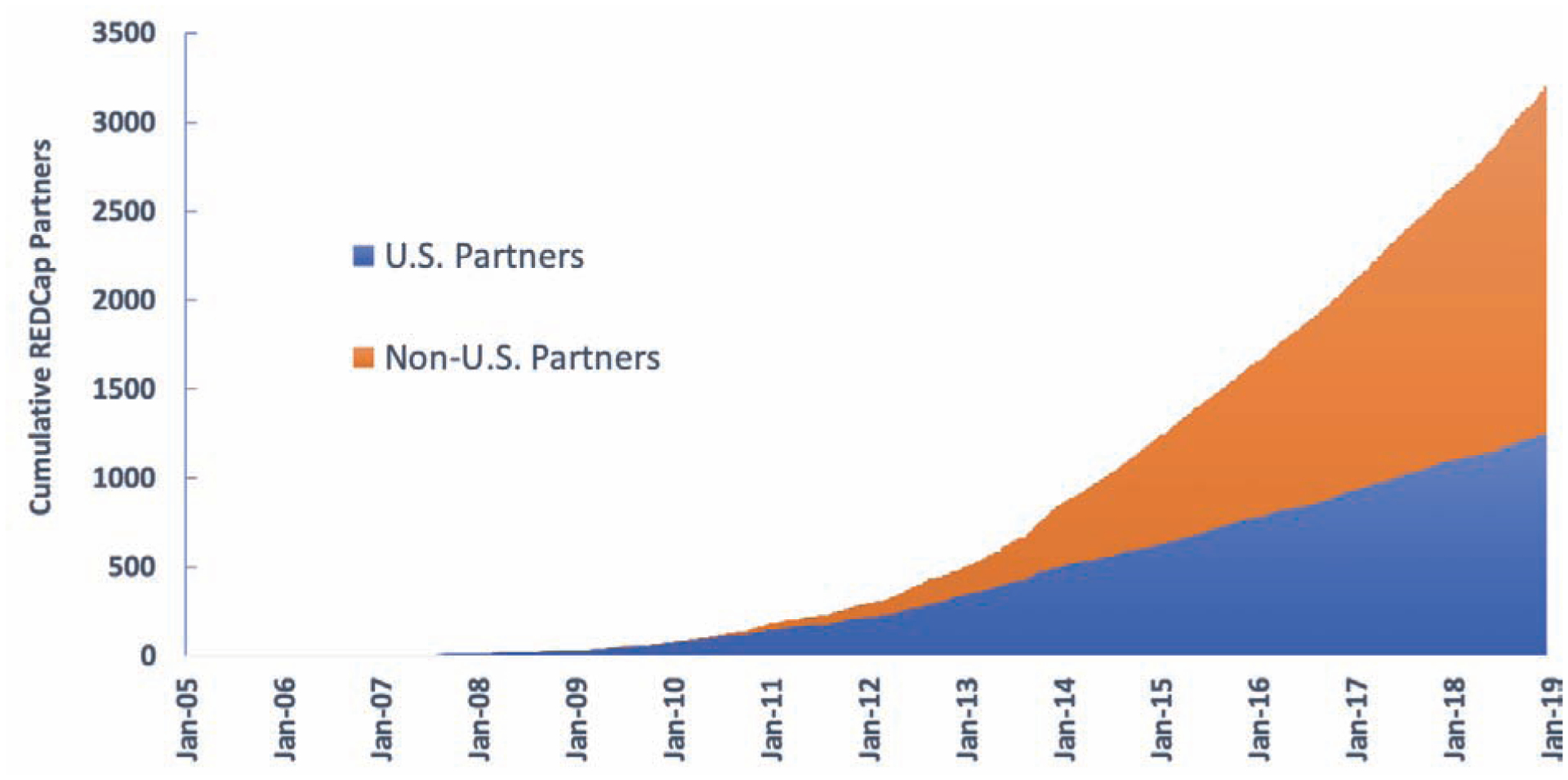
Growth of REDCap Consortium per Year.
Table 1:
Number of licensed REDCap Consortium partners by country or territory as of December 19, 2018
| Argentina (21) | El Salvador (1) | Macedonia (2) | Rwanda (2) |
| Australia (137) | Estonia (2) | Madagascar (1) | Saudi Arabia (17) |
| Austria (9) | Ethiopia (7) | Malawi (9) | Senegal (1) |
| Azerbaijan (1) | Faroe Islands (1) | Malaysia (5) | Serbia (3) |
| Bahrain (1) | Finland (5) | Mali (3) | Sierra Leone (1) |
| Bangladesh (11) | France (77) | Malta (1) | Singapore (18) |
| Barbados (1) | Gabon (1) | Mauritius (1) | Slovakia (2) |
| Belgium (18) | Gambia (1) | Mexico (17) | Slovenia (3) |
| Benin (1) | Germany (92) | Mongolia (2) | Solomon Islands (1) |
| Bhutan (1) | Ghana(15) | Morocco (1) | South Africa (45) |
| Bolivia (2) | Greece (9) | Mozambique (4) | South Korea (26) |
| Bosnia and Herzegovina (1) | Guatemala (6) | Myanmar/Burma (1) | Spain (77) |
| Botswana (2) | Haiti (1) | Namibia (2) | Sri Lanka (7) |
| Brazil (139) | Honduras (1) | Nepal (3) | Swaziland (1) |
| Bulgaria (1) | Hungary (7) | Netherlands (15) | Sweden (19) |
| Burkina Faso (3) | Iceland (2) | New Zealand (14) | Switzerland (63) |
| Cambodia (3) | India (43) | Nicaragua (1) | Taiwan (17) |
| Cameroon (4) | Indonesia (5) | Niger (1) | Tanzania (8) |
| Canada(156) | Ireland (11) | Nigeria (18) | Thailand (17) |
| Chile (19) | Israel (14) | Norway (4) | Tunisia (1) |
| China (104) | Italy (100) | Oman(l) | Turkey (19) |
| Colombia (36) | Jamaica (3) | Pakistan (4) | Uganda (15) |
| Congo, Dem Republic (1) | Japan (48) | Panama (1) | Ukraine (9) |
| Costa Rica (2) | Jordan (3) | Papua New Guinea (1) | United Arab Emirates (3) |
| Cote d’lvoire (1) | Kazakhstan (2) | Peru (5) | United Kingdom (141) |
| Croatia (2) | Kenya(22) | Philippines (8) | United States (1249) |
| Cyprus (2) | Kuwait (5) | Poland (13) | Uruguay (4) |
| Czech Republic (9) | Latvia (1) | Portugal (7) | Vanuatu (1) |
| Denmark (18) | Lebanon (2) | Puerto Rico (7) | Venezuela (2) |
| Dominican Republic (6) | Lesotho (1) | Qatar (2) | Vietnam (16) |
| Ecuador (5) | Lithuania (1) | Romania (6) | Zambia (4) |
| Egypt (32) | Luxembourg (1) | Russian Federation (11) | Zimbabwe (5) |
Figure 3:

Web of Science Treemap Visualization showing the number of papers citing REDCap by Research Area. This graphic was generated on 12/19/2018 and represents the top 15 of 105 relevant research areas by frequency of occurrence mapped to 6233 total papers citing REDCap between 2009 and 2018. This bibliometric report shows the diversity of scientific impact for the REDCap platform.
