Abstract
Strand displacement reactions are widely used in DNA nanotechnology as a building block for engineering molecular computers and machines. Here, we demonstrate that strand displacement-based probes can be triggered by RNA expressed in mammalian cells, thus taking a step towards adapting the DNA nanotechnology toolbox to a cellular environment. We systematically compare different probe architectures in order to identify a design that works robustly in living cells. Our optimized strand displacement probe combines chemically modified nucleic acids that enhance stability to degradation by cellular nucleases with structural elements that improve probe retention in the cytoplasm. We visualize probe binding to individual mRNA carrying 96 repeats of a target sequence in the 3’UTR. We find that RNA counts based on live cell imaging using a strand displacement probe are comparable to counts from independent measurement based on fluorescence in situ hybridization experiments. We used probes with scrambled toeholds and scrambled binding domains to demonstrate that target recognition indeed occurs through toehold-mediated strand displacement. Our results demonstrate that strand displacement probes can work reliably in mammalian cells and lay the groundwork for future applications of such probes for live-cell imaging and molecular computing.
Dynamic DNA nanotechnology has been remarkably successful at creating molecular robots1–3, computational circuits4–10 or reconfigurable nanostructures11–15 using only DNA as an engineering material. Underlying this success is the predictability of Watson Crick base pairing, the low cost and wide availability of synthetic DNA and a focus on cell-free settings which allows testing and optimization of engineered devices without interference of evolved biomolecules. Still, it is tempting to ask whether some of the design principles that enabled scaling up the complexity of cell-free dynamic DNA systems could be adapted to the cellular environment.
A vast majority of dynamic DNA devices built to date rely on the same basic molecular primitive, toehold-mediated DNA strand displacement (DSD)16. DSD is a competitive hybridization reaction whereby an incoming single strand of DNA (“input”) displaces an incumbent (“output”) from a complementary binding partner. This reaction is initiated by hybridization of the incoming strand to a complementary single-stranded “toe-hold” and proceeds through a branch migration step, eventually resulting in the release of the output. Multiple strand displacement reactions can be cascaded, enabling the design of multi-step networks. The ubiquity and simplicity of the DSD mechanism make it an obvious candidate for adaptation to a cellular setting.
Recent work by Hemphill and Deiters17 showed that DSD-based logic gates could be delivered to mammalian cells and could be triggered by cellular microRNA. The Mirkin group18 used DSD-based fluorescent reporters attached to nanoparticles to detect and visualize cellular RNA. However, in these studies progress of reactions was measured only at the cellular level rather than at the level of individual molecules. In our own work19, we showed that logic gates containing chemically modified nucleic acids could be delivered to cells and could be triggered by independently delivered synthetic inputs. We further showed that multiple logic gates could be co-localized on an mRNA scaffold, enabling us to visualize gate activation at molecular resolution. Still, the mRNA served only as a scaffold rather than as the input to the reaction. Here, we demonstrate that a strand displacement reaction can be triggered directly by an RNA transcript expressed in mammalian cells. We use a fluorescent nucleic acid strand displacement-based reporter together with live-cell microscopy to visualize reactions at the level of individual mRNA molecules.
For our studies we used HT1080–96X cells, a variant of human fibroblastoma cells (HT1080) which were engineered by the Raj and Tsourkas labs20,21 to express a GFP mRNA carrying 96 tandem repeats of “probe target” sequences (each 50-nucleotides (nt) long) in the 3’-UTR. Colocalization of up to 96 probes on the same transcript can dramatically increase the signal to background ratio. For negative control experiments, we used HT1080- cells expressing GFP mRNA without target sites in the 3’UTR. We validated target expression using fluorescence in situ hybridization (FISH) with probes designed to be complementary to 24 nt within the repeat target sequence. Single transcripts expressed in HT1080–96X cells were detectable as bright fluorescent spots (mean: 58.26 mRNA per cell) while the HT1080-control cells showed primarily background fluorescence (mean 17.3 mRNA per cell; see Fig. S1).
