Abstract
Amphiphilic DNA block copolymers have been utilized in preparing self-assembled amphiphilic structures in aqueous solution. These block copolymers usually contain specifically designed hydrophobic regions, and typically assemble under near-physiological conditions. Here, we report self-assembly of spherical micelles and one-dimensional nanorods under acidic conditions from cholesterol-conjugated DNA strands (Cholesterol-DNA). Further study also revealed that the nanorods were hierarchically assembled from the micelle nanostructures. The morphology of the nanorod assemblies can be tuned by altering solution condition and the design of Cholesterol-DNA. The self-assembly of Cholesterol-DNA nanostructures under acidic conditions and the discovery of the relationship between the nanorods and the micelles can provide new insights for future design of self-assemblies of amphiphilic DNA block copolymers.
Graphical Abstract
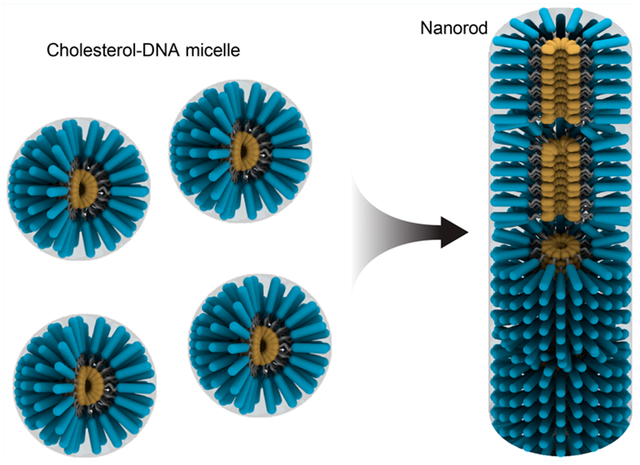
Over the past few decades, DNA has drawn considerable interest as a promising and versatile material for nanotechnology. The unique 4-digit code pairing characteristic of DNA endows it with the ability for constructing well-defined, precisely controlled nanostructures with high programmability and versatility.1,2 The self-assembly of DNA nanostructures mainly relies on the base pairing between different strands with delicately designed sequences. Many methods of DNA nanostructure construction have been developed and widely utilized for various applications.3–5 In addition, conjugation of DNA strands to other molecules, polymers, or nanostructures generates DNA block copolymers (DBC) with new interactions during assembly, such as hydrophobic, electrostatic, and π-π interactions.6 These special interactions can result in orthogonal bindings and long-range morphology control in DBC self-assembly, and lead to a new generation of DNA nanostructures.7–11
One particular kind of DBCs is the amphiphilic DNA copolymer. In amphiphilic DNA block copolymers, DNA strands mainly serve as the hydrophilic parts, while the conjugated molecules, such as synthetic polymers, fluorescent dyes, and lipids, usually serve as the hydrophobic parts.12–14 These copolymers combine the favorable properties of hydrophilic DNA and hydrophobic molecules in aqueous solution, and may offer the possibility of evolving completely new functions.15 Amphiphilic DBCs have been utilized to assemble one-dimensional (1D), two-dimensional (2D), and three-dimensional (3D) nanostructures with tunable size and shape.16–18 Previous work has demonstrated that during the DBC assembly, many unique properties may occur. For example, Wang and co-workers constructed a DBC whose assembly could undergo reversible phase transitions between spherical micelles and long nanofibers, which demonstrated the possibility of controlling DBC nanostructures’ morphology beyond the sample preparation step.19 Liu et al. utilized thermally responsive DBCs to construct nanostructures which could alter the morphology and property with a temperature trigger.20,21 With these unique properties, amphiphilic DBCs have been widely employed in research and many applications have been reported, such as diagnostics,22 programmable nanoreactors,23 and, especially, drug delivery.24–26 Similar to some other delivery systems such as DNA modified liposomes,27 DBC nanostructures also exhibit low toxicity and high reproducibility, thus making them a potent agent for a next generation targeted therapeutic approach.28
Cholesterol-conjugated DNA strands (Cholesterol-DNA) were usually used as the anchor in the process of detection and dynamic motion, especially when involving membranes.29–32 It has been reported that Cholesterol-DNA with variable modifications could form micellular structures.33,34 Block copolymers that combine Cholesterol-DNA together with other polymers can also produce micelles and single crystals.35,36 Here, we discovered that Cholesterol-DNA with specific sequences can also form spherical micelles and nanorods under acidic conditions. The morphology of the nanostructure can be tuned by pH, salt concentration, DNA sequence, and the length of connecting spacers. Further study revealed that the nanorod assembly was closely linked to the initial formation of micelles. Our work provides new design principles for DBC self-assembly, and can be useful for the construction of new DBC nanostructures for applications (e.g., drug delivery) at different conditions.
