Abstract
Autosomal recessive hereditary spastic paraplegia (ARHSP) with thin corpus callosum is a distinct and usually severe form of complex HSP classified as SPG11. The gene for SPG11 was localized to chromosome 15q13-q15 and recently mutations in Spatacsin (KIAA1840) were shown to cause the majority of SPG11 cases.
We analysed the 40 coding exons of this gene in the probands from 8 families with complex ARHSP, four of these families had a thin corpus callosum. Three families were identified with novel mutations in the SPG11 gene. One family was of Asian origin with a homozygous nonsense mutation and had a very severe phenotype without a thin corpus callosum. In the other two English families the parents were unrelated and the mutations were compound heterozygotes. In these two families the phenotype was mild and both probands had a thin corpus callosum. Given the likely mechanism of action of the mutations in the Spatacsin gene we discuss the probable genotype phenotype correlations in these families. This study confirms the frequent occurrence of Spatacsin mutations in complex ARHSP with genotype phenotype effects and exposes the spectrum of clinical heterogeneity in SPG11.
Introduction
The hereditary spastic paraplegias (HSPs) are a large group of inherited neurological disorders characterized by slowly progressive spasticity affecting the lower limbs first and then upper limbs. They are classified genetically into autosomal dominant, autosomal recessive and X-linked HSP where a total of 15 disease genes have been identified. Clinically they are separated into pure HSP, where the spastic paraplegia is the only clinical manifestation or complicated HSP when it is accompanied by additional neurological such as cognitive decline, ataxia or peripheral neuropathy 1,2.
Autosomal recessive hereditary spastic paraplegia with thin corpus callosum (ARHSP, MIM 604360) is one of the more prevalent forms of complex ARHSP and is characterized by progressive spastic paraparesis, cognitive decline, axonal neuropathy and a thin corpus callosum 3. ARHSP with a thin corpus callosum was first linked to the SPG11 locus on chromosome 15q13-q15 in families from the Mediterranean basin 4 and confirmed in different ethnicities 5–7 and the locus refined to 6cM 8 in one family group and 2.93cM in another 9.
Recently 10 different mutations in KIAA1840 (SPG11; 604360) were described in 11 ARHSP-TCC families. One of this novel mutations (p.M245VfsX) was also identified in two affected members from a consanguineous Italian HSP-TCC family10. And later on, additional 27 novel SPG11 mutations, including frameshift, nonsense and splice site mutations have been reported in ARHSP families with and without TCC 11,12. Being to date a total of 41 HSP families reported with SPG11 mutations. The KIAA1840 gene has 7332 open reading frame base pairs and encodes a 2443 amino acid protein (Spatacsin) of unknown function that is predicted to contain a Glycosyl hydroxylase F1 signature, a Leucine zipper, a short coiled coil domain, a Myb domain, and four transmembrane domains (TM). Spatacsin is expressed ubiquitously in the nervous system but most prominently in the cerebellum, cerebral cortex, hippocampus and pineal gland 13. Although SPG11 mutations are frequent in ARHSP-TCC families, a number of families with ARHSP with and without TCC were found to be negative for the Spatacsin gene and a further locus has been identified 14.
We identified 8 ARHSP families from the UK and Asia. These families had been evaluated in detail and varied in their clinical severity and phenotype. Four of the probands had a thin corpus callosum on MRI. We sequenced the 40 coding exons and flanking introns of the SPG11 gene in our patient group.
Methods
Patients
Informed consent was obtained from all participants in this study. Eight ARHSP families were studied, four had thinning of the entire corpus callosum, two mild thinning of the body of the corpus callosum and four families had consanguineous parents. A summary of the clinical details and examination findings are given in Table 1. Clinical examination of parents of each case where available showed no neurological deficit; parental consanguinity was present in four families.
Table 1.
Clinical features of all SPG11 cases with mutations in the (Spatacsin) KIAA1840 gene.