Next, we turned to the design of strand displacement probes for target mRNA detection. Our initial probe design (SDProbe_v0) was closely modeled on “reporter” probes used in cell-free settings. The probe comprised of a longer fluorophore-labeled strand (26 nt long) and a shorter, partially complementary quencher-labeled blocking strand. The sequence of the longer strand was fully complementary to a 26 nt section of the repeat target site in the GFP mRNA 3’-UTR (Fig. S2a). The 26 nt section in the target site was chosen such that it is primarily in single-stranded form (based on computational secondary structure predictions) and can bind to the complementary single-stranded toehold domain of the probe. We varied the length of the blocking strand to create probes with single-stranded toehold and double-stranded stem domains of varying lengths ((t,ds) = 14,12: 12nt stem-14nt toehold, (t,ds) = 10,16: 16nt stem-10nt toehold, (t,ds) = 8,18: 18nt stem-8nt toehold). Probes were synthesized with 2’O-methylated ribonucleotides to make them more resistant to intracellular degradation19,22,23.
Probe binding to the target begins with the (reversible) hybridization of the toehold domain, proceeds through a three-way branch migration and eventually results in the complete displacement of the quencher labeled strand. The displacement of the quencher-labeled strand results in unquenching of the fluorophore and an increase in fluorescence (see Fig. S2a).
In live-cell imaging experiments, the probes with 12 nt and 16 nt double-stranded stems were primarily localized in the nucleus (see Fig. S2b), consistent with earlier reports on the nuclear localization of short synthetic double stranded nucleic acid complexes24 and imaging probes25. Complexes with shorter stem domains are more prone to spontaneously dissociate and, moreover, Exportin 5-mediated nuclear export of short double-helical RNAs requires a double-stranded stem region greater than 14 nucleotides26. The probes with 18 nt double-stranded stem were primarily localized in the cytoplasm rather than the nucleus, but, unfortunately, appeared to be in the form of cytoplasmic aggregates rather than binding specifically to the mRNA targets (Fig. S2b). Such aggregates were observed in prior studies using molecular probes and are likely the result of lysosomal capture of probes delivered by electroporation24,27.
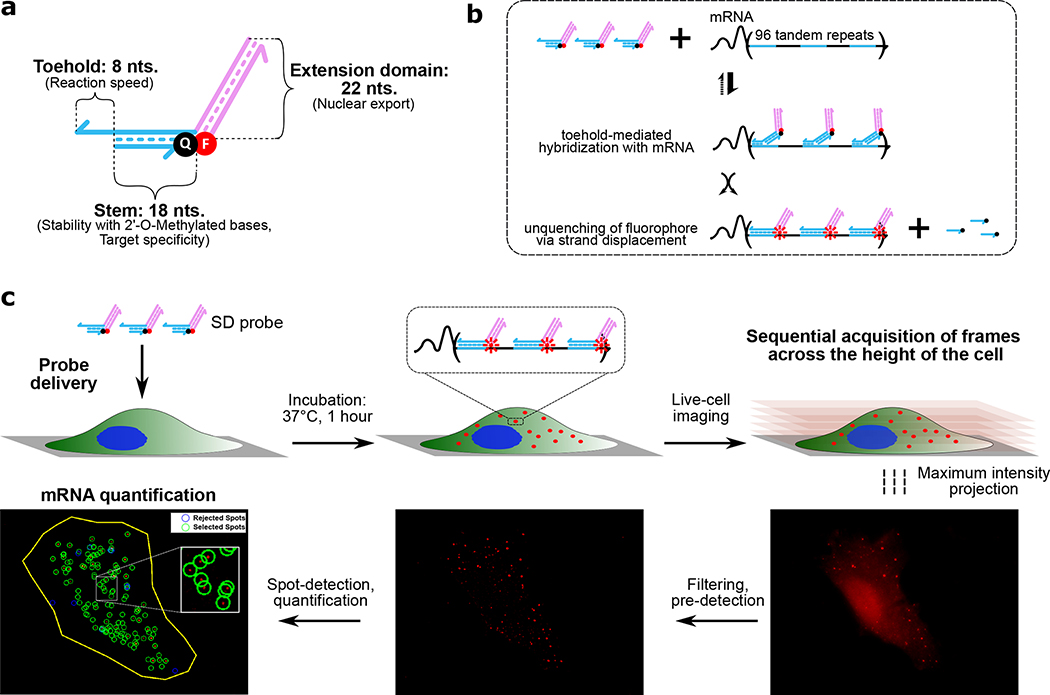
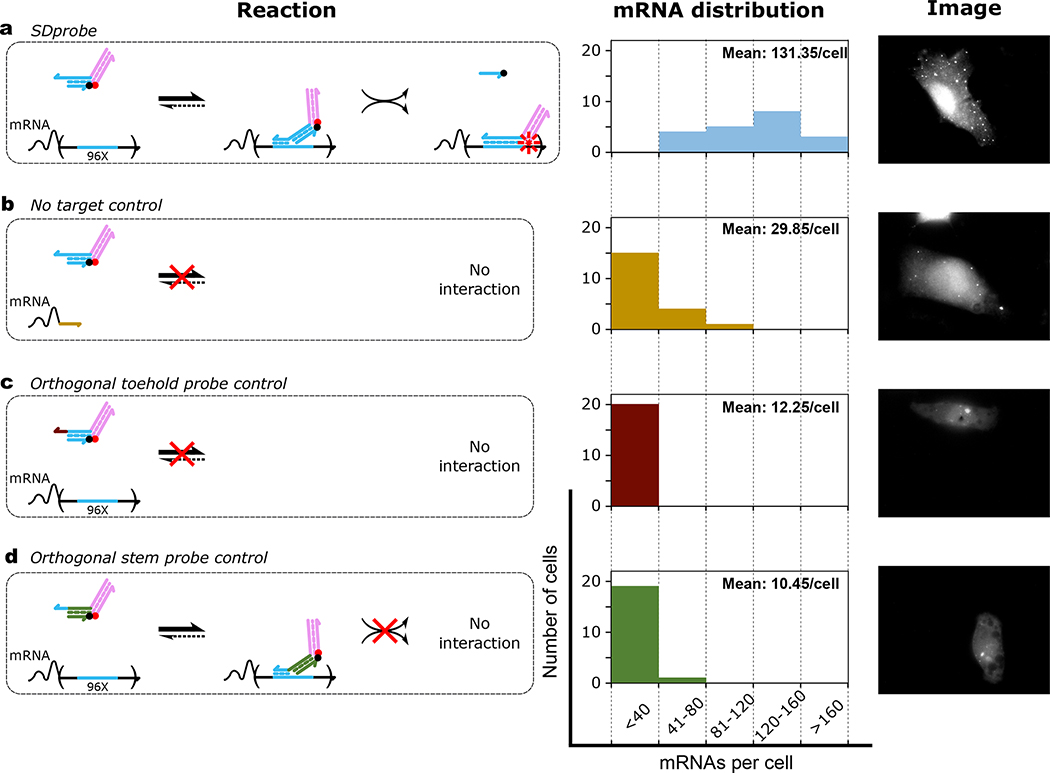