RESULTS
The Cholesterol-DNA we used in this paper consists of 3 parts: hydrophilic DNA, internal linker, and hydrophobic cholesterol (Figure 1A). With a specific GA-rich DNA sequence, we found that the Cholesterol-DNA could form micelles (Figure 1B) and nanorods (Figure 1C) under acidic conditions. The products varied among spherical micelles, short nanorods, and micrometer-long nanorods (Figure 1D), depending on the buffer conditions, assembly protocols, and the design of the DNA sequences and the linkers.
Figure 1.
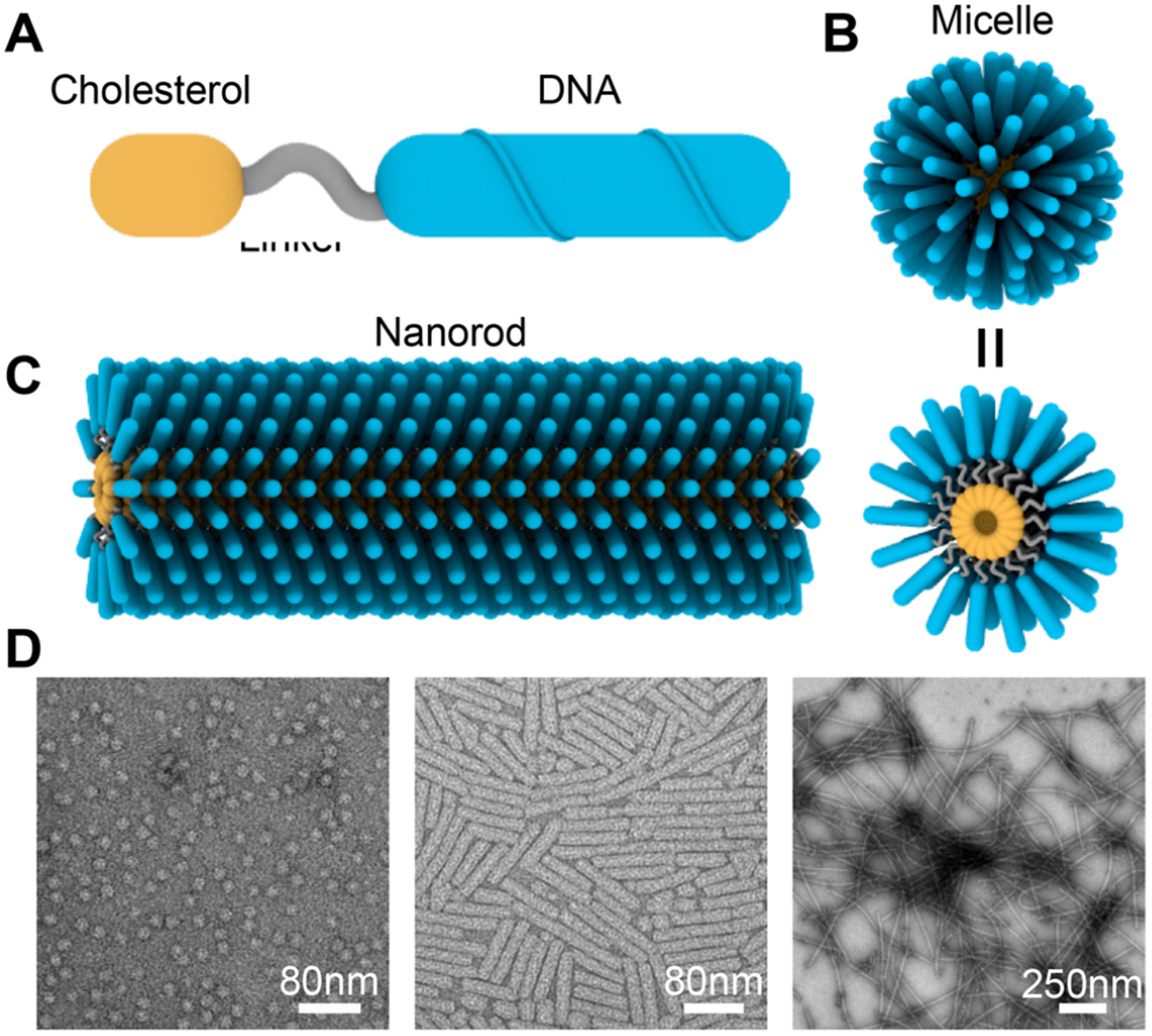
Micelles and nanorods self-assembled from Cholesterol-DNA. (A) Design of a Cholesterol-DNA. (B) Schematic of spherical micelles formed by Cholesterol-DNA. (C) Schematic of a nanorod assembled from Cholesterol-DNA. (D) Representative TEM images of different types of products, including spherical micelles, short nanorods, and long nanorods.