| Family/Origin | Inheritance | Mutation | Age at onset (symptoms) | Age at exam | Severity | Gait | Ataxia | Reflexes | Plantars | Neuropathy | Cognitive problems | Sphincter problems | MRI | EMG/NCS | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 Iran | Parents first cousins | c..398delG/p.C133LfsX154 in homozygous (exon2) | 17 (walking) | 27 | Severe | Wheelchair | yes | +++ | Ext/ Ext | Axonal | moderate | Yes | Mild TCC, cortical and cerebellar atrophy, no TCC or WM abnormalities | mild axonal neuropathy | spastic dysartria, Jerky pursuit, hypometric saccades. Slow tonge |
| Family 2 England | Parents not related | Compound heterozygous. c.C5623T/p.Q1875X (exon 30) +c.7155- 7156 insAAAC/ p.K2386Qfs X2393 (exon40) | 27 (dragging her left leg) | 34 | Mild | Mild spastic gait. No stick | No | ++ | Ext/ Ext | Mild vibration loss | Mild | No | TCC with white matter abnormalities | No | Hypometric saccades |
| Family 3 England | Parents not related | Compound heterozygous. c.2834+1G>T (exon 15) and c.6754+4insTG (exon 36) | 13 (walking) | 18 | Mild | Mild spastic gait. No stick | Mild | ++ | Ext/ Ext | No | Mild | No | TCC with white matter abnormalities | Normal | Hypometric saccades, poor upgaze, spastic dysarthria, slow tongue |
| Family 4 India | Parents first cousins | Sequence variant p.A59V of unknown significance. Identified as a heterozygote in one control. | 13 (walking) | 24 | Severe | Wheelchair | Severe | +++ | Ext/ Ext | Axonal | Mild | Severe | No/borderline TCC, Cortical and cerebellar atrophy | Axonal neuroapthy | Severe dystonia in the limbs and torticollis, spastic dysarthria, ophthalmoplegia, hypometric saccades. Slow tonge |
| Affected cousin | Parents first cousins | N/A Died in early childhood |
Early childhood | N/A | severe | Wheelchair | Severe | N/A | N/A | N/A | Development delay | N/A | Not done | Not done | Died young |
| Affected cousin | Parents first cousins | N/A | 2 | 3 | Severe | Wheelchair | Severe | ++ | Ext/ Ext | No | Development delay | N/A | CT head normal age 3 | Normal | Rotatory nystagmus and severe tremor |
TCC = thin corpus callosum. N/A = not available. Ext = extensor. For reflexes, + = normal, ++ = brisk, +++ = very brisk often with clonus.
The diagnosis of complex HSP was made clinically; acquired causes of complex HSP were excluded as far as possible with imaging of the brain and spinal cord, lumbar puncture, white cell enzymes, long chain fatty acids and in four families a muscle biopsy. Spastic paraplegia was present in all cases along with cognitive decline, sensory or motor neuropathy, ataxia, speech and swallowing problems. In addition two probands had seizures, one had cervical dystonia, six had jerky pursuit and slow saccades, and one had a skew deviation and poor upgaze.
Molecular Analysis
DNA was extracted by a standard phenol chloroform method. PCR was employed to analyse all 40 exons and flanking introns of the KIAA1840 gene. PCR analysis was performed using 10pmol of both forward and reverse genomic primers (primer sequences are available on request), 10 μl of FastStart Taq DNA polymerase (www.roche-applied-science.com) and a 57td52 thermo-cycling program that consists in a denaturation cycle of one minute at 94ºC, following by 15cycles of 94ºC for 30s, 57ºC for 30s, and 72ºC for 30s; 16 cycles of 94ºC for 30s, 57ºC for 30s dropping 0.3ºC each cycle, and 72ºC for 30s; 14 cycles of 94ºC for 30s, 52ºC for 30s, 72ºC for 30s, and a final extension cycle of 72 ºC for 5 minutes. Then each purified product was sequenced using forward or reverse primers with Applied Biosystems BigDye terminator v3.1 sequencing chemistry as per the manufacturer’s instructions; the resulting reactions were resolved either on an ABI3730XL or an ABI3100 genetic analyser (Applied Biosystems, Foster city, CA) and analysed with Sequencer software (Gene Codes Corporation, Ann Arbor, MI). All mutations were verified by sequencing in both directions and sequencing of specific exons in the proband and other family members was performed to test segregation of the mutations within families. All mutations are numbered from the ATG start site of the SPG11 (KIAA1840) gene, accession number NP_079413. Mutation position was labelled according to the standard nomenclature from den Dunnen and Antonarakis 15.
