Abstract
Patient-derived 3D organoids show great promise for understanding patient heterogeneity and chemotherapy response in human-derived tissue. The combination of organoid culture techniques with mass spectrometry imaging provide a label-free methodology for characterizing drug penetration, patient-specific response, and drug biotransformation. However, current methods used to grow tumor organoids employ extracellular matrices which can produce small molecule background signal during mass spectrometry imaging analysis. Here, we develop a method to isolate 3D human tumor organoids out of a Matrigel extracellular matrix into gelatin mass spectrometry compatible microarrays for high-throughput MS imaging analysis. The alignment of multiple organoids in the same z-axis is essential for sectioning organoids together and for maintaining reproducible sample preparation on a single glass slide for up to hundreds of organoids. This method successfully removes organoids from extracellular matrix interference and provides an organized array for high-throughput imaging analysis to easily identify organoids by eye for area selection and further analysis. With this method, mass spectrometry imaging can be readily applied to organoid systems for pre-clinical drug development and personalized medicine research initiatives.
Keywords: MALDI-MSI, Organoids, High-throughput, Microwells, Drug Discovery
Introduction
Tumor organoids represent an important 3D cell culture strategy in pre-clinical drug discovery because large numbers (10s–1000s) of patient-derived organoids can be generated from a resected human tumor tissue sample.1 When grown in the laboratory, these small (100–800μm) organoids contain all or most of the types of cells that are present in the original tumor2. They also closely recapitulate in vivo-like tissue architecture containing the genetic mutation drivers and morphological features of the cancer cells in the original tumor.3–6 Organoids have been made from a variety of tumor types including colorectal7–9, pancreas10, 11, bladder12, prostate5, and breast cancers.13 Large numbers of organoid samples can be produced from a single patient, so large patient organoid biobanks can be generated that also contain correlated patient data and chemotherapy response.5, 14, 15 These biobanks can facilitate drug development and personalized medicine. Organoids are advantageous for drug discovery because of the relatively short culture time and large number of samples available for high-throughput drug testing, compared with the time needed to generate and test animal models of cancer. Additionally, organoids possess stromal cell types. This is particularly relevant for pancreatic cancer, where the stromal cells play a critical role in drug response and tumor progression.16–18 Organoids provide insights into patient and tumor heterogeneity in drug response in in pre-clinical drug development and can be used to design personalized medicine strategies to effectively tailor treatment regimens.10, 19 In this manuscript, we describe sample preparation strategies for applying matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) to a large number of organoids as a label-free methodology to image the localization of biomolecules.
MSI is a powerful technology that can image hundreds to thousands of molecules in a single experiment label free, as each molecule is tentatively identified based on its mass to charge ratio (m/z value) with confirmation of molecular structure typically provided with tandem mass spectrometry.20–22 MSI is an important analytical technique in drug discovery because it can image drug penetration and biomolecule distribution in organoids.7, 23–25 Here, we apply MALDI MSI to organoids and describe sample preparation strategies to improve throughput of mass spectrometry analysis of organoid systems. Imaging of intact organoids can account for cellular heterogeneity in tumor subpopulations present in organoids, contrary to methods that require tissue homogenization for analysis.19, 26 Organoids are formed by either a mechanical or enzymatic digestion of whole tissues into small tissue blocks, and then embedding these tissue blocks in a 3D matrix such as Matrigel, a basement membrane secreted by Engelbreth-Holm-Swarm mouse sarcoma.1, 27, 28 This material is a natural extracellular matrix-based hydrogel that allows organoids to develop epithelial-like layers, budding structures, and lumens. Matrigel is commonly used for tumor organoid formation because it better resembles the extracellular environment, due to the inclusion of many common extracellular matrix proteins and growth factors, which in turn increases the physiological relevance of the organoid tissue to the in vivo system of interest.11, 28 While Matrigel is ideal for 3D growth, these extracellular matrix components can sequester small molecules and peptides from the Engelbreth-Holm-Swarm mouse sarcoma secretions that can interfere with mass spectrometry signals in this analysis.30 Additionally, batch-to-batch variation in the production of Matrigel can introduce batch-dependent variation in these background signals. 31, 32
In this manuscript, we show that these interfering signals prevent MSI from being directly applied to organoids cultured in Matrigel. We then describe a centrifugation method to remove organoids from the Matrigel and transfer them into a gelatin microwell microarray format compatible for MSI. This microarray format contains multiple 800μm microwells which align organoids on the same z-axis.35, 36 This sample preparation step is important for creating very thin sections needed for MSI, with each section containing an array of organoids. The microwell mold is commercially available, so this technique can be easily adapted for labs that regularly perform MSI. This sample preparation technique increases sample throughput and allows for stacking of multiple organoids on a single slide for reproducible sample preparation and high-throughput MSI analysis.