The design of all Cholesterol-DNAs is included in the SI Table S1. The initial design of Cholesterol-DNA contains an 18-base (18B) DNA strand with a GA-rich sequence and a triethylene glycol (TEG) linker (SI Figure S1). Micelle-like structures were observed at pH 3.6. We then carefully investigated the effect of salt concentration at this pH (Figure 2A and SI Figure S2). Two ions commonly used in DNA self-assembly were chosen for the investigation: sodium cation (Na+) and magnesium cation (Mg2+). Four groups of Na+ and Mg2+ concentrations were tested. All samples were subjected to an annealing process from 37 to 29 °C (see SI for more details). Mg2+ appeared crucial in the assembly of nanorods, as all groups without Mg2+ did not show the formation of nanorods. With sodium and magnesium ion concentration increasing, more spherical micelles or nanorods were observed, which indicated that the cations were necessary for the assembly of the relatively compact DNA shells. However, at relatively higher salt concentrations, the numbers of micelles and nanorods decrease, which may be due to the disruption of the electrostatics balance of DNA parts.
Figure 2.
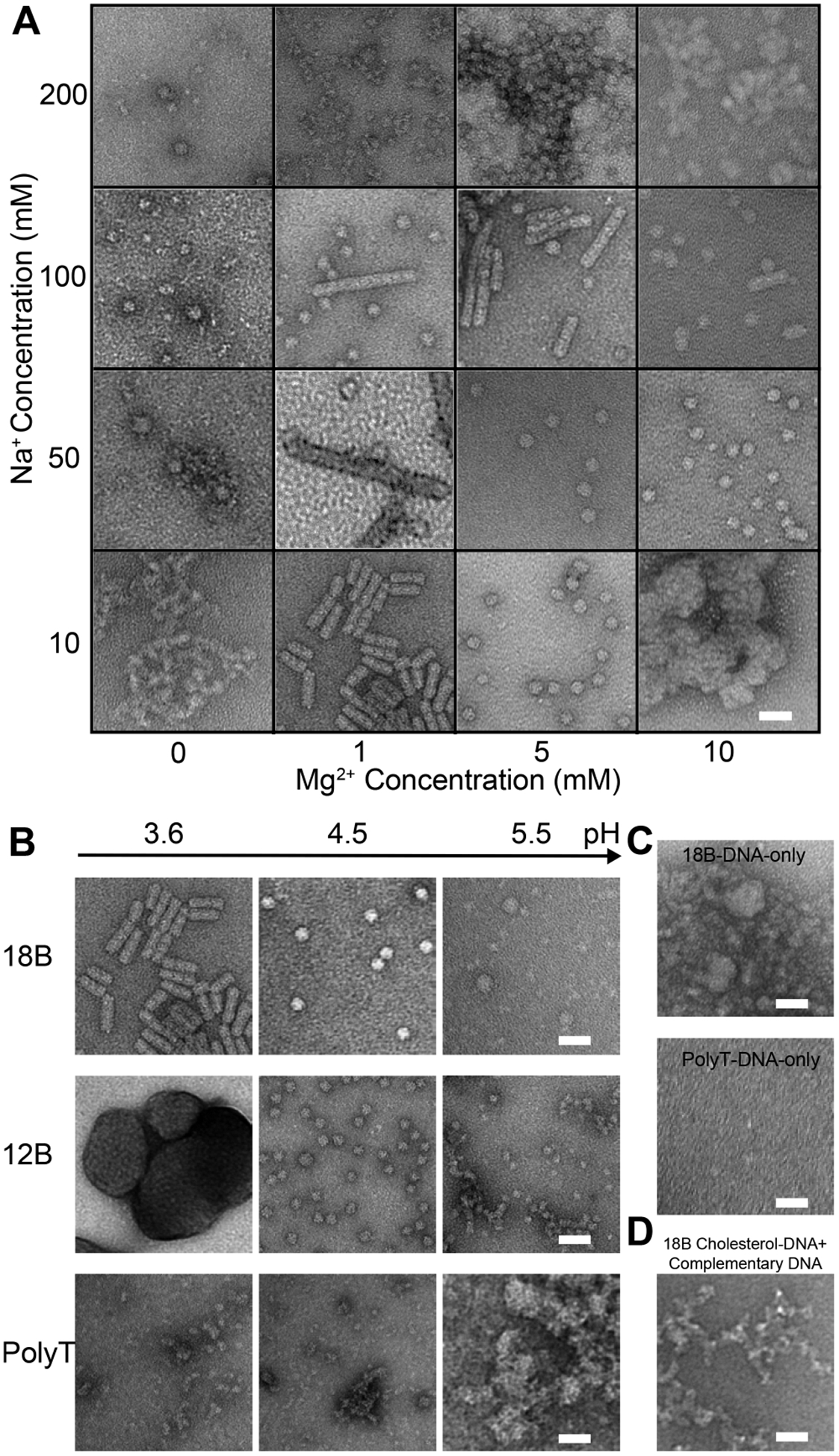
Effects of different factors on Cholesterol-DNA assembly. (A) TEM images of Cholesterol-DNA assembled with different salt concentrations. (B) TEM images of the assembly of 18B, 12B, and PolyT under different pH. (C) TEM images of the assembly result of DNA strands. The upper image shows the assembly from the 18B GA-rich DNA, and the lower image shows the assembly from the 18-base PolyT DNA. (D) TEM image of the assembly result of 18B Cholesterol-DNA annealed in the presence of the complementary strand of its DNA part. The scale bars are 40 nm.