Results
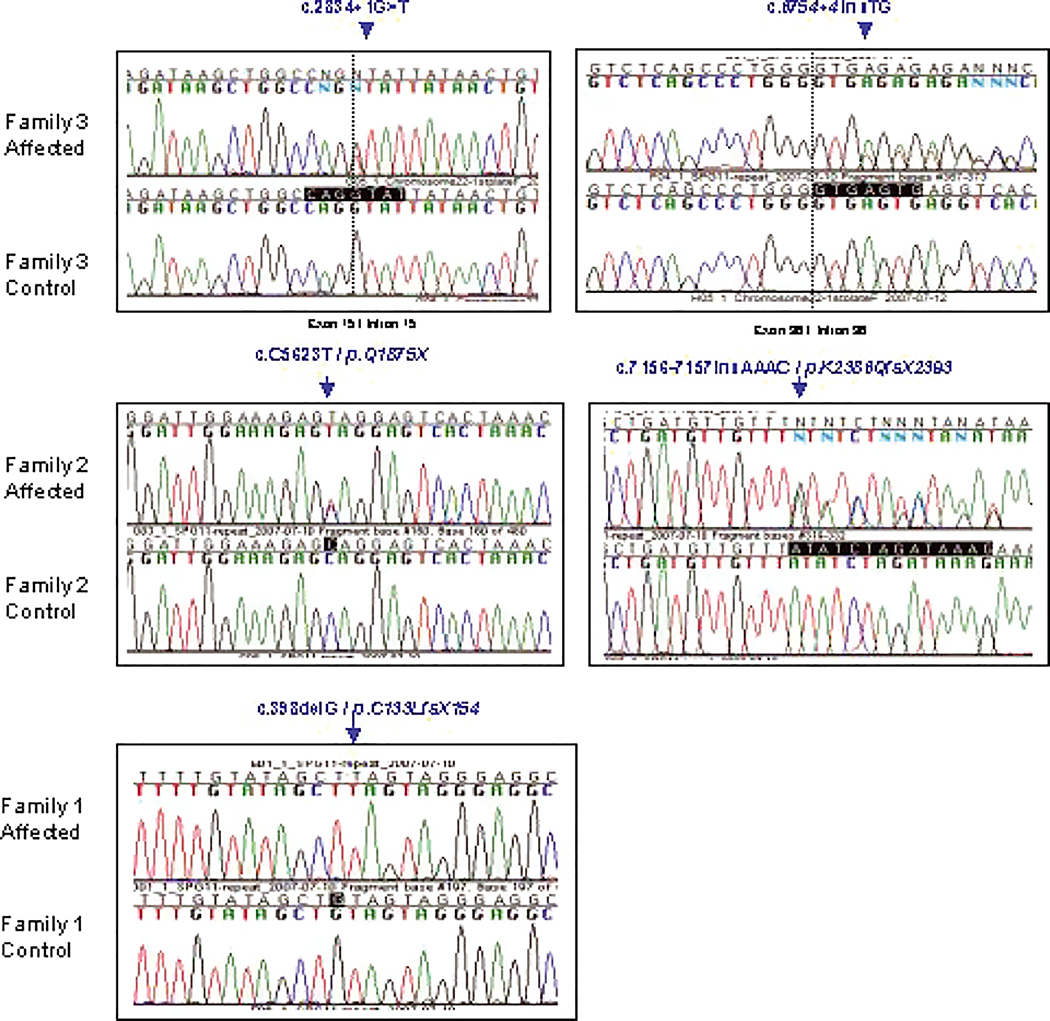
We identified five novel Spatacsin mutations, of which three (p.C133LfsX154, p.Q1875X and p.K2386QfsX2393) lead to a truncated protein and two, c.2834+1G>T and c.6754+4insTG, were found to be located at the splice donor site in intron 15 and intron 36 respectively (Figure 1A for chromatograms and Figure 1B for family trees). A sixth missense Spatacsin variant (p.A59V) was identified in an Indian family with severe ARHSP (Table 1), a normal/borderline thin corpus callosum with cerebral and cerebellar atrophy. This sequence change was found as a heterozygous change in one control, overall estimated frequency 0.58% and therefore is only a sequence variant.
Figure 1.
Spatacsin (KIAA1840) sequence analysis. Electropherograms showing the mutations identified in the three SPG11 families with controls. The arrow indicated the position of the mutation or frameshift change.