To improve in vivo strand displacement kinetics and facilitate nuclear escape, we further modified the probe design to separate functional elements responsible for control of localization and mRNA sensing25. The redesigned strand displacement probe (SDProbe_v1) was a nicked partially double stranded complex with an interaction domain and an extension domain (Fig.1a). The interaction domain can bind to the target site at an 8-nt toehold and release the 3’-quencher-labeled strand (18 nt) through strand displacement (Fig. 1b). The extension domain consists of a 22 nucleotide long double stranded region with a 5’-fluorophore labeled strand. The reaction kinetics of SDProbe_v1 was first characterized in vitro in OPTI-MEM cell culture media (Fig. S3). Probes were successfully activated via displacement of the fluorophore-labeled strand upon addition of a short synthetic input strand with the same sequence as the target site. The increase in signal was linearly proportional to the amount of input added. Upon delivery to HT1080–96X cells via electroporation, SDProbe_v1 constructs were transported to the cytoplasm where they bound to the 3’-UTR of GFP mRNAs, observable as distinct fluorescent spots throughout the cells (Mean: 131.35 mRNA per cell; Fig.1c, Fig.2a). In contrast, when these probes were electroporated to HT1080-control cells, there were significantly fewer observed spots (Mean: 29.85 mRNA per cell; Fig.2b). These results showed that SDprobe_v1 probes can be used to detect intracellular mRNAs via the multiplexed 3’-UTR target sites. The residual fluorescent spots observed in the HT1080-control cells were possibly due to partial degradation of the probes in the intracellular environment. Additional modifications to the backbone (e.g. phosphorothioate bonds) or sugar moieties (e.g. locked nucleic acids) could help to further reduce degradation effects. The slight increase in the number of observed spots in Fig. 2b compared to Fig. 2c and Fig. 2d, is likely due to variation in transfection efficiency with electroporation or similar differences between the two cell lines we used in our study28,29. Still, all the control cases (Fig. 2b, c and d) showed statistically significant lower observed spot counts than was the case for the intended probe and target combination (Fig. 2a). While the number of transcripts detected with live cells were higher compared to transcripts detected through FISH on fixed cells (Fig. S1), the positive-to-control spot ratios were similar.
Figure 1. Probe design, reaction mechanism and experimental workflow.
a) Domain-level schematic of SDprobe_v1: Individual functional domains are annotated with their respective primary features in parentheses. Half-arrows indicate 3’-ends of the corresponding strands. The fluorophore and quencher are denoted by red and black circles respectively. b) Schematic of probe interaction with the 3’-UTR target sites (blue domains). Probes bind to the target sites via the short toehold domain, which initiates a 3-way branch migration step, ultimately displacing the quencher-labeled strand and thereby