The importance of cations in the Cholesterol-DNA assembly suggested that the interactions among DNA strands could have played a significant role in the micelle/nanorod formation process, since it is known that cations affect the DNA hybridization. To understand the effect of DNA interaction, we investigated Cholesterol-DNA assembly with different DNA sequences, in the presence of 50 mM Na+ and 1 mM Mg2+. We specifically designed three additional Cholesterol-DNAs with different DNA sequence: a 12-base (12B) GA-rich, a 24-base (24B) GA-rich sequence, and an 18-T sequence (PolyT), which was expected to greatly reduce DNA-DNA interactions. The assembly of 12B, 24B, and PolyT were compared to the assembly of 18B at pH from 3.1 to 5.5 (Figure 2B and SI Figures S3, S4). As expected, the PolyT Cholesterol-DNA assembly failed to produce any ordered structures, while 12B, 18B, and 24B all generated ordered micelles/nanorods at specific pH, proving the interaction of the GA-rich sequence was crucial for the Cholesterol-DNA assembly. The 12B, 18B, and 24B Cholesterol-DNA mainly assembled into ordered micelle/nanorod nanostructures under more acidic conditions, from pH 3.1 to 4.5. However, no well-defined nanostructure was formed under pH more than 5.5 for all samples. Both 12B and 24B produced only spherical micelles, but no nanorod formation. The pH window for 24B to form micelles was larger than the window for 12B, suggesting that the longer DNA sequence of 24B could stabilize the micelle formation. Nonetheless, the nanorod formation was only observed in the 18B sample. The 18B contains 78% guanine and adenine. To test how the GA content affects the assembly of the Cholesterol-DNA, we designed an 18B-high GA with 100% GA content and an 18B-low GA with 56% GA content. The assembly of these two Cholesterol-DNAs were carried out at pH 3.1, 4.0, and 5.0, and were compared to the assembly of the original 18B (SI Figure S5). At pH 4.0 and 5.0, clear micellular structures could be observed for the 18B-high GA, but they tended to form large aggregates. When pH was reduced to 3.1, the micellular aggregates morphed to large structures without well-defined morphology. For the 18B-low GA, which we expected to have lower level of interaction between the DNA strands, the assembly produced a mixture of micelles and nonstructured aggregates at pH 3.1, and only aggregates at pH 4.0 and 5.0. Our result further demonstrated that the assembly of Cholesterol-DNAs was affected by the interaction of GA-rich DNA strands, and the formation of the nanorods requires more stringent sequences than the micelles.
Additional experiments were performed to further confirm the observation that DNA interaction was important for the Cholesterol-DNA assembly. First, a DNA strand without cholesterol and the linker (18B-DNA-only) were annealed under different pH from 3.1 to 5.5 (Figure 2C and SI Figure S6). Although the assembly did not produce ordered micelles or nanorods, aggregation was observed in all samples. Similarly, the assembly of a 12B-DNA-only and a 24B-DNA-only also resulted in aggregation (SI Figure S6). In contrast, the assembly of a PolyT-DNA-only showed no formation of any assembly (Figure 2C and SI Figure S6). This outcome suggested that the GA-rich sequences have stronger interstrand interactions at the low pH, and was consistent with previous results that 12B, 18B, and 24B could form well-defined nanostructures while PolyT could not. Then another experiment was carried out by annealing 18B in the presence of a complementary DNA strand. The assembly led to the formation of random aggregation (Figure 2D), since the micelle/nanorod formation of 18B was disrupted by the complementary DNA’s interaction with the 18B DNA. The aforementioned experiments indicated the mechanism of the Cholesterol-DNA assembled micelles/nanorods: On the one hand, the hydrophobic interactions between cholesterols is a key driving force, because without cholesterol, DNA itself cannot form well-defined nanostructures. On the other hand, the assembly is also regulated by the interactions between DNA strands. These interactions are largely affected by the solution composition, causing the varying results of the final products.