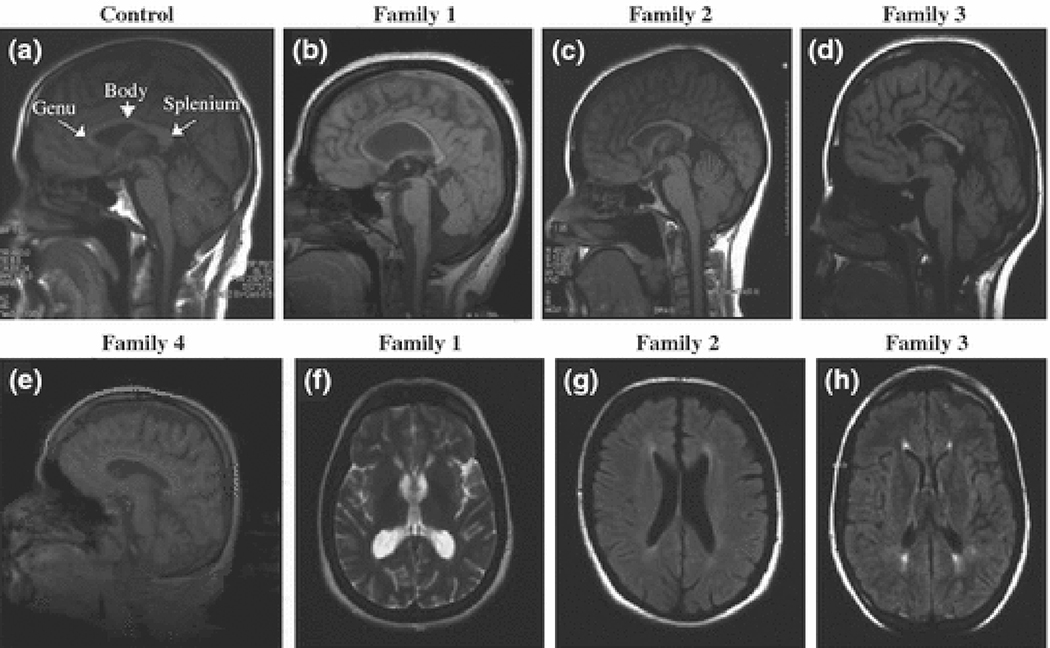
Family 1 was originally from Iran and had a severe ARHSP where the parents were first cousins. The proband presented in the early teens and progressed rapidly with a severe ataxia and lower limb spasticity, speech and swallowing problems, jerky pursuit, gaze evoked nystagmus, hypometric saccades, cognitive problems, urinary problems and limb pain. MRI showed enlarged ventricles with possible thinning of the body of the corpus callosum which is stretched due to ventricular enlargement and cerebrum atrophy along with cerebellar atrophy, with no white matter abnormalities (Figure 2). This patient was found to carry a homozygous “G” deletion located at 398 nucleotide position which leads a frameshift that creates a premature codon stop at 154 amino acid position (p.C133LfsX154).
Figure 2.
T1 (b–e, g, h) and T2 weighted (2f) MRI brain scans of the probands from families 1, 2, 3 and 4. (a) shows a control MRI scan with the different parts of a normal labelled corpus callosum. Sagittal sections (c and d) show considerable thinning of the corpus callosum. (d) and (e) show mild thinning of the body as well as cortical and cerebellar atrophy. There are white matter abnormalities around the ventricles in images from families 2 (g) and 3 (h), but not family 1(f) and 4.
Families 2 and 3 were both mildly affected with ARHSP, they originated from England, their parents were not known to be related. The duration of disease was six and four years respectively. In both families disease was caused by compound heterozygous Spatacsin gene mutations (c.2834+1G>T and c.6754+4insTG in family 3; p.Q1875X + p.K2386QfsX2393 in family 2). The wild type nucleotide of both splice site mutations is highly conserved in the respective consensus sequences of other mammalian species. In both cases there was a thin corpus callosum with peri-ventricular and non-specific white matter abnormalities (Figure 2). Family 2 was the most mildly affected. She was a good athlete and only presented at the age of 27 after her mother noticed she was dragging her left leg, later she also had problems with her right leg. Examination revealed spasticity in her lower limbs, mild cognitive decline and hypometric saccades. Her disease is very slowly progressive and she is able to carry out a normal job.
The proband in Family 3 was also mildly affected with a slowly progressive spastic ataxia. She presented at the age of 13 with walking problems but was independent in all activities and able to complete a college course. Over the next five years her lower limb spasticity progressed slowly and later she developed increased tone in her upper limbs and mild cognitive difficulties, ataxia, mild spastic dysarthria, hypometric saccades, poor upgaze and her tongue movements were slow. She had no neuropathy (Table 1 and Figure 2).
DISCUSSION
Recently it has been reported that mutations in SPG11 are a major cause of familial spastic paraplegia11,12. We have screened eight families with complex HSP cases and provide data that further confirms SPG11 as the most frequent mutated gene in ARHSP. Out of the eight complex ARHSP families we analysed, three had Spatacsin mutations and a further family (family 4) had a missense variant, p.A59V, that was identified as a heterozygote in one control case. However further analysis of this variant in control and HSP population are required in order to evaluate the role of this variant in the SPG11 phenotype.