unquenching the fluorophore. Co-localization of multiple fluorophores on a single mRNA significantly increases the signal. c) Experimental workflow for detection of mRNAs using strand displacement probes: Probes are delivered via electroporation to cultured cells. Cells are incubated for 1 hour at 37°C allowing the probes to interact with mRNA target sites. Co-localization of up to 96 probes per mRNA generates a bright fluorescent spot detectable with fluorescence microscopy. After incubation, sequential image frames are collected at every 0.3 um z-distance across the cell height and the acquired frames are combined to generate maximum intensity projections representing the entire cell volume. These images are processed using the FISH-Quant33 MATLAB tool; a filtering step performs background signal correction and spots are pre-detected based on a threshold signal cut-off. The pre-detected spots are then fitted and examined for accuracy followed by spot quantification, where the spots are either selected or rejected based on signal intensity and elimination of false-positives. The selected spots represent the number of detected mRNAs per cell.
Figure 2. Intracellular mRNA detection using strand displacement probes with extension domains (SDProbe_v1).
Each row demonstrates the reaction mechanism for interaction of probes with mRNAs (left), histogram of detected mRNAs per cell (middle) and representative image (right). For quantifying the distribution of detected mRNAs per cell, 20 randomly selected cells were imaged for each of the four different conditions: probes were electroporated to a) HT1080–96X cells expressing GFP mRNAs with 96 tandem repeats of probe target sites in the 3’-UTR, b) HT1080-control cells expressing GFP mRNAs without probe target sites. HT1080–96x cells were electroporated with SD control probes similar to SDProbe (SDProbe_v1) but with c) a toehold orthogonal to the corresponding section of the probe target sequence, and d) a double stranded stem domain orthogonal to the probe target sequence. For each case, the mean number of spots detected per cell is shown as inset to the histograms.
Further control experiments were performed with probe variants having toehold sequences or double-stranded stem domains orthogonal to the target sites. These probes were designed to confirm that probe activation indeed proceeds through toehold-mediated strand displacement rather than through spontaneous probe dissociation or partial degradation followed by hybridization to that target. For both controls, we observed low levels of probe activation (Mean: 12.25 mRNA per cell and 10.45 mRNA per cell respectively) in HT1080–96X, suggesting that the probes had minimal non-specific interaction within the intracellular environment (Fig.2C, D).