Organoids represent a fascinating platform for personalized medicine, particularly because the tissue features are preserved in vitro. This makes them ideal candidates for applying MSI to acquire chemical information about cellular heterogeneity within the tumor. Additionally, for organoids used in the drug development pipeline, MSI can be used to image drug penetration, metabolism, and pharmacodynamic markers in the organoids. The method described here can expand the application of MSI studies to organoid systems in a high-throughput fashion.
Experimental Section
Organoid Sample Preparation
Human tissue was collected with informed consent. All studies were approved by the Institutional Review Board at the University of Wisconsin-Madison. Surgically resected pancreatic cancer tissue was placed in cold chelation buffer on ice for one hour. The tissue was washed with phosphate buffered saline (PBS) and digested at 37°C in DMEM/F12 medium (Invitrogen, Thermo Fischer Scientific, Waltham, MA) containing 1 mg/mL collagenase (Sigma-Aldrich, St. Louis, MO), 0.125 mg/mL dispase (Invitrogen, Thermo Fischer Scientific, Waltham, MA), 10% Fetal Bovine Serum (FBS) (Gibco, Thermo Fischer, Waltham, MA), and 1% penicillin streptomycin solution (Gibco, Thermo Fischer Scientific, Waltham, MA) for 2–3 hours with intermittent shaking. The resulting cell macro-suspension was rinsed in PBS, re-suspended in 1:1 DMEM/F12: Matrigel, plated in 50 μL droplets, and allowed to solidify at 37°C, 5% CO2 in a cell incubator. Once solidified, droplets were overlaid with DMEM/F12 supplemented with 7% FBS, 20 μM Y-27632 (Sigma-Aldrich, St. Louis, MO), 50 ng/ml EGF (Invitrogen, Thermo Fischer Scientific, Waltham, MA), RSPO-conditioned medium (homemade) and 1% penicillin-streptomycin.
Drug Treatment
72 hours prior to organoid isolation, media was replaced with fresh media containing 10 μM 5-fluorouracil and 85 μM gemcitabine (5-FU + GEM). Doses were selected to replicate clinically relevant peak plasma concentrations.37 24 hours later, gemcitabine was removed from cultures to simulate single bolus delivery, while 5-FU exposure was maintained throughout the experiment to simulate daily oral delivery. Drugs were obtained from the University of Wisconsin Carbone Cancer Center Pharmacy.
Gelatin Microarray Preparation
3D Petri Dishes® Microtissues Molds (Sigma-Aldrich, St. Louis, MO) purchased for use in 12-well plate for both large (800μm) microwells and small (200 μm) microwells, were used to pre-cast the gelatin microwell molds. 150mg/mL gelatin (Difco, Becton Dickinson, Franklin Lakes, NJ) dissolved in distilled water was heated to 37°C and then 500μL of dissolved gelatin was pipetted into each mold, ensuring to fill the mold and that no bubbles are in the mold. The filled molds were kept at 4°C for 10min, or until solid, and then were carefully removed from the mold and placed into 15mm x 15mm x 5mm Tissue-Tek Cryomold (Sakura, Tokyo, Japan) for centrifugation.