The mutations described here bring the total number of Spatacsin mutations described thus far to 42. Mutations are located across the gene and have been described in patients from Asia and Europe, this report is the first published on patients from Iran and England. Spatacsin contains 40 exons and the reported mutations are located in several exons across the gene where the majority of mutations are in single families. Frameshift or nonsense changes are the most frequent mutations reported in the SPG11 gene, however five splice site mutations have been recently identified 11,12 and additional two have been here reported, suggesting that splice site mutations may also be frequent in the SPG11 disease.
The function of Spatacsin is unknown, but the presence of a Leucine zipper motif and a Myb domain suggest involvement in the regulation of gene expression. The presence of transmembrane domains also suggests that Spatacsin may be involved in trafficking, a mechanism implicated in a number of other forms of HSP (SPG3A (ATL1), SPG10 (KIF5A), SPG7 (PGN), SPG4 (Spastin), SPG20 (Spartin) 16.
The clinical features in the families with Spatacsin varied considerably. Family 1 was severely affected with a complex spastic paraplegia that rapidly progressed from onset. Families 2 and 3 were mildly affected. In particular family 2, where the proband had minimal symptoms and signs (Table 1). Interestingly both families with the mild phenotype had a thin corpus callosum on MRI and white matte abnormalities whereas in the severely affected from family 1, there was only very mild corpus callosum body thinning perhaps mainly due to enlarged ventricles and cortical and cerebellar atrophy seen and no abnormal white matter changes (Figure 2). The severe ARHSP family 4 also had normal/borderline corpus callosum body thinning, cortical and cerebellar atrophy.
These data suggest that there is significant clinical heterogeneity in cases with SPG11 mutations and that imaging of the brain may reveal varying effects on the corpus callosum. The diffuse white matter changes frequently reported in SPG11 10–12 may also be absent. Although we describe only three cases of genetically proven SPG11, there is possibly a genotype phenotype effect. The reported Spatacsin mutations are stop, splice or frameshift indicating the likely mechanism to be nonsense mediated mRNA decay (NMD), resulting in haploinsufficiency 17. The severely affected from family 1 has a homozygous deletion in exon 2 of the SPG11 gene suggesting haploinsufficiency and the little active protein that escapes this degradation will be very short, abnormally truncated. The other two families have compound heterozygous stop, frameshift or splice site mutations in exons 30 and 40 in the most mildly affected from family 2 and in introns 15 and 36 in mildly affected from family 3. Again, most protein will be degraded by NMD but residual protein will likely be more active as it will be longer. To confirm the genetic mechanisms cell lines and brain tissue will be essential in the future and further detailed clinical studies will help to define these genotype phenotype correlations.
In summary, we present further evidences that mutations in the SPG11 gene are responsible for a proportion of familial spastic paraplegia cases. We also show that there is significant clinical heterogeneity in mutant families and some have no thinning of the corpus callosum. Finally we speculate on the mechanism of action of the Spatacsin gene and discuss the genotype phenotype correlations.
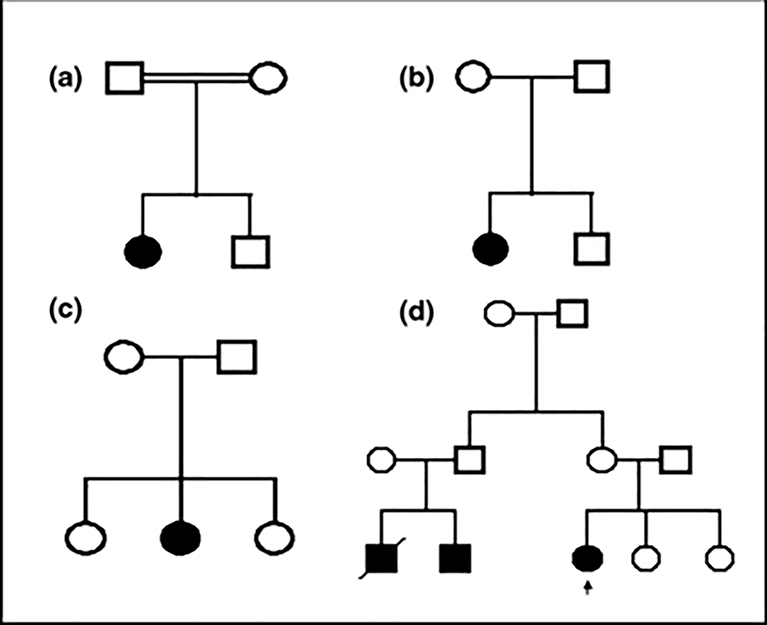
Figure 3.