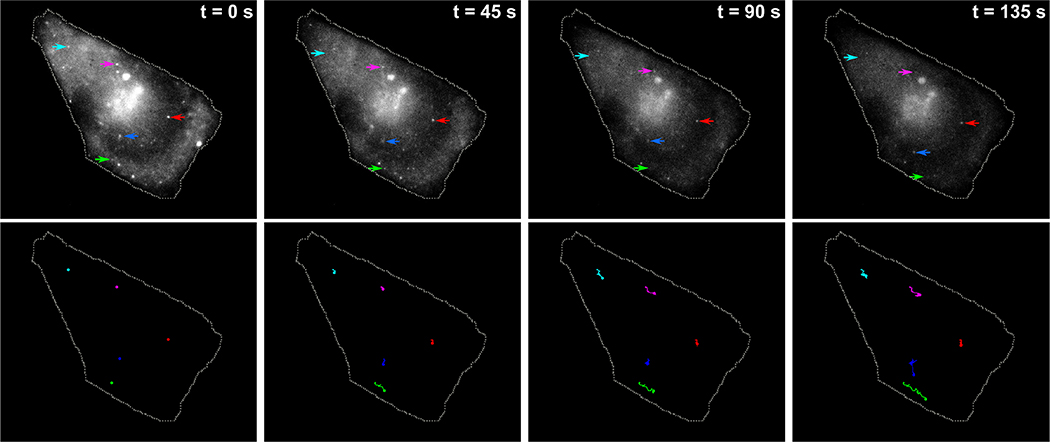
To further explore the potential of our probes for live-cell imaging applications, we used them to track target mRNA dynamics. Using time-lapse microscopy we followed the movement of individual target mRNAs over the course of 3 minutes (Supplementary movie S1) observing a range of behaviors from highly confined mRNAs to mRNAs exhibiting significant intracellular movement, similar to what was observed previously using other live cell imaging probes25,30 (Fig.3).
Figure 3. Visualizing intracellular mRNA movement using strand displacement probes.
Five separate target mRNAs were tracked over a period of 150 seconds using time-lapse imaging. Top row consists of a montage of four frames showing positions of five specific mRNAs (indicated by five distinctly colored arrows) at different timepoints. The trajectories of the corresponding mRNAs up to that time-point are shown in the bottom row.
Our main motivation for this study was to characterize strand displacement in living cells and demonstrate that cellular RNA could serve as an input to a strand displacement probe, thus laying the foundation for adapting more sophisticated strand displacement-based devices from the test tube to the cell. However, with further technical improvements, this kind of approach can be used as a more generalizable platform for single-molecule imaging in live cells. Particularly, the very high specificity of strand displacement probes even to single nucleotide variants makes this an intriguing technology31,32.
However, so far, we have only tested our approach in cells that were genetically modified to express an mRNA with 96 tandem repeats of a target sequence and imaging of endogenous mRNAs still needs to be demonstrated.
To conclude, we have shown that strand displacement probes can effectively detect mRNAs with engineered 3’UTR sequence repeats in the cytoplasm of mammalian cells. In the future, multiple probes detecting different intracellular mRNAs could be connected through downstream molecular logic. The simplicity, scalability and robustness of circuit design with strand displacement opens up the possibilities of a wide range of applications for sensing and computation in living cells.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Arjun Raj and Andrew Tsourkas for sharing the HT1080–96X and HT1080-GFP cell lines used in this study.
Funding Sources
No competing financial interests have been declared. This work was supported by NIH Award 1-R01-CA207029–01A1 to GS.