Centrifugation of Organoids in Microarrays
Drug-treated and control organoids embedded in Matrigel were washed with 1mL 1X PBS for 3 times. The organoids in Matrigel were transferred into a 15mL centrifuge tube with 3mL of PBS, and then centrifuged at 1000 rotations per minute (rpm) for 5 minutes at 4°C. A wide-bore transfer pipette was then used to transfer the pellet of organoids in PBS into the microarray and then the cryomold with organoids is placed on top of a 50mL conical tube and spun for 5 minutes at 1000 rpm at 4°C. Following organoids settling into the microwells of the microarray, excess PBS was removed from the lip of the microarray. Organoids in the microarray were flash frozen on dry ice. Warm gelatin was poured into the mold to embed the microarrays for sectioning and MSI sample preparations. The samples were flash frozen and then stored at −80°C.
Sample Preparation for MALDI MSI
Flash frozen organoid microarrays were sectioned into 12 μm sections using a cryostat set at −20°C. Tissue sections were thaw-mounted onto standard glass microscope slides. Dihydroxybenzoic acid (DHB) at 40mg/mL (in 50:50 methanol:water and 0.1% trifluoracetic acid) was applied using an automated TM Sprayer (HTX Technologies LLC, Carrboro, NC, USA). The matrix TM Sprayer conditions for DHB were: nozzle temperature of 80°C, flow rate of 0.05mL/min, 24 passes, 3 mm track spacing (rotate and offset) at a nozzle velocity of 1250 mm/min.
MALDI-Orbitrap MSI
A MALDI-Orbitrap mass spectrometer (Thermo Fischer Scientific, Waltham, MA), equipped with an N2 laser (spot diameter of 75 μm) was used in positive ion mode for imaging of the organoid microarrays. Imaging was performed using a mass range of m/z 50–1000 and a mass resolution of 60,000 (at m/z 400). The organoid array region to be imaged and the raster step size were controlled using the LTQ Tune software (Thermo Fischer Scientific, Waltham, MA) and the instrument methods were created using Xcalibur (Thermo Fischer Scientific, Waltham, MA). To generate images, the spectra were collected at 75 μm pixel intervals in both the x and y dimensions across the sample.
MALDI MSI Data Processing
MS data was processed using Xcalibur (Thermo Fischer Scientific, Waltham, MA) and ImageQuest (Thermo Fischer Scientific, Waltham, MA). MS data (in .raw format) was exported into imzmL format prior to uploading into MSiReader.38, 39 All images were normalized to total ion current and peaks were extracted from organoid regions using a ratio of peaks found in more than 10% of the organoids, but less than 5% of the background noise (consisting of matrix and gelatin). Generated images were also manually checked. MSiReader data shows both treated and untreated organoids on the same relative intensity scale – ranging from 0– 100% scale. MS data as a non-centroided imzmL format is loaded into SCiLS software 2019 (SCiLS Lab Software, Bruker Daltonics, Bremen, Germany) for statistical analysis. Using SCiLS, we performed bisecting-k-means clustering to segment out the organoids from the microwells, which is a combination of hierarchical clustering and k-means algorithms. We also performed Receiver Operator Characteristic analysis and t-test analysis in SCiLS to compare treated and control organoids for a comparison of m/z signals. METLIN metabolite database (Scripps Center for Metabolomics, La Jolla, CA) was used for accurate mass matching and lipid assignment with 5 ppm tolerance.40
Results and Discussion
Matrigel Interference
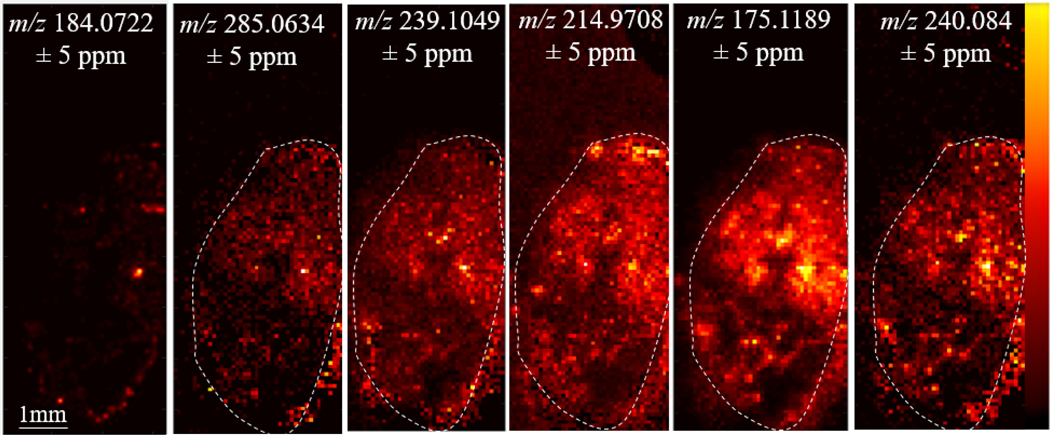
Matrigel, basement membrane secreted by Engelbreth-Holm-Swarm mouse sarcoma, was used to embed and culture 3D organoids to maintain in vivo morphology.28 These Matrigel hydrogels were cultured in 35mm glass-bottom dishes and then washed with 1X PBS 3 times prior to harvesting the hydrogel for MSI. With Matrigel embedding, organoids are at multiple z-axis depths throughout the gel, thus, only a few organoids can be sectioned at a specific sample depth in the hydrogel. This limits the throughput of the analysis because multiple sections of the hydrogel must be analyzed to obtain larger populations of organoids. Additionally, because Matrigel is basement membrane, created from a cell line, secreted small molecules and peptides from the cell line can be sequestered in the protein mixture. In Figure 1, we imaged the organoids embedded in the Matrigel, which was then embedded in gelatin for stabilization during sectioning and for mass spectrometry analysis. A phosphatidylcholine head group peak, m/z 184.0722, a typical signal from the cell membrane, shows the placement of organoids within a 12μm thick section of Matrigel (Figure 1). The other m/z values in Figure 1 are used to demonstrate that while these signals are present in the organoids, they are also present in the Matrigel hydrogel. A thin white line is included in the figure to outline the Matrigel hydrogel boundary within the embedded gelatin. Gelatin is considered an MSI compatible material for small molecule and lipid analysis.41, 42 Supplemental Table 1 contains a list of all background peaks detected from the Matrigel, which are also found in the organoids themselves. These results from organoids embedded in Matrigel hydrogels also limit the number of organoid samples for MSI analysis. From this data, we conclude that the Matrigel itself creates confounding background noise for organoid MSI. To successfully image metabolite and peptide signals from the organoids themselves, organoids need to be removed from the surrounding Matrigel.
Figure 1. Organoids in Matrigel:
m/z 184.0722 is a peak resulting from a phosphatidyl choline head group which indicates where organoid cells are located in the Matrigel material. The white dotted line outlines Matrigel. m/z values 285.0634, 239.1049, 214.9708, 175.1189, and 240.0840 represent examples of small molecule background signal in the Matrigel. This background interference can complicate MSI analysis.