The “REDCap Consortium” is a community comprising academic, non-profit, and government institutions that have adopted REDCap and the cadre of software administrators at those sites who manage local installations of the platform and support local investigators using the software. In general, REDCap Consortium partner distribution tends to correlate with population density and regions of the world where biomedical research is supported. Exceptions include areas where U.S. export control laws prohibit licensing and also regions of the world where internet connectivity is extremely limited or where local government imposed internet accessibility restrictions reduce discoverability of the program. We describe here our phased model for developing and sharing research software and growing a community of adopting sites. In addition, we share lessons learned during the evolution of the REDCap software platform and the REDCap Consortium over the lifecycle of the program. While there are likely unique elements associated with REDCap and the REDCap Consortium model, we believe the core principles are generalizable and can benefit any software development team or sharing consortium.
Building and Supporting the REDCap Consortium: A Phased Approach
Phase 1: Developing REDCap at Vanderbilt University (1 site: ~ 2004–2005)
REDCap was developed in 2004 to provide Vanderbilt University clinical and translational researchers with a straightforward approach to create data management plans while implementing a secure, compliant data management system for their research studies. The institution recognized a need for a centralized data collection and management platform that could support diverse research domains without requiring study-specific programming support. A web-based solution provided researchers with an easy-to-access tool that was compliant with the U.S. Health Insurance Portability and Accountability Act of 1996 (HIPAA) security requirements [49]. Research teams appreciated the fact that using the REDCap system created an opportunity to “own” the data management plan, speed development of study-specific data collection interfaces, and greatly reduce budget requirements for data management. During this phase, the software’s defining features included a metadata-driven process for developing data capture case report forms, a basic data export module, user rights functionality, and extensive data and operations logging necessary for HIPAA compliance. The REDCap system data architecture was originally flat, using a set of five replicable database tables for each project as previously described (for scalability, we later moved to an entity-attribute-value data model utilizing global tables in a single MySQL database) [26]. Local support at the time included only the primary faculty lead who served as part-time developer and part-time end-user trainer. Funding support for initial development came from institutional sources as well as the NIH supported General Clinical Research Center (GCRC)[50].
Challenges:
Building novel informatics tools required trust and investment from the local research community. Early stages of software development included extensive work sessions with users and rapid cycle development designed to translate stated concerns and needs into software modifications and improvements. Building an initial base of users at Vanderbilt was laborious and required establishing trust among the research community that the software would be supported over a long period of time. Early adopters were invaluable at helping improve the software through feature suggestions and by championing the REDCap system to peers. In developing the software platform, we focused on an agile delivery method where practical solutions to investigator needs were implemented with rapid turnaround rather than ‘perfect architecture.’ This approach allowed us to address the needs of our institutional investors while obtaining rapid user feedback on feature implementation. We adhered to two strict principles when meeting with research stakeholders: 1) never promise a new feature or timeline, as this unnecessarily overcommits the development team and potentially places researchers at risk if features/timelines cannot be delivered; and 2) always promise to build new features/functions in a way that ensures backward compatibility with all existing projects.
Benefits:
Although the initial development of REDCap required networking and multiple small interactions with users, it led to close ties with a local research community and permitted nimble and receptive software development that focused on accommodating dozens of local projects. This close relationship with the user base enabled us to build a resource well-grounded in the practical needs of clinical research teams and began a culture of collaboration that is the foundation of the existing REDCap community. Building “institutional buy-in” was relatively easy once the model was proven and researchers began to voluntarily adopt REDCap. Migrating to a centralized, HIPAA-compliant data collection system with comprehensive audit trails and secure user controls was recognized as a way to protect researchers, volunteers, and the institution. Empowering local research teams to create and manage their own REDCap projects reduced per-project cost and ultimately allowed Vanderbilt to provide REDCap at no cost to any student, staff, or faculty member for any research project.
Phase 2: Recruiting Software Collaborators (3 sites: ~ 2005–2006)
By 2005, REDCap adoption had accelerated at Vanderbilt and we learned that other institutions faced the same challenge supporting their local research communities with centralized, HIPAA-compliant solutions for data planning, collection, and management. We were approached by another university (University of Puerto Rico) and offered to share REDCap on the condition they would contribute 5% of a software developer’s time in exchange for receiving the REDCap code and consultations with the Vanderbilt faculty lead. The University of Puerto Rico countered by volunteering 30% effort of a programmer to express their full support of the collaborative venture. After 12 months, the sharing and remote collaboration model had worked so well that a second site (Medical College of Wisconsin) joined under the same reciprocal programming agreement. The Vanderbilt faculty lead and collaborating software developers communicated via weekly Skype calls and transported code by e-mail (e.g., zip files) with little regard for formal versioning, relying instead on active communication and straightforward date-based or as-needed code versioning. Project management included defining and testing program components built at Vanderbilt and at the two partner institutions. Extracting the codebase for installation at partner institutions required programming effort at Vanderbilt but did not require extensive documentation given the frequency of communication among developers and a similar technical infrastructure in each destination environment. Local context access control was abstracted and enabled using lightweight directory access protocol (LDAP) system methods.
Challenges:
Extracting REDCap code for use at other institutions required time and effort. Existing code had to be modified to support alternate site-specific configurations. Vanderbilt resources were limited and the effort of supporting partner sites resulted in an overall slowdown of feature programming as well as local end-user training and promotion. Maintaining high expectations for feature development by Vanderbilt research teams while growing REDCap’s early consortium of adopters forced us to define how to prioritize and balance local and outside interests.
Benefits:
Expanding our programming team (remotely) provided invaluable feedback and ideas for solving complex programming tasks. In addition to programming expertise, we gained a biostatistician who provided guidance on refining our data export module (i.e., enabling REDCap to export data into common statistical packages). Implementing the software at three sites allowed for needs-based assessment and abstraction of user authentication methods, which despite implementation challenges led to a more flexible resource. Installations and collaborators at multiple institutions provided all participating investigators (including Vanderbilt research teams) with confidence of REDCap’s future longevity, a critical consideration given multi-year grant commitments and need for stability in research projects.
Phase 3: Creating a Small Consortium of Partners (4–10 sites; ~ 2006–2008)
By mid-2006, the three-site REDCap collaborative established in Phase 2 had proven so productive that we decided to establish a true consortium by expanding to ten sites. The newly created CTSA institutions offered opportunities for additional local financial support and expectations for collaboration within the national research community. The REDCap faculty lead at Vanderbilt presented the REDCap model at national meetings and personally contacted 10 colleagues from GCRC/CTSA networks to adopt and join the program. New “partner” institutions would receive the REDCap software and support at no cost, but with the stipulation that they would provide an in-kind contribution of 5% personnel effort (not necessarily a programmer) to support the program and consortium. A simple “Academic End-User License Agreement” was drafted to recognize Vanderbilt’s legal ownership of the software.
The Vanderbilt team grew to include a full-time programmer and a part-time consortium support staff member (25%) in addition to the REDCap faculty lead, who began to focus on higher-level management of the project. The team hosted a weekly webinar via Skype to check the status of implementation across all partner sites, organize site-to-site help and support where appropriate, and discuss interesting use cases, end-user training, researcher marketing methods, challenges, and new feature requests. Partners provided weekly reports of site evaluation metrics (e.g., number of projects, research end users) and meeting summary reports were distributed via e-mail immediately after the weekly calls. When applicable, developers exchanged code through direct file transfers (e.g., e-mailed zip files) and maintained rudimentary versioning via weekly (or as-needed) REDCap software releases.