Footnotes
ASSOCIATED CONTENT
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- (1).Wickham SFJ; Bath J; Katsuda Y; Endo M; Hidaka K; Sugiyama H; Turberfield AJ A DNA-Based Molecular Motor That Can Navigate a Network of Tracks. Nat. Nanotechnol. 2012, 7 (3), 169–173. [DOI] [PubMed] [Google Scholar]
- (2).Thubagere AJ; Li W; Johnson RF; Chen Z; Doroudi S; Lee YL; Izatt G; Wittman S; Srinivas N; Woods D; et al. A Cargo-Sorting DNA Robot. Science (80-. ). 2017, 357 (6356). [DOI] [PubMed] [Google Scholar]
- (3).Lund K; Manzo AJ; Dabby N; Michelotti N; Johnson-Buck A; Nangreave J; Taylor S; Pei R; Stojanovic MN; Walter NG; et al. Molecular Robots Guided by Prescriptive Landscapes. Nature 2010, 465 (7295), 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Seelig G; Soloveichik D; Zhang DY; Winfree E Enzyme-Free Nucleic Acid Logic Circuits. Science 2006, 314 (5805), 1585–1588. [DOI] [PubMed] [Google Scholar]
- (5).Qian L; Winfree E Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332 (6034), 1196–1201. [DOI] [PubMed] [Google Scholar]
- (6).Chen Y-J; Dalchau N; Srinivas N; Phillips A; Cardelli L; Soloveichik D; Seelig G Programmable Chemical Controllers Made from DNA. Nat. Nanotechnol. 2013, 8 (10), 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Srinivas N; Parkin J; Seelig G; Winfree E; Soloveichik D Enzyme-Free Nucleic Acid Dynamical Systems. Science (80-. ). 2017, 358 (6369). [DOI] [PubMed] [Google Scholar]
- (8).Chatterjee G; Dalchau N; Muscat RA; Phillips A; Seelig G A Spatially Localized Architecture for Fast and Modular DNA Computing. Nat. Nanotechnol. 2017, 12 (9), 920–927. [DOI] [PubMed] [Google Scholar]
- (9).Zadorin AS; Rondelez Y; Gines G; Dilhas V; Urtel G; Zambrano A; Galas JC; Estevez-Torres A Synthesis and Materialization of a Reaction-Diffusion French Flag Pattern. Nat. Chem. 2017, 9 (10), 990–996. [DOI] [PubMed] [Google Scholar]
- (10).Kim J; Yin P; Green AA Ribocomputing: Cellular Logic Computation Using RNA Devices. Biochemistry. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dietz H; Douglas SM; Shih WM Folding DNA into Twisted and Curved Nanoscale Shapes. Science (80-. ) 2009, 325 (5941), 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Steinhauer C; Jungmann R; Sobey TL; Simmel FC; Tinnefeld P DNA Origami as a Nanoscopic Ruler for Super-Resolution Microscopy. Angew. Chem. Int. Ed. Engl. 2009, 48 (47), 8870–8873. [DOI] [PubMed] [Google Scholar]
- (13).Marras AE; Zhou L; Su H-J; Castro CE Programmable Motion of DNA Origami Mechanisms. Proc. Natl. Acad. Sci. 2015, 112 (3), 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Funke JJ; Dietz H Placing Molecules with Bohr Radius Resolution Using DNA Origami. Nat. Nanotechnol. 2016, 11 (1), 47–52. [DOI] [PubMed] [Google Scholar]
- (15).Douglas SM; Bachelet I; Church GM A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science (80-. ). 2012, 335 (6070), 831–834. [DOI] [PubMed] [Google Scholar]
- (16).Zhang DY; Seelig G Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat. Chem. 2011, 3 (2), 103–113. [DOI] [PubMed] [Google Scholar]
- (17).Hemphill J; Deiters A DNA Computation in Mammalian Cells: MicroRNA Logic Operations. J. Am. Chem. Soc. 2013. [DOI] [PubMed] [Google Scholar]