Microarray Method Development
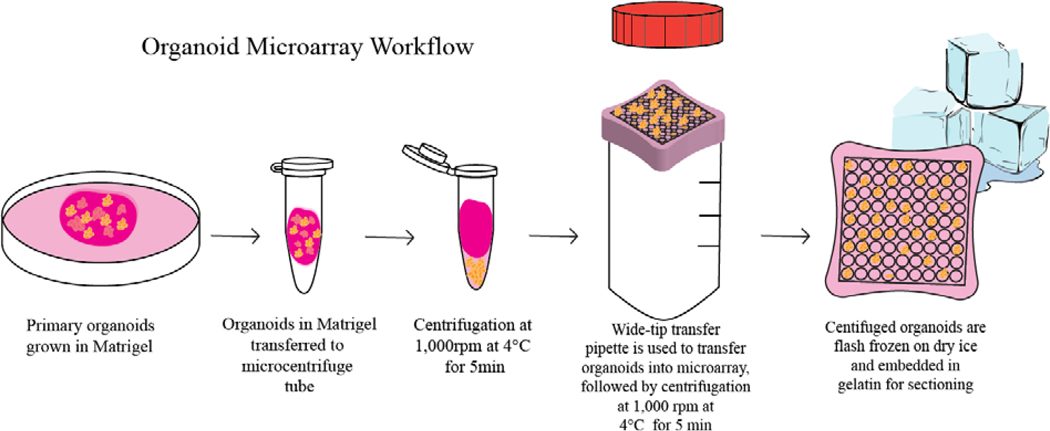
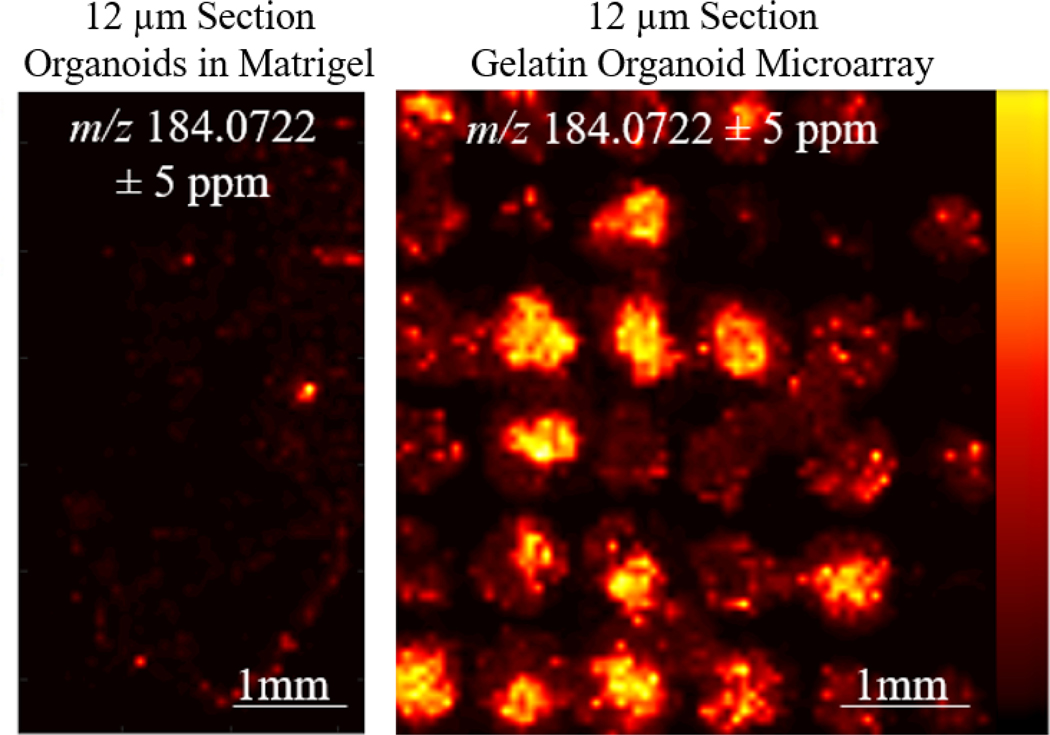
To combat the Matrigel background signals, we developed a method to centrifuge out organoids from the Matrigel at 4°C. At this temperature, Matrigel transitions from a solid into a liquid, and using centrifugation at 1000 rpm for 5 minutes at 4°C, we successfully separate the organoids from the Matrigel liquid. A wide-bore plastic transfer pipette was used to transfer organoids into the gelatin microarray. The organoids were then centrifuged by placing the Tissue Tek cryomold on top of an open 50mL conical tube. The centrifugation of organoids in the gelatin microarrays allows them to settle into the 800μm microwells. While not all the microwells were filled by organoids, the microwells in the array were all present in the same z-axis. A brief overview of this modified workflow is described in Figure 2. Additionally, because of the grid architecture of the microarray, finding organoids can be done by eye. In previous MSI studies of organoids, organoid serial sections underwent Hematoxylin & Eosin staining to locate the organoids in sectioned samples.7 The main advantage of this microarray method is the increase in organoid sampling, which is a result of aligning more organoids on the same z-axis for sectioning, as shown in Figure 3. This is a more high-throughput approach because more organoids can be analyzed in each section. Additionally, the same sample preparation steps, such as sectioning and matrix application, are duplicated across many organoids, resulting in improved reproducibility and less variability between organoids due to preparation. Day-to-day variations from the mass spectrometer and mass calibration are also minimized. Alignment of multiple organoids on the same z-axis also reduces sample preparation time, as organoids do not have to be sectioned one-by-one. Without z-plane alignment, multiple sections need to be taken at all depths of the Matrigel hydrogel to capture all the organoids present in the sample.