Challenges:
Additional partners meant more time spent managing groups and less time working on needs specific to Vanderbilt research teams. Developing formal installation documentation placed a high burden on the Vanderbilt REDCap group, as we had to design a generalizable resource while responding to many customized support requests from adopting institutions. Additional partners also brought a diversity of opinions on the software’s trajectory. Adopting and adhering to larger program goals for REDCap required diplomacy in the face of individual project interests but was necessary for long-term success. Organizing remote volunteer work required extensive managerial oversight and led to occasional delays in program deliverables. We had to set expectations early regarding the level of support that Vanderbilt was able to provide: a “zero-fee” REDCap Consortium membership meant we could only provide support to our partners’ informatics teams, not their end users. Partner sites had to accept responsibility for training and supporting their own REDCap end users. Critical software enhancements, including an overhaul of the data architecture, streamlined workflow, and refinement of user experience, led to a more stable and robust platform, but also slowed down our work enabling new features for specific use cases.
Benefits:
The culture shift of thinking and acting like a consortium (as opposed to single groups acting on behalf of their own sites) began during this phase. A sense of community developed within the individuals participating in the weekly all-hands teleconferences. By collectively sharing and discussing use cases (e.g., local projects using or considering use of REDCap) on the weekly teleconferences, all Consortium REDCap administrators learned together how to optimally configure the platform to support diverse research studies. Although other electronic data capture systems existed (e.g., local homegrown systems, large commercial systems like Medidata Rave and Qualtrics), the fact that REDCap was easily configurable for diverse research study requirements and had a rapidly expanding group of distributed experts across research-centric academic medical centers increased the platform’s reputation and led to greater uptake and adoption by new Consortium partners. The up-front expectation for in-kind contribution from adopting sites allowed the Vanderbilt team to request and expect help from external experts. In turn, sites that actively contributed to REDCap development felt a positive sense of ownership towards the project. Remote collaboration with partners across the country was intellectually stimulating and provided an expanded expertise base. Additionally, having more partner sites enhanced REDCap’s reputation and credibility, while more end users provided invaluable feedback that ultimately enabled feature prioritization.
Phase 4: Creating a Broader National Presence (~ 50 sites; ~ 2008–2009)
In 2008, having found success with our ten-site model, we initiated a wider call for participation by targeting similar academic institutions at national meetings and other venues. The CTSA community was growing and presented a pool of potential new partners as many institutions were grappling with “last mile” clinical research informatics support at the enterprise level. Organizations that adopted REDCap during this Phase were typically U.S.-based academic medical centers, though we also began working with a few highly motivated international partners from Japan, Singapore, and Canada. The REDCap application grew to include more features for end users and administrators and development of an online case report form editor and data dictionary upload mechanism, further simplifying creation of projects.
As the Consortium grew, we dropped the request for 5% effort contributions requirement from participating sites due to management complexity and instead expanded the on-site Vanderbilt team to include a second programmer. However, partner site experts enjoyed the opportunity to collaborate with likeminded informatics specialists across the country, thereby maintaining the original community spirit. To encourage continued Consortium activity, we declared that actively contributing site representatives would have the ‘loudest voice’ in new feature prioritization. Consortium growth increased participation in the weekly all-hands calls, prompting purchase of licensed webinar software (GoToMeeting), and Vanderbilt set up an official REDCap administrators’ email listserv for asynchronous e-mail communication. Formal code versioning was enabled, and we began using a hosted intranet solution (TRAC) for code distribution, wiki communication, and bug tracking/reporting. The first REDCap Consortium Committee was formed to review technical gaps associated with setting up REDCap in a validated environment, with REDCap technical developers subsequently using the analysis to prioritize related feature development.
As the consortium grew and additional institutional resources were committed to the program, we were encouraged by institutional leaders to further solidify and formalize inter-institutional license agreements. Vanderbilt’s technology transfer office assisted in creating a more sophisticated end-user license agreement (EULA) for use in on-boarding new sites. Eligible organizations could join the REDCap Consortium (and download the REDCap source code) by agreeing to the terms with Vanderbilt University. The paper EULA documents required signature by institutional officials on both sides, which introduced a two-to-three-week administrative turnaround time for joining the Consortium. A public website was created to let new potential partner sites know about the platform and consortium model (https://projectredcap.org).
Challenges:
The no-cost licensing agreement required vetting by approximately 50 sites before finalization, but ultimately became the single accepted End User Licensing Agreement (EULA) for REDCap. Vanderbilt declined any individual REDCap EULA negotiations, as the maintenance of a growing number of divergent agreements would place an excessive burden on Vanderbilt’s technology transfer office. Increasing the number of Consortium partners also increased the amount of support required to run the Consortium, hampering the effectiveness of the small Vanderbilt REDCap team. Although the Vanderbilt team could provide only a small amount of Consortium support, appeals to our Consortium members to help answer questions via the listserv without prompting helped cultivate a distributed support mechanism where Consortium partners supported each other. Requests for non-technical support from interested potential partners became overbearing (e.g., “we are considering REDCap, but our IT team needs you to complete a 20-page vendor questionnaire”), and we eventually had to create a policy denying most requests due to time constraints. More installations of the REDCap software at top-tier medical centers brought additional security scrutiny, forcing our technical team to develop standard operating procedures with rapid turnaround times for assessing, patching, and distributing versioned code to partner sites whenever a security vulnerability was discovered and reported.
Benefits:
The uptake and use of REDCap at many institutions provided great scientific and collaborative return on investment. During 2008 and 2009, our earliest phase of formally tracking return on investment for REDCap support at Vanderbilt, our local research teams cited REDCap’s value in securing 23 new funded/awarded projects across numerous biomedical research areas (e.g., clinical pharmacology, vaccine and treatment evaluation, epilepsy monitoring, mental health, emergency department heart failure risk stratification, genetic markers and predictive risk modeling, nutrition, pediatric obesity, asthma, cystic fibrosis). We were also encouraged by similar success metrics and diverse use cases reported in ad hoc fashion by other Consortium REDCap administrators. By using REDCap, many research users across the U.S. were able to work on a local data management platform also used locally by peers at other institutions, which promoted resource sharing while reducing training costs. Institutional Review Boards (IRBs) and Privacy Offices became aware of the platform and were supportive of our goal to provide research teams with “an easy way to do the right thing” regarding data collection and management. Research users at local institutions (including Vanderbilt) were increasingly confident adopting the platform for individual studies given widespread national use and recommendations from local compliance offices.