Pedigrees of the families with Spatacsin mutations and variants. Arrow indicates the proband. Family 1 = A, Family 2 = B, Family 3 = C and Family 4 = D in the text and Table 1.
Acknowledgements
We are grateful to the Medical Research Council (MRC) for their support. HH holds an MRC clinician scientist fellowship. This work was supported in part by the Intramural Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, USA. This work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. We also thank the families involved for their help with our work.
REFERENCES
- 1.Fink JK: Hereditary spastic paraplegia. Curr Neurol Neurosci Rep 2006; 6: 65–76. [DOI] [PubMed] [Google Scholar]
- 2.Harding AE: Classification of the hereditary ataxias and paraplegias. Lancet 1983; 1: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 3.Honda Y, Hino H: [Familial cases presenting spastic paraparesis, mental disturbance and thinning of corpus callosum]. Rinsho Shinkeigaku 1994; 34: 190–191. [PubMed] [Google Scholar]
- 4.Martinez Murillo F, Kobayashi H, Pegoraro E et al. : Genetic localization of a new locus for recessive familial spastic paraparesis to 15q13–15. Neurology 1999; 53: 50–56. [DOI] [PubMed] [Google Scholar]
- 5.Brockmann K, Simpson MA, Faber A, Bonnemann C, Crosby AH, Gartner J: Complicated hereditary spastic paraplegia with thin corpus callosum (HSP-TCC) and childhood onset. Neuropediatrics 2005; 36: 274–278. [DOI] [PubMed] [Google Scholar]
- 6.Casali C, Valente EM, Bertini E et al. : Clinical and genetic studies in hereditary spastic paraplegia with thin corpus callosum. Neurology 2004; 62: 262–268. [DOI] [PubMed] [Google Scholar]
- 7.Lossos A, Stevanin G, Meiner V et al. : Hereditary spastic paraplegia with thin corpus callosum: reduction of the SPG11 interval and evidence for further genetic heterogeneity. Arch Neurol 2006; 63: 756–760. [DOI] [PubMed] [Google Scholar]
- 8.Stevanin G, Montagna G, Azzedine H et al. : Spastic paraplegia with thin corpus callosum: description of 20 new families, refinement of the SPG11 locus, candidate gene analysis and evidence of genetic heterogeneity. Neurogenetics 2006; 7: 149–156. [DOI] [PubMed] [Google Scholar]
- 9.Olmez A, Uyanik G, Ozgul RK et al. : Further clinical and genetic characterization of SPG11: hereditary spastic paraplegia with thin corpus callosum. Neuropediatrics 2006; 37: 59–66. [DOI] [PubMed] [Google Scholar]
- 10.Del Bo R, Di Fonzo A, Ghezzi S et al. : SPG11: a consistent clinical phenotype in a family with homozygous Spatacsin truncating mutation. Neurogenetics 2007; 8: 301–305. [DOI] [PubMed] [Google Scholar]
- 11.Hehr U, Bauer P, Winner B et al. : Long-term course and mutational spectrum of spatacsin-linked spastic paraplegia. Ann Neurol 2007; 62: 656–665. [DOI] [PubMed] [Google Scholar]
- 12.Stevanin G, Azzedine H, Denora P et al. : Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 2007. [DOI] [PubMed] [Google Scholar]
- 13.Stevanin G, Santorelli FM, Azzedine H et al. : Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 2007; 39: 366–372. [DOI] [PubMed] [Google Scholar]
- 14.Al-Yahyaee S, Al-Gazali LI, De Jonghe P et al. : A novel locus for hereditary spastic paraplegia with thin corpus callosum and epilepsy. Neurology 2006; 66: 1230–1234. [DOI] [PubMed] [Google Scholar]
- 15.den Dunnen JT, Antonarakis SE: Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000; 15: 7–12. [DOI] [PubMed] [Google Scholar]
- 16.Crosby AH, Proukakis C: Is the transportation highway the right road for hereditary spastic paraplegia? Am J Hum Genet 2002; 71: 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker M, Mackenzie IR, Pickering-Brown SM et al. : Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006; 442: 916–919. [DOI] [PubMed] [Google Scholar]