Figure 2. Organoid Microarray Workflow:
Primary organoids are first grown in Matrigel. Media is removed, and organoids embedded in Matrigel are washed with 1X PBS three times and transferred to a microcentrifuge tube. The microcentrifuge tube is then centrifuged at 1,000 rpm at 4°C for 5 min to pellet out the organoids in the bottom of the microcentrifuge tube. The Matrigel is removed from the top of the tube, and then using a wide-bore pipette, organoids are transferred to the gelatin microwell, which is the nest in a cryomold sitting on top of a 50mL conical tube. Centrifugation is then applied at 1,000 rpm at 4°C for min to ensure that organoids lie evenly within the microwells. Excess PBS is removed following centrifugation, and the gelatin-embedded organoids are flash frozen on dry ice and stored at −80°C prior to sectioning.
Figure 3. Gelatin Organoid Microarray:
(Right) High-throughput MSI analysis of organoids can be performed in a gelatin microarray as a direct result of alignment in the same z-axis prior to sectioning compared to direct sectioning in Matrigel (left).
Drug Treatment in the Organoids
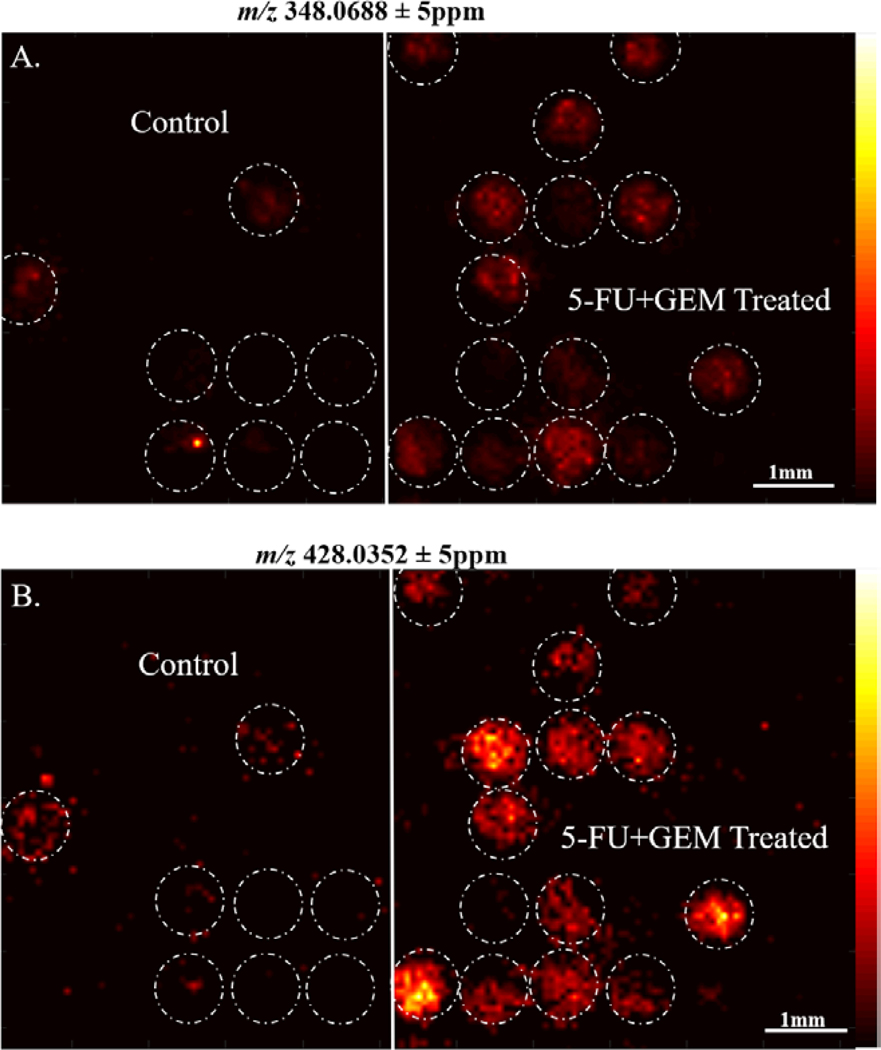
Using this microarray method, we tested organoid response to a combination of 5-Fluorouracil (5-FU) and Gemcitabine (GEM), which is a common combination therapy given to pancreatic cancer patients. Organoids do not fall into all microwells in the array, so we first performed a bisecting-k-means algorithm, shown in Supplemental Figure 1, to reveal 1) the localization of organoids within the 800μm well platform and 2) the statistical variance in detected molecular compositions between organoids. The result of this segmentation analysis is an interactive binary hierarchical tree, which contains nested sub regions based on spectra unique to these specific regions. This bisecting k-means analysis reveals the chemical heterogeneity in the organoid composition, which is expected as organoids can contain multiple types of cells with distinct molecular compositions. This data shows the segmentation of the organoids based on chemical similarity. Data was analyzed using