- (18).Briley WE; Bondy MH; Randeria PS; Dupper TJ; Mirkin CA Quantification and Real-Time Tracking of RNA in Live Cells Using Sticky-Flares. Proc. Natl. Acad. Sci. 2015, 112 (31), 9591–9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Groves B; Chen YJ; Zurla C; Pochekailov S; Kirschman JL; Santangelo PJ; Seelig G Computing in Mammalian Cells with Nucleic Acid Strand Exchange. Nat. Nanotechnol. 2016, 11 (3), 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang X; Song Y; Shah AY; Lekova V; Raj A; Huang L; Behlke M a; Tsourkas, A. Quantitative Assessment of Ratiometric Bimolecular Beacons as a Tool for Imaging Single Engineered RNA Transcripts and Measuring Gene Expression in Living Cells. Nucleic Acids Res. 2013, 41 (15), e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yang Y; Chen M; Krueger CJ; Tsourkas A; Chen AK Quantifying Gene Expression in Living Cells with Ratiometric Bimolecular Beacons. In Methods in Molecular Biology; 2018; Vol. 1649, pp 231–242. [DOI] [PubMed] [Google Scholar]
- (22).Bramsen JB; Laursen MB; Nielsen AF; Hansen TB; Bus C; Langkjoør N; Babu BR; Højland T; Abramov M; Van Aerschot A; et al. A Large-Scale Chemical Modification Screen Identifies Design Rules to Generate siRNAs with High Activity, High Stability and Low Toxicity. Nucleic Acids Res. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Shukla S; Sumaria CS; Pradeepkumar PI Exploring Chemical Modifications for siRNA Therapeutics: A Structural and Functional Outlook. ChemMedChem 2010, 5 (3), 328–349. [DOI] [PubMed] [Google Scholar]
- (24).Ohrt T; Muetze J; Svoboda P; Schwille P Intracellular Localization and Routing of miRNA and RNAi Pathway Components. Curr. Top. Med. Chem. 2012, 12 (2), 79–88. [DOI] [PubMed] [Google Scholar]
- (25).Zhang X; Zajac AL; Huang L; Behlke MA; Tsourkas A Imaging the Directed Transport of Single Engineered RNA Transcripts in Real-Time Using Ratiometric Bimolecular Beacons. PLoS One 2014, 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gwizdek C; Ossareh-Nazari B; Brownawell AM; Doglio A; Bertrand E; Macara IG; Dargemont C Exportin-5 Mediates Nuclear Export of Minihelix-Containing RNAs. J. Biol. Chem. 2003, 278 (8), 5505–5508. [DOI] [PubMed] [Google Scholar]
- (27).Chen AK; Behlke MA; Tsourkas A Efficient Cytosolic Delivery of Molecular Beacon Conjugates and Flow Cytometric Analysis of Target RNA. Nucleic Acids Res. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Maurisse R; De Semir D; Emamekhoo H; Bedayat B; Abdolmohammadi A; Parsi H; Gruenert DC Comparative Transfection of DNA into Primary and Transformed Mammalian Cells from Different Lineages. BMC Biotechnol. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jordan ET; Collins M; Terefe J; Ugozzoli L; Rubio T Optimizing Electroporation Conditions in Primary and Other Difficult-to-Transfect Cells. J. Biomol. Tech. 2008. [PMC free article] [PubMed] [Google Scholar]
- (30).Santangelo PJ; Nitin N; Bao G Direct Visualization of mRNA Colocalization with Mitochondria in Living Cells Using Molecular Beacons. J. Biomed. Opt. 2013, 10 (4), 44025. [DOI] [PubMed] [Google Scholar]
- (31).Chen SX; Seelig G An Engineered Kinetic Amplification Mechanism for Single Nucleotide Variant Discrimination by DNA Hybridization Probes. J. Am. Chem. Soc. 2016, 138 (15), 5076–5086. [DOI] [PubMed] [Google Scholar]
- (32).Li Q; Luan G; Guo Q; Liang J A New Class of Homogeneous Nucleic Acid Probes Based on Specific Displacement Hybridization. Nucleic Acids Res. 2002, 30 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mueller F; Senecal A; Tantale K; Marie-Nelly H; Ly N; Collin O; Basyuk E; Bertrand E; Darzacq X; Zimmer C FISH-Quant: Automatic Counting of Transcripts in 3D FISH Images. Nature Methods. 2013, pp 277–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.