Phase 5: Building a Diverse Community (~ 500 sites; ~ 2009–2012)
Interest in REDCap spiked once approximately 50 partners had joined the Consortium and membership trended upwards in 2009. We published a descriptive paper about the REDCap platform to support research teams needing to cite data management capacity in grants and methods sections for research publications[26]. We solidified our membership processes during the early stages of Phase 5, enabling our existing REDCap support team to manage the growth. A rapidly growing user base drove functionality requests and use cases to unprecedented rates. The REDCap development team at Vanderbilt designed new software modules to facilitate clinical trials (e.g., participant randomization, data query workflow) and support direct data capture from patients via user-facing surveys. We continued support via consortium listserv communication and added a weekly programmer-led “Technical Assistance Call” to assist with complex technical help requests. We created a “shared instrument library” where REDCap users at any institution could download and use REDCap-reviewed versions of validated data collection instruments [51]. We built features to support localization for those in non-U.S. environments (e.g., local data validation types and language abstraction to allow partners to translate REDCap into non-English languages). Data collection and software configuration resources were maintained by two new consortium-driven workgroups, the REDCap Library Oversight Committee and the Field Validation Group. We also built a “plug-in” development framework that allowed local developers to add custom REDCap functionality for individual projects. An initial set of restful API methods were developed and disseminated to enable back-end, record level data transactions for local teams desiring to build interoperability between REDCap and other systems. Initial API functionality included basic data import and export features, but later evolved to include more sophisticated methods (e.g., project creation, project metadata editing, project users and user rights management).
The first REDCap administrators’ conference was held at Vanderbilt University in 2009 to enable face-to-face interaction among Consortium partners, and Consortium sites volunteered to host the meeting in subsequent years. We established ‘friendly’ Consortium competitions (e.g., a traveling trophy for highly productive partners; meeting-based competitions for best REDCap use cases or plug-in modules) to promote comradery, sharing, and a sense of contribution among new and seasoned Consortium members. As the consortium grew, the focus and format of weekly all-hands meetings changed to remain relevant to a broader audience. We incorporated fewer open-microphone discussions and focused more on “what’s new and what’s next” software demonstrations.
During the course of growing the Consortium, we occasionally received requests from research groups at institutions that lacked local capacity to support REDCap installation and provide ongoing support for the technical infrastructure. In order to respond to this need, we established the Vanderbilt Data Coordinating Center Core (VDCC). The Core model enabled us to provide hosting services to users outside of Vanderbilt whose needs went beyond standard REDCap infrastructure, allowed diversification of our financial support model, and introduced the REDCap platform to more diverse users (including industry based research teams). Hosting fees from the Core model allowed us to expand our REDCap team to include a dedicated community-facing support person for training and Consortium support.
Challenges:
Growing numbers of stakeholders and users limited our ability to be as responsive to individual project needs. We attempted to mitigate this by creating small topic-specific consortium working groups. More partners led to even greater demand for just-in-time information requests from REDCap administrators supporting local installations and user training. To maintain our no-cost distribution and support model, we asked consortium partners to be even more proactive sharing local communication strategies and training materials with one another. The increasing visibility of REDCap led our Vanderbilt Center for Technology Transfer and Commercialization to question our ‘no cost’ consortium model, ultimately resulting in hiring of a consulting firm to assess various models of commercialization. Internal discussions and business use case evaluations required considerable time commitment from the Vanderbilt team, but we ultimately settled on agreement with the technology transfer office that we would continue to support and grow the consortium with no-cost software distribution and limited support for academic, non-profit, and government institutions while formulating a commercial-based hosting service for industry sponsored trials.
Benefits:
Increased growth and visibility of the REDCap Consortium stimulated additional interest in the software platform among non-profit institutions, government stakeholders, and international research centers. By the end of 2012, we were supporting more than 500 Consortium partners. As always, new partners brought greater numbers of end users, and more users brought additional use cases which ultimately helped us further evolve the program. Enhanced API functionality prompted research informatics teams to begin building interoperable systems leveraging “REDCap as a Platform.” Numbers of publications referencing REDCap grew each year, providing an important metric for evaluating scientific impact (Figure 3). The addition of a yearly in-person REDCap Conference enabled the REDCap Consortium to become a professional home for many applied informatics experts.
Phase 6: Expanding Globally (> 500 sites; ~ 2013–2018)
In recent years, we have seen exponential growth in the number of U.S. and international REDCap partners. In 2018 alone, we added 575 new REDCap partners with approximately 2.75 international members for each U.S. request. More than 20% of REDCap partners are now located in countries designated as low-and middle-income by the World Bank. New telephone and SMS-based survey features, “offline” functionality via the REDCap mobile application for coordinators, and a separate mobile application (MyCap, https://www.projectmycap.org) for participants have enabled research data collection in settings unsuited to traditional web-based applications. International partnership has influenced the addition of new REDCap features including enhanced validation for country-specific data types, language abstraction enabling REDCap rendering in non-English languages, data transfer methods optimized for settings with poor network connectivity, and features in support of non-U.S. regulatory requirements (e.g., the European Union’s General Data Protection Regulation). Recent work with major cloud vendors on deployment scripts and automated REDCap upgrade methods have created an opportunity for partners to set up REDCap installations in the cloud, thereby eliminating the need for on-site hardware and reducing local systems administrator support. REDCap’s back-end architecture, all of which runs on open source components, is very flexible and does not force strict requirements for general implementation. REDCap can operate “onpremise” (on the local institution’s hardware servers or virtualized servers) or on various cloud-based infrastructures (e.g., AWS, Azure, Google Cloud). For virtualized setups, REDCap may run as a traditional virtualized web server (e.g., VMware, VirtualBox) or as a containerized service (e.g., Docker, Kubernetes). As new server technologies have developed over the past decade, we have observed a gradual increase among REDCap partner institutions adopting virtualized environments running REDCap, and in more recent years, containerized solutions for supporting the software platform. Minor revisions to the REDCap EULA have clarified licensing for non-U.S. partners and cloud-based installations.
The number of REDCap end users has grown proportionately, creating additional demand for generalized REDCap training. We have streamlined end-user training opportunities by increasing help text and embedded videos within the software package and by creating a formal 5-week Massive Open Online Course (MOOC) on data management using REDCap as a training framework. Consortium members continue to provide cross-institutional support for one another through sharing resources (e.g., end-user local training material repository), participating in formal presentations (e.g., monthly REDCapU sessions design to support REDCap domain knowledge and career growth of local informatics champions), and contribution to Consortium Committees. New Committees have formed around shareable training materials, software plug-in development, and electronic health record systems integration. Funding support for REDCap has become increasingly diverse, including support from institutional sources, the CTSA, the Vanderbilt Data Core model, and other NIH-funded grants. We also expanded the Data Core model beyond basic REDCap hosting to offer support for REDCap custom programming. To accommodate the continued growth of the Consortium, the Vanderbilt REDCap team has expanded its staffing to five full-time developers and two dedicated community support people, with the addition of other associated faculty and custom add-on developers from the Data Core.
Challenges:
Given the Consortium growth rate, the Vanderbilt team could no longer maintain a close connection to each new partner site and groups that were not proactive in their use of the Consortium mailing list were less likely to integrate with the existing community. We upgraded from a mailing list to a web-based, question-and-answer-style support forum software to provide the community with improved collaborative workspaces with searchable support archives. The need for additional communication mechanisms (e.g., persistent forum software vs. simple listservs, dedicated monthly consortium calls for Europe/Africa and Asia/Oceania time zones, expanded educational content for annual REDCap conference) increased need for administrative support. For-profit software vendors also began to regard the REDCap community as a potential market, attempting to sell alternate software solutions.
Benefits:
As the Consortium has grown, so has the sophistication and influence of our Consortium partners. Experts in informatics, biostatistics, data warehousing, clinical systems, trial operations, standards and ontologies, security methods, computer science, global health, mobile systems, library science, and other domains participate in the consortium and consult with the development team as needed. Our increasing international partner base in resource-limited areas has prompted our teams to build a REDCap mobile app to allow offline data collection when internet connectivity is not present. The growth of the REDCap Consortium and user base has created opportunities to collaborate with other consortia (e.g., PhenX [52,53], Medical Data Models [54], BioPortal [55], PROMIS [56]) to integrate standardized terminologies and instruments. Portability of project metadata across REDCap installations has allowed new types of sharing and collaboration. During the Ebola outbreak in 2014, we saw a rapid mobilization of REDCap partner sites sharing methods for surveillance data collection in both academia and state governments, setting the stage for additional attention and adoption across U.S. state health departments, the U.S. Department of Veterans Affairs and other government agencies. A large partnership has made it possible to gain attention and support to build integration and interoperability with other software platforms and cloud-based ecosystems [57]. Regional relevance (e.g., having many REDCap partners in a single country or geographic region) has prompted several of our international partners to organize and self-fund regional annual or biannual regional REDCap conferences [58,59].
Discussion
The REDCap model has been successful to date, growing from a single site to a global consortium over the course of approximately 13 years. The iterative software design philosophy and incremental consortium growth fostered a collaborative spirit during REDCap’s early days, which has been sustained through the active participation of thousands of REDCap administrators in REDCap calls, mailing lists, working groups, and conferences. Software feature requests and guidance from active research teams have enabled the platform to grow from a rudimentary feature set (data capture, data export, logging) to a rich set of modules capable of supporting diverse research operations on almost any scale. More than six thousand journal articles have cited use of the software platform in conducting studies during the period between 2004 and 2018 [60]. While bibliometric capture and analysis methods (Figure 3) provide evidence of value for REDCap in diverse scientific research domains, the platform is used at Vanderbilt and across REDCap Consortium partner institutions for many other use cases (Figure 4), including operational support, quality improvement projects, and even as a sub-component for larger systems and workflow architecture. For example, Vanderbilt’s IRB system uses REDCap as a central component for protocol submissions and our research data warehousing teams have built system exchange mechanisms allowing REDCap to serve as a final data delivery mechanism.
Figure 4:
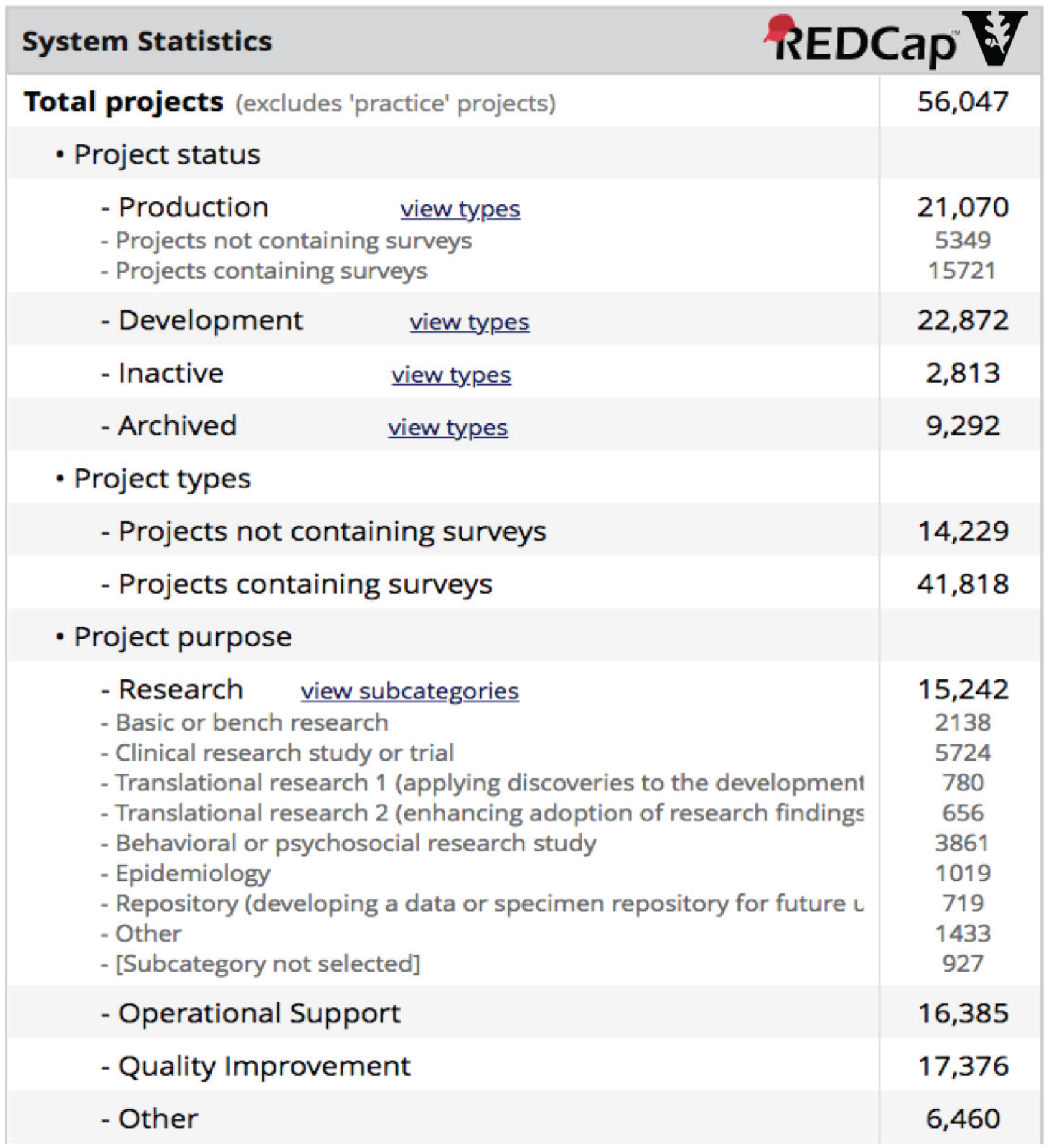
System activity and metrics reporting screen from Vanderbilt’s primary REDCap installation. Notably, research project numbers show the platform’s utility for basic research use cases and also the full spectrum of clinical and translational research. Operational and Quality Improvement project numbers also highlight the diversity of use at Vanderbilt.
The REDCap platform and Consortium represent only a single model in terms of software development and dissemination. Alternate software distribution models exist (e.g., i2b2 [61], OpenMRS [62], and Vivo [63]) and software projects may find such models instructive as well. Nevertheless, we believe lessons learned from the REDCap program over the past 13 years can serve others interested in creating and disseminating informatics software through a consortium approach. We present below lessons learned in key areas of software development, dissemination, and evolutionary growth:
Initial Platform Development
Make a difference locally first. Start with a good idea solving a known important problem, based on well-researched needs of a community of targeted users. The “build small, evaluate, and evolve” development approach works well in environments where needs and priorities change over time.
Build ‘measure as you go’ audit trails and evaluation dashboards into the software platform, enabling rapid objective assessment of impact for new and existing functional modules. Allow evaluation to guide evolution and do not be afraid to “fail early, fail small” in initial stages of development.
Avoid obsessing over technical back-end architecture and methods, striving instead for a “good enough” approach that recognizes users’ priorities and can evolve over time as needed.
Never leave users behind when developing new functions and features – backward compatibility is essential for evolving platforms.
Building External Collaborations
Choose initial partners carefully based on level of perceived commitment and ability to add value through contributed resources, expertise, or increase of the platform user base. Be prepared and willing to sacrifice local development timelines and priorities to support the growth of the consortium, and to evolve the consortium support model over time.
Build trust by under-promising and over-delivering. Never make a hard commitment regarding new functionality or timelines before release.
Avoid obsessing about studying, selecting, and perfecting consortium communication methods (tasks that can be particularly tempting for technical teams). Adopt simple tools and methods, then devote all energy possible to collaborate on the intended platform development.
As mistakes are made or unanticipated problems are encountered (e.g., a new software bug or found security vulnerability), own them quickly and make all attempts to remedy with consortium partners. This approach increases trust in the software platform and the consortium itself.
Evolving the Platform
Define and adhere to a program development philosophy.
Define a workable feature prioritization model and be honest with consortium partners about the fact that there will always be more great ideas than time and resources to implement great ideas. Be prepared for criticism and help the development team understand necessity of staying focused and saying ‘no’ or ‘not yet’ to feature requests.
Create flexibility when disseminating new functional modules – do not assume all teams will operate like your team or that they will wish to implement all modules.
Be opportunistic. Never turn down an opportunity to promote tools or services. Evolving platforms benefit greatly from ongoing promotion to inform and reengage users.
While the REDCap software development and consortium growth model have been successful, core components of our model may not be suited to all groups. Foremost, although REDCap is available at no cost, it is not open source software. Eligible partners must be non-profit or government institutions (not individuals), comply with U.S. export regulations, and agree to the EULA terms, which include not sharing the codebase with third parties and not offering REDCap “reseller” services. The EULA restrictions had several motivating factors. Many institutional leaders showed greater willingness to adopt software with a closed codebase, given the sensitive nature of the data stored on REDCap platforms (such as individually identifiable health information) and the perceived security risk of open source code. Given the extensive Vanderbilt investment required for code generalization and sharing, we also wanted to ensure early Consortium partners were committed to be active participants rather than “software window shoppers.” Finally, this licensing model ensured REDCap’s sustainability as a software platform. By limiting Consortium membership to non-profit and government organizations, Vanderbilt reserved the right to market REDCap in the commercial space and also to perform fee-based hosting for research teams in organizations not able or willing to join the REDCap consortium. This provided a stable source of software development funding that was independent from grant-driven requirements.
The Vanderbilt team also maintains control of the REDCap software development trajectory, although we frequently seek input on features and prioritization from the Consortium. As an academic medical center and main consumer of REDCap, we must remain responsive to national and international research priorities to support our own research community. The mixed funding model we use to support REDCap also requires that we prioritize development in areas consistent with the larger funding initiatives (e.g., PCORI grant initiatives led to many features and functions necessary for participant engagement and patient reported data). While there is a limit to core software development and support, we have created platform features (e.g., restful API services, sustained plug-in architecture) that enable any REDCap Consortium partner to build and share ‘add-on’ features that might be necessary to support individual study needs.
Conclusion
The REDCap Consortium has proven a successful model for software development and dissemination. Key features of the model have been early strategic partnering followed by managed growth and support of our community in stages. Although the REDCap Consortium sharing model is far from a definitive or authoritative example, we believe our observations and experiences will serve others wishing to build and disseminate software platforms.
Highlights:
The Research Electronic Data Capture (REDCap) data platform launched in 2004
The REDCap Consortium is the community of REDCap administrators and organizations
The REDCap Consortium grew from local to international impact in six phases
REDCap partners include 3207 organizations in 128 countries as of December 2018
Our consortium-building lessons should help the research informatics community
Acknowledgments
The REDCap platform and consortium is supported by NIH/NCATS UL1 TR002243. We also gratefully acknowledge Mr. Chad Lightner for figure and graphical design support, Ms. Nan Kennedy for editing and reference support, Mr. Alan Bentley and Mr. Peter Rousos for REDCap licensing support, and Mr. Adam Lewis for publication and citation support. Finally, we acknowledge and express gratitude to all Administrators from the REDCap consortium for sharing innovative ideas and participating in activities that contribute to the empowerment of our REDCap development team and research teams around the world.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Embi PJ, Clinical research informatics: survey of recent advances and trends in a maturing field, Yearb. Med. Inform 8 (2013) 178–184. [PubMed] [Google Scholar]
- [2].Embi PJ, Payne PRO, Advancing methodologies in Clinical Research Informatics (CRI): Foundational work for a maturing field, J. Biomed. Inform 52 (2014) 1–3. doi: 10.1016/j.jbi.2014.10.007. [DOI] [PubMed] [Google Scholar]
- [3].Kahn MG, Weng C, Clinical research informatics: a conceptual perspective, J. Am. Med. Inform. Assoc 19 (2012) e36–e42. doi: 10.1136/amiajnl-2012-000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pober JS, Neuhauser CS, Pober JM, Obstacles facing translational research in academic medical centers, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 15 (2001) 2303–2313. doi: 10.1096/fj.01-0540lsf. [DOI] [PubMed] [Google Scholar]
- [5].Sung NS, Crowley WF, Genel M, Salber P, Sandy L, Sherwood LM, Johnson SB, Catanese V, Tilson H, Getz K, Larson EL, Scheinberg D, Reece EA, Slavkin H, Dobs A, Grebb J, Martinez RA, Korn A, Rimoin D, Central challenges facing the national clinical research enterprise, JAMA. 289 (2003) 1278–1287. [DOI] [PubMed] [Google Scholar]
- [6].May L, Zatorski C, Vora D, The Role of Technology in Translational Science, World Med. Health Policy 5 (2013) 389–394. doi: 10.1002/wmh3.69. [DOI] [Google Scholar]
- [7].Fahy BG, Balke CW, Umberger GH, Talbert J, Canales DN, Steltenkamp CL, Conigliaro J, Crossing the chasm: information technology to biomedical informatics, J. Investig. Med. Off. Publ. Am. Fed. Clin. Res 59 (2011) 768–779. doi: 10.231/JIM.0b013e31821452bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sheetz MP, Can small institutes address some problems facing biomedical researchers?, Mol. Biol. Cell 25 (2014) 3267–3269. doi: 10.1091/mbc.E14-05-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Etz RS, Hahn KA, Gonzalez MM, Crabtree BF, Stange KC, Practice-Based Innovations: More Relevant and Transportable Than NIH-funded Studies, J. Am. Board Fam. Med 27 (2014) 738–739. doi: 10.3122/jabfm.2014.06.140053. [DOI] [PubMed] [Google Scholar]
- [10].Committee to Review the Clinical and Translational Science Awards Program at the National Center for Advancing Translational Sciences, Board on Health Sciences Policy, Institute of Medicine, The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research, National Academies Press (US), Washington (DC), 2013. http://www.ncbi.nlm.nih.gov/books/NBK144067/ (accessed December 10, 2014). [PubMed] [Google Scholar]
- [11].National Institutes of Health, Institutional Clinical and Translational Science Award RFA-TR-14-009, (2014). http://grants.nih.gov/grants/guide/rfa-files/RFA-TR-14-009.html (accessed December 10, 2014).
- [12].Zerhouni EA, Clinical research at a crossroads: the NIH roadmap, J. Investig. Med. Off. Publ. Am. Fed. Clin. Res 54 (2006) 171–173. [DOI] [PubMed] [Google Scholar]
- [13].Guerrero LR, Nakazono T, Davidson PL, NIH Career Development Awards in Clinical and Translational Science Award Institutions: Distinguishing Characteristics of Top Performing Sites, Clin. Transl. Sci (2014). doi: 10.1111/cts.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS, Launching PCORnet, a national patient-centered clinical research network, J. Am. Med. Inform. Assoc. JAMIA 21 (2014) 578–582. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rose LM, Everts M, Heller C, Burke C, Hafer N, Steele S, Academic Medical Product Development: An Emerging Alliance of Technology Transfer Organizations and the CTSA, Clin. Transl. Sci 7 (2014) 456–464. doi: 10.1111/cts.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Masys DR, Harris PA, Fearn PA, Kohane IS, Designing a Public Square for Research Computing, Sci. Transl. Med 4 (2012) 149fs32. doi: 10.1126/scitranslmed.3004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].CD2H, CD2H. (n.d.). https://ctsa.ncats.nih.gov/cd2h/ (accessed December 17, 2018).
- [18].CTSA Program Collaborative Innovation Awards, CTSA Program Collab. Innov. Awards (n.d.). https://ncats.nih.gov/ctsa/projects/ccia (accessed December 17, 2018).
- [19].Reis SE, Berglund L, Bernard GR, Califf RM, Fitzgerald GA, Johnson PC, Reengineering the national clinical and translational research enterprise: the strategic plan of the National Clinical and Translational Science Awards Consortium, Acad. Med. J. Assoc. Am. Med. Coll 85 (2010) 463–469. doi: 10.1097/ACM.0b013e3181ccc877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ravichandran T, Organizational assimilation of complex technologies: an empirical study of component-based software development, IEEE Trans. Eng. Manag 52 (2005) 249–268. doi: 10.1109/TEM.2005.844925. [DOI] [Google Scholar]
- [21].Noll J, Beecham S, Richardson I, Global Software Development and Collaboration: Barriers and Solutions, ACM Inroads. 1 (2011) 66–78. doi: 10.1145/1835428.1835445. [DOI] [Google Scholar]
- [22].Sherif K, Vinze A, Barriers to adoption of software reuse: A qualitative study, Inf. Manage 41 (2003) 159–175. doi: 10.1016/S0378-7206(03)00045-4. [DOI] [Google Scholar]
- [23].Sherif K, Vinze A, A Qualitative Model for Barriers to Software Reuse Adoption, in: Proc. 20th Int. Conf. Inf. Syst., Association for Information Systems, Atlanta, GA, USA, 1999: pp. 47–64. http://dl.acm.org/citation.cfm?id=352925.352930 (accessed November 16, 2015). [Google Scholar]
- [24].Nadan CH, Software Licensing in the 21st Century: Are Software Licenses Really Sales, and How Will the Software Industry Respond, AIPLA Q. J 32 (2004) 555. [Google Scholar]
- [25].Goldman R, Gabriel RP, Innovation Happens Elsewhere: Open Source as Business Strategy, Morgan Kaufmann, 2005. [Google Scholar]
- [26].Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG, Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support, J. Biomed. Inform 42 (2009) 377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Petraitiene R, Petraitis V, Hope WW, Mickiene D, Kelaher AM, Murray HA, Mya-San C, Hughes JE, Cotton MP, Bacher J, Walsh TJ, Cerebrospinal Fluid and Plasma (1→3)-β-d-Glucan as Surrogate Markers for Detection and Monitoring of Therapeutic Response in Experimental Hematogenous Candida Meningoencephalitis, Antimicrob. Agents Chemother. 52 (2008) 4121–4129. doi: 10.1128/AAC.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lense MD, Shivers CM, Dykens EM, (A)musicality in Williams syndrome: examining relationships among auditory perception, musical skill, and emotional responsiveness to music, Front. Psychol 4 (2013) 525. doi: 10.3389/fpsyg.2013.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mason M, Light J, Campbell L, Keyser-Marcus L, Crewe S, Way T, Saunders H, King L, Zaharakis NM, McHenry C, Peer Network Counseling with Urban Adolescents: A Randomized Controlled Trial with Moderate Substance Users, J. Subst. Abuse Treat 58 (2015) 16–24. doi: 10.1016/j.jsat.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Slusher TM, Olusanya BO, Vreman HJ, Brearley AM, Vaucher YE, Lund TC, Wong RJ, Emokpae AA, Stevenson DK, A Randomized Trial of Phototherapy with Filtered Sunlight in African Neonates, N. Engl. J. Med 373 (2015) 1115–1124. doi: 10.1056/NEJMoa1501074. [DOI] [PubMed] [Google Scholar]
- [31].Pang X, Kozlowski N, Wu S, Jiang M, Huang Y, Mao P, Liu X, He W, Huang C, Li Y, Zhang H, Construction and management of ARDS/sepsis registry with REDCap, J. Thorac. Dis 6 (2014) 1293–1299. doi: 10.3978/j.issn.2072-1439.2014.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].da Silva KR, Costa R, Crevelari ES, Lacerda MS, de Moraes Albertini CM, Filho MM, Santana JE, Vissoci JRN, Pietrobon R, Barros JV, Glocal clinical registries: pacemaker registry design and implementation for global and local integration--methodology and case study, PloS One. 8 (2013) e71090. doi: 10.1371/journal.pone.0071090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Backus LI, Gavrilov S, Loomis TP, Halloran JP, Phillips BR, Belperio PS, Mole LA, Clinical Case Registries: simultaneous local and national disease registries for population quality management, J. Am. Med. Inform. Assoc. JAMIA 16 (2009) 775–783. doi: 10.1197/jamia.M3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bellgard M, Beroud C, Parkinson K, Harris T, Ayme S, Baynam G, Weeramanthri T, Dawkins H, Hunter A, Dispelling myths about rare disease registry system development, Source Code Biol. Med 8 (2013) 21. doi: 10.1186/1751-0473-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Randolph AG, Vaughn F, Sullivan R, Rubinson L, Thompson BT, Yoon G, Smoot E, Rice TW, Loftis LL, Helfaer M, Doctor A, Paden M, Flori H, Babbitt C, Graciano AL, Gedeit R, Sanders RC, Giuliano JS, Zimmerman J, Uyeki TM, Pediatric Acute Lung Injury and Sepsis Investigator’s Network and the National Heart, Lung, and Blood Institute ARDS Clinical Trials Network, Critically ill children during the 2009–2010 influenza pandemic in the United States, Pediatrics. 128 (2011) e1450–1458. doi: 10.1542/peds.2011-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hastings-Tolsma M, Emeis C, McFarlin B, Schmiege S., 2012. Task Analysis: A Report of Midwifery Practice, American Midwifery Certification Board, n.d. [Google Scholar]