In our study of TEM images, we noticed an interesting phenomenon that the 18B nanorods contained uniform units separated by small but visible gaps (Figure 3A). This led us to hypothesize that the nanorods were hierarchically assembled from preformed micelles and the gap might be the result of micelles not fully merged together. Additional experiments were performed to investigate this hypothesis. First, we tested longer spacers in the linker domain of the 18B Cholesterol-DNA: one with a single spacer (18B1S) and the other with two spacers (18B2S). Like 18B assembly, both 18B1S and 18B2S produced well-formed nanorods (Figure 3B and C). The lengths of these nanorods were longer than the 18B nanorods (SI Figures S7 and S8). The 18B1S nanorods could grow to several micrometers in length (Figure 3C and SI Figure S9). We measured the average widths of 18B, 18B1S, and 182S nanorods in TEM images and confirmed that the diameters of the nanorods increased with longer spacers (SI Figure S10). In addition, the periodic gaps in the 18B1S and 18B2S nanorods became more pronounced than the 18B nanorods. To rule out the possibility that the nanorod formation was induced by the drying procedure used in TEM imaging, we also imaged the 18B1S nanorod and 18B2S nanorod via liquid-phase atomic force microscopy (AFM) imaging, which confirmed that the nanorods were formed in the solution (SI Figure S11). A time-course study on the growth of the 18B1S nanorods confirmed the hypothesis of hierarchical assembly process (Figure 3 and SI Figure S12). The micelles formed with 5 min and we started to notice the merging of micelles at ~10 min. After 30 min, well-formed nanorods appear in the images and became the dominant product over time, but the gaps were clearly visible in the final products. We suspected that the gaps might be a transitional state and would eventually disappear if given more time. However, no visible changes for the nanorods were observed after long incubations of preassembled 18B1S nanorods for up to 15 days (SI Figure S13). To find out the critical micelle concertation (CMC) for 181S, we also tested the assembly at different concentrations (SI Figure S14). The nanorod formation disappeared at about 5 μM concentration and the spherical micelles were no longer detectable at concentrations lower than 1 μM.
Figure 3.
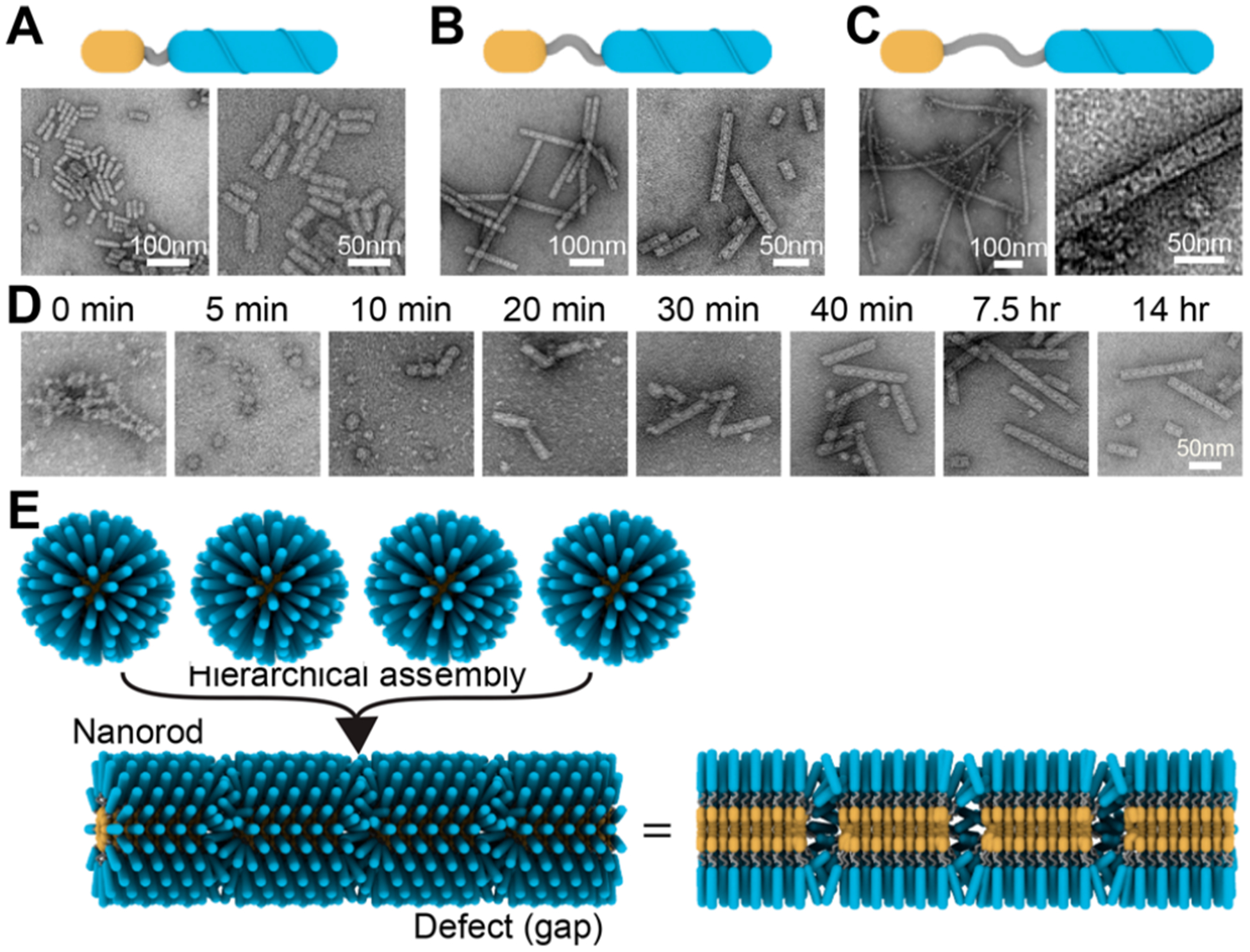
Investigation of the assembly mechanism. (A) to (C) Assembly of Cholesterol-DNA with different length linkers. (A) Schematic and TEM images of the assembly result of the 18B with a short linker. (B) Schematic and TEM images of the assembly result of the 18B1S with a median length linker. (B) Schematic and TEM images of the assembly result of the 18B2S with a long linker. (D) TEM images of the assembly products of 18B1S at different time. (E) Proposed hierarchical assembly of micelles leads to the formation of the nanorods and the visible periodic gaps in the nanorods.