SCiLS software to compare peaks between the control and 5-FU + GEM treated organoids. Supplemental Table 2 contains a complete list of significantly different m/z values between the control and treated organoids. In Figure 4, we demonstrate differences in intensity of m/z peaks 348.0688 and 428.0352, which are both increased in 5-FU + GEM treated organoids. These peaks tentatively match within 5ppm accuracy to adenosine monophosphate (AMP), and adenosine diphosphate (ADP), respectively. This increase in ADP and AMP in the treated organoids is likely due to patient response from the therapy from application of the combination drug treatment and could indicate a decrease in cell viability in treated organoids.43–45 Further MS/MS confirmation of these molecules would be need to confirm this. In this analysis, we were unable to detect either 5-FU or GEM parent drugs or known drug active metabolites in the organoids, as shown in Supplemental Figure 2. The lack of signal of GEM and 5-FU in the organoid composition can likely be attributed to removal of the GEM drug after 24 hours, and the metabolism and/or excretion of the 5-FU drug.37 We did detect an accumulation of excretion of the inactive metabolite, 5-flurouridine (FUdr), in the areas adjacent to the organoids, shown in Supplemental Figure 2. Despite not detecting the parent drugs and active drug metabolites in the organoids, we do identify the presence of significantly changed metabolites between treated and control organoids that correlate with the effectiveness of this treatment on these organoids, and drug excretion products surrounding the organoids. Further biological characterization of this patient response indicates that these organoids are responsive to 5-FU+GEM combination therapy regimen.46
Figure 4. Chemotherapy Treatment Alters Organoid Metabolites:
A. m/z 348.0688, tentatively identified as AMP, is significantly increased in 5-FU+GEM treated organoids versus controls B. m/z 428.0352, tentatively identified as ADP, is also significantly increased in 5-FU+GEM treated organoids versus controls.
Expanding into Smaller 200μm Microwell Microarrays