- [37].Sewell JL, Day LW, Tuot DS, Alvarez R, Yu A, Chen AH, A brief, low-cost intervention improves the quality of ambulatory gastroenterology consultation notes, Am. J. Med 126 (2013) 732–738. doi: 10.1016/j.amjmed.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McCoy AB, Waitman LR, Lewis JB, Wright JA, Choma DP, Miller RA, Peterson JF, A framework for evaluating the appropriateness of clinical decision support alerts and responses, J. Am. Med. Inform. Assoc. JAMIA 19 (2012) 346–352. doi: 10.1136/amiajnl-2011-000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gates JD, Arabian S, Biddinger P, Blansfield J, Burke P, Chung S, Fischer J, Friedman F, Gervasini A, Goralnick E, Gupta A, Larentzakis A, McMahon M, Mella J, Michaud Y, Mooney D, Rabinovici R, Sweet D, Ulrich A, Velmahos G, Weber C, Yaffe MB, The initial response to the Boston marathon bombing: lessons learned to prepare for the next disaster, Ann. Surg 260 (2014) 960–966. doi: 10.1097/SLA.0000000000000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Archer KR, Coronado RA, Haug CM, Vanston SW, Devin CJ, Fonnesbeck CJ, Aaronson OS, Cheng JS, Skolasky RL, Riley LH, Wegener ST, A comparative effectiveness trial of postoperative management for lumbar spine surgery: changing behavior through physical therapy (CBPT) study protocol, BMC Musculoskelet. Disord 15 (2014). doi: 10.1186/1471-2474-15-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Collins SP, Lindsell CJ, Yealy DM, Maron DJ, Naftilan AJ, McPherson JA, Storrow AB, A Comparison of Criterion Standard Methods to Diagnose Acute Heart Failure, Congest. Heart Fail. Greenwich Conn 18 (2012) 262–271. doi: 10.1111/j.1751-7133.2012.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Choi BKC, Pak AWP, A method for comparing and combining cost-of-illness studies: an example from cardiovascular disease, Chronic Dis. Can 23 (2002) 47–57. [PubMed] [Google Scholar]
- [43].Hartmann AS, Thomas JJ, Greenberg JL, Matheny NL, Wilhelm S, A Comparison of Self-Esteem and Perfectionism in Anorexia Nervosa and Body Dysmorphic Disorder, J. Nerv. Ment. Dis 202 (2014) 883–888. doi: 10.1097/NMD.0000000000000215. [DOI] [PubMed] [Google Scholar]
- [44].Smith SM, Balise RR, Norton C, Chen MM, Flesher AN, Guardino AE, A feasibility study to evaluate breast cancer patients’ knowledge of their diagnosis and treatment, Patient Educ. Couns 89 (2012) 321–329. doi: 10.1016/j.pec.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Becevic M, Boren S, Mutrux R, Shah Z, Banerjee S, User Satisfaction With Telehealth: Study of Patients, Providers, and Coordinators, Health Care Manag. 34 (2015) 337–349. doi: 10.1097/HCM.0000000000000081. [DOI] [PubMed] [Google Scholar]
- [46].Anand V, Spalding SJ, Leveraging Electronic Tablets and a Readily Available Data Capture Platform to Assess Chronic Pain in Children: The PROBE system, Stud. Health Technol. Inform 216 (2015) 554–558. [PubMed] [Google Scholar]
- [47].Lyon JA, Garcia-Milian R, Norton HF, Tennant MR, The use of Research Electronic Data Capture (REDCap) software to create a database of librarian-mediated literature searches, Med. Ref. Serv. Q 33 (2014) 241–252. doi: 10.1080/02763869.2014.925379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Klipin M, Mare I, Hazelhurst S, Kramer B, The process of installing REDCap, a web based database supporting biomedical research: the first year, Appl. Clin. Inform 5 (2014) 916–929. doi: 10.4338/ACI-2014-06-CR-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Modifications to the HIPAA Privacy, Security, Enforcement, and Breach Notification Rules Under the Health Information Technology for Economic and Clinical Health Act and the Genetic Information Nondiscrimination Act; Other Modifications to the HIPAA Rules, 2013. [PubMed]
- [50].Nathan DG, Nathan DM, Eulogy for the clinical research center, J. Clin. Invest 126 (2016)2388–2391. doi: 10.1172/JCI88381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Obeid JS, McGraw CA, Minor BL, Conde JG, Pawluk R, Lin M, Wang J, Banks SR, Hemphill SA, Taylor R, Harris PA, Procurement of shared data instruments for Research Electronic Data Capture (REDCap), J. Biomed. Inform 46 (2013) 259–265. doi: 10.1016/j.jbi.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hendershot T, Pan H, Haines J, Harlan WR, Marazita ML, McCarty CA, Ramos EM, Hamilton CM, Using the PhenX Toolkit to Add Standard Measures to a Study, Curr. Protoc. Hum. Genet. Editor. Board Jonathan Haines Al 86 (2015) 1.21.1–1.21.17. doi: 10.1002/0471142905.hg0121s86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McCarty CA, Huggins W, Aiello AE, Bilder RM, Hariri A, Jernigan TL, Newman E, Sanghera DK, Strauman TJ, Zeng Y, Ramos EM, Junkins HA, PhenX RISING network, PhenX RISING: real world implementation and sharing of PhenX measures, BMC Med. Genomics 7 (2014) 16. doi: 10.1186/1755-8794-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Medical Data Models Portal, (n.d.). https://medical-data-models.org/.
- [55].Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA, BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications, Nucleic Acids Res. 39 (2011) W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bevans M, Ross A, Cella D, Patient-Reported Outcomes Measurement Information System (PROMIS®): Efficient, Standardized Tools to Measure Self-Reported Health and Quality of Life, Nurs. Outlook 62 (2014) 339–345. doi: 10.1016/j.outlook.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].This repository contains AWS CloudFormation templates to automatically deploy a REDCap environment that adheres to AWS architectural best practices: vanderbilt-redcap/redcap-aws-cloudformation, Vanderbilt REDCap Group, 2018. https://github.com/vanderbilt-redcap/redcap-aws-cloudformation (accessed December 17, 2018).
- [58].Japan REDCap Consortium - JREC - - Home, (n.d.). https://www.facebook.com/japan.redcap/ (accessed December 17, 2018).
- [59].First Latin American and Brazilian REDCapCon, (n.d.). http://www.redcapbrasil.com.br/redcapcon/en/redcapcon/en/ (accessed December 17, 2018).
- [60].Citations – REDCap, (n.d.). https://projectredcap.org/resources/citations/ (accessed December 17, 2018).
- [61].Natter MD, Quan J, Ortiz DM, Bousvaros A, Ilowite NT, Inman CJ, Marsolo K, McMurry AJ, Sandborg CI, Schanberg LE, Wallace CA, Warren RW, Weber GM, Mandl KD, An i2b2-based, generalizable, open source, self-scaling chronic disease registry, J. Am. Med. Inform. Assoc. JAMIA 20 (2013) 172–179. doi: 10.1136/amiajnl-2012-001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mohammed-Rajput NA, Smith DC, Mamlin B, Biondich P, Doebbeling BN, OpenMRS A Global Medical Records System Collaborative: Factors Influencing Successful Implementation, AMIA. Annu. Symp. Proc 2011 (2011) 960–968. [PMC free article] [PubMed] [Google Scholar]
- [63].Borner K, Ding Y, Conlon M, Corson-Rikert J, VIVO: A Semantic Approach to Scholarly Networking and Discovery, Morgan & Claypool Publishers, 2012. [Google Scholar]