Based on the experimental results, we concluded that the nanorods with gaps were hierarchically assembled from the micelles and were stable (Figure 3E). The visible black gaps in TEM images indicated that the merging of micelles were not “complete”, perhaps due to the size difference between the small cholesterol hydrophilic core and hydrophobic DNA strands.37 Compared with other amphiphilic DBCs with larger hydrophobic parts and nanofibers with no defects, Cholesterol-DNA’s hydrophobic core is rather small in size.7,19 Therefore, during the merging process, the interactions between the relatively larger hydrophilic DNA strands could hinder the small micelle hydrophobic cores from merging with others. In other words, the formation of a Cholesterol-DNA nanorod with a continuous, narrow hydrophobic core (without gaps) would need to pack DNA strands more compactly and have to pay a greater energy penalty, and the formation of gaps was a response to reduce the energy penalty.
CONCLUSION
In summary, we report self-assembly of micelles and nanorods from Cholesterol-DNA under acidic conditions. We carefully investigated the influence of salt concentration, pH, DNA sequence, and internal spacer on the assembly. Furthermore, we designed several experiments to explore the mechanism of the Cholesterol-DNA assembly and discovered the unique hierarchical assembly process of Cholesterol-DNA nanorods. We believe the relatively small volume of the cholesterol hydrophobic cores and the interactions between DNA strands led to the formation of gaps in the nanorods. Our work not only further proves the versatility of amphiphilic DBCs and enriches the family of amphiphilic DBC nanostructures, but also can provide insights on new design principles and assembly mechanisms for future studies.
Supplementary Material
ACKNOWLEDGMENTS
Y.K. acknowledges NIH support under grant 1R21AI135753-01. R.P. thanks the support by China Scholarship Council (No.201706130095). The authors also thank Hong Yi for her help with electron microscope characterization. The EM data described here was gathered on the TEM supported by the Atlanta Clinical and Translational Science Institute (ACTSI) award UL1TR000454.
ABBREVIATIONS
- DBC
DNA block copolymer
- TEG
Triethylene glycol
- PBS
Phosphate-buffered saline
- TEM
Transmission electron microscopy
- AFM
Atomic force microscopy
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00322.
Cholesterol-DNA designs, methods for nanostructures preparation and characterization, additional images, and analysis (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Ke Y, Castro C, and Choi JH (2018) Structural DNA Nanotechnology: Artificial Nanostructures for Biomedical Research. Annu. Rev. Biomed. Eng 20, 375–401. [DOI] [PubMed] [Google Scholar]
- (2).Wang P, Meyer TA, Pan V, Dutta PK, and Ke Y (2017) The Beauty and Utility of DNA Origami. Chem. 2, 359–382. [Google Scholar]
- (3).Winfree E, Liu F, Wenzler LA, and Seeman NC (1998) Design and Self-Assembly of Two-Dimensional DNA Crystals. Nature 394, 539–544. [DOI] [PubMed] [Google Scholar]
- (4).Rothemund PWK (2006) Folding DNA to Create Nanoscale Shapes and Patterns. Nature 440, 297–302. [DOI] [PubMed] [Google Scholar]
- (5).Ong LL, Hanikel N, Yaghi OK, Grun C, Strauss MT, Bron P, Lai-Kee-Him J, Schueder F, Wang B, Wang P, et al. (2017) Programmable Self-Assembly of Three-Dimensional Nanostructures from 10,000 Unique Components. Nature 552, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mai Y, and Eisenberg A (2012) Self-Assembly of Block Copolymers. Chem. Soc. Rev 41, 5969–5985. [DOI] [PubMed] [Google Scholar]
- (7).Bousmail D, Chidchob P, and Sleiman HF (2018) Cyanine-Mediated DNA Nanofiber Growth with Controlled Dimensionality. J. Am. Chem. Soc 140, 9518–9530. [DOI] [PubMed] [Google Scholar]
- (8).Serpell CJ, Edwardson TGW, Chidchob P, Carneiro KMM, and Sleiman HF (2014) Precision Polymers and 3D DNA Nanostructures: Emergent Assemblies from New Parameter Space -Supporting Information General Considerations. J. Am. Chem. Soc 136, 15767–15774. [DOI] [PubMed] [Google Scholar]
- (9).Qi H, Ghodousi M, Du Y, Grun C, Bae H, Yin P, and Khademhosseini A (2013) DNA-Directed Self-Assembly of Shape-Controlled Hydrogels. Nat. Commun 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jiang T, Meyer TA, Modlin C, Zuo X, Conticello VP, and Ke Y (2017) Structurally Ordered Nanowire Formation from Co-Assembly of DNA Origami and Collagen-Mimetic Peptides. J. Am. Chem. Soc 139, 14025–14028. [DOI] [PubMed] [Google Scholar]
- (11).Wilks TR, Bath J, De Vries JW, Raymond JE, Herrmann A, Turberfield AJ, and O’Reilly RK (2013) Giant Surfactants” Created by the Fast and Efficient Functionalization of a DNA Tetrahedron with a Temperature-Responsive Polymer. ACS Nano 7, 8561–8572. [DOI] [PubMed] [Google Scholar]
- (12).Li Z, Zhang Y, Fullhart P, and Mirkin CA (2004) Reversible and Chemically Programmable Micelle Assembly with DNA Block-Copolymer Amphiphiles. Nano Lett. 4, 1055–1058. [Google Scholar]
- (13).Chien MP, Thompson MP, and Gianneschi NC (2011) DNA-Nanoparticle Micelles as Supramolecular Fluorogenic Substrates Enabling Catalytic Signal Amplification and Detection by DNAzyme Probes. Chem. Commun 47, 167–169. [DOI] [PubMed] [Google Scholar]
- (14).Wang Y, Wu C, Chen T, Sun H, Cansiz S, Zhang L, Cui C, Hou W, Wu Y, Wan S, et al. (2016) DNA Micelle Flares: A Study of the Basic Properties That Contribute to Enhanced Stability and Binding Affinity in Complex Biological Systems. Chem. Sci 7, 6041–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zimmermann J, Kwak M, Musser AJ, and Herrmann A (2011) Amphiphilic DNA Block Copolymers: Nucleic Acid-Polymer Hybrid Materials for Diagnostics and Biomedicine In Bioconjugation Protocols: Strategies and Methods, Methods in Molecular Biology (Mark SS, Ed.) pp 239–266, Chapter 15, V 751, Springer Science & Business Media, Berlin. [DOI] [PubMed] [Google Scholar]
- (16).Lu Y, Yue Z, Xie J, Wang W, Zhu H, Zhang E, and Cao Z (2018) Micelles with Ultralow Critical Micelle Concentration as Carriers for Drug Delivery. Nat. Biomed. Eng 2, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhou C, Zhang Y, Dong Y, Wu F, Wang D, Xin L, and Liu D (2016) Precisely Controlled 2D Free-Floating Nanosheets of Amphiphilic Molecules through Frame-Guided Assembly. Adv. Mater 28, 9819–9823. [DOI] [PubMed] [Google Scholar]
- (18).Dong Y, Sun Y, Wang L, Wang D, Zhou T, Yang Z, Chen Z, Wang Q, Fan Q, and Liu D (2014) Frame-Guided Assembly of Vesicles with Programmed Geometry and Dimensions. Angew. Chem., Int. Ed 53, 2607–2610. [DOI] [PubMed] [Google Scholar]
- (19).Wang L, Feng Y, Yang Z, He YM, Fan QH, and Liu D (2012) Reversibly Controlled Morphology Transformation of an Amphiphilic DNA-Dendron Hybrid. Chem. Commun 48, 3715–3717. [DOI] [PubMed] [Google Scholar]
- (20).Zhao Z, Chen C, Dong Y, Yang Z, Fan QH, and Liu D (2014) Thermally Triggered Frame-Guided Assembly. Angew. Chem., Int. Ed 53, 13468–13470. [DOI] [PubMed] [Google Scholar]
- (21).Wu F, Zhao Z, Chen C, Cao T, Li C, Shao Y, Zhang Y, Qiu D, Shi Q, Fan QH, et al. (2018) Self-Collapsing of Single Molecular Poly-Propylene Oxide (PPO) in a 3D DNA Network. Small 14, 1703426. [DOI] [PubMed] [Google Scholar]
- (22).Yang CJ, Pinto M, Schanze K, and Tan W (2005) Direct Synthesis of an Oligonucleotide-Poly-(Phenylene Ethynylene) Conjugate with a Precise One-to-One Molecular Ratio. Angew. Chem., Int. Ed 44, 2572–2576. [DOI] [PubMed] [Google Scholar]
- (23).Trinh T, Chidchob P, Bazzi HS, and Sleiman HF (2016) DNA Micelles as Nanoreactors: Efficient DNA Functionalization with Hydrophobic Organic Molecules. Chem. Commun 52, 10914–10917. [DOI] [PubMed] [Google Scholar]
- (24).Wang H, Wang Y, Wang Y, Hu J, Li T, Liu H, Zhang Q, and Cheng Y (2015) Self-Assembled Fluorodendrimers Combine the Features of Lipid and Polymeric Vectors in Gene Delivery. Angew. Chem., Int. Ed 54, 11647–11651. [DOI] [PubMed] [Google Scholar]
- (25).Tan X, Li BB, Lu X, Jia F, Santori C, Menon P, Li H, Zhang B, Zhao JJ, and Zhang Ke (2015) Light-Triggered, Self-Immolative Nucleic Acid-Drug Nanostructures. J. Am. Chem. Soc 137, 6112–6115. [DOI] [PubMed] [Google Scholar]
- (26).Chan MS, Tam DY, Dai Z, Liu LS, Ho JWT, Chan ML, Xu D, Wong MS, Tin C, and Lo PK (2016) Mitochondrial Delivery of Therapeutic Agents by Amphiphilic DNA Nanocarriers. Small 12, 770–781. [DOI] [PubMed] [Google Scholar]
- (27).Banga RJ, Chernyak N, Narayan SP, Nguyen ST, and Mirkin CA (2014) Liposomal Spherical Nucleic Acids. J. Am. Chem. Soc 136, 9866–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Pack DW, Hoffman AS, Pun S, and Stayton PS (2005) Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discovery 4, 581–593. [DOI] [PubMed] [Google Scholar]
- (29).Stengel G, Simonsson L, Campbell RA, and Höök F (2008) Determinants for Membrane Fusion Induced by Cholesterol-Modified DNA Zippers. J. Phys. Chem. B 112, 8264–8274. [DOI] [PubMed] [Google Scholar]
- (30).Kocabey S, Kempter S, List J, Xing Y, Bae W, Schiffels D, Shih WM, Simmel FC, and Liedl T (2015) Membrane-Assisted Growth of DNA Origami Nanostructure Arrays. ACS Nano 9, 3530–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Johnson-Buck A, Jiang S, Yan H, and Walter NG (2014) DNA-Cholesterol Barges as Programmable Membrane-Exploring Agents. ACS Nano 8, 5641–5649. [DOI] [PubMed] [Google Scholar]
- (32).Gunnarsson A, Jönsson P, Marie R, Tegenfeldt JO, and Höök F (2008) Single-Molecule Detection and Mismatch Discrimination of Unlabeled DNA Targets. Nano Lett. 8, 183–188. [DOI] [PubMed] [Google Scholar]
- (33).Magnusson JP, Fernández-Trillo F, Sicilia G, Spain SG, and Alexander C (2014) Programmed assembly of polymer-DNA conjugate nanoparticles with optical readout and sequence-specific activation of biorecognition. Nanoscale 6, 2368–2374. [DOI] [PubMed] [Google Scholar]
- (34).Banchelli M, Gambinossi F, Durand A, Caminati G, Brown T, Berti D, and Baglioni P (2010) Modulation of Density and Orientation of Amphiphilic DNA on Phospholipid Membranes. II. Vesicles. J. Phys. Chem. B 114, 7348–7358. [DOI] [PubMed] [Google Scholar]
- (35).Choi KM, Kwon IC, and Ahn HJ (2013) Self-Assembled Amphiphilic DNA-Cholesterol/DNA-Peptide Hybrid Duplexes with Liposome-like Structure for Doxorubicin Delivery. Biomaterials 34, 4183–4190. [DOI] [PubMed] [Google Scholar]
- (36).Brady RA, Brooks NJ, Foderà V, Cicuta P, and Di Michele L (2018) Amphiphilic-DNA Platform for the Design of Crystalline Frameworks with Programmable Structure and Functionality. J. Am. Chem. Soc 140, 15384–15392. [DOI] [PubMed] [Google Scholar]
- (37).Lombardo D, Kiselev MA, Magazù S, and Calandra P (2015) Amphiphiles Self-Assembly: Basic Concepts and Future Perspectives of Supramolecular Approaches. Adv. Condens. Matter Phys 2015, 151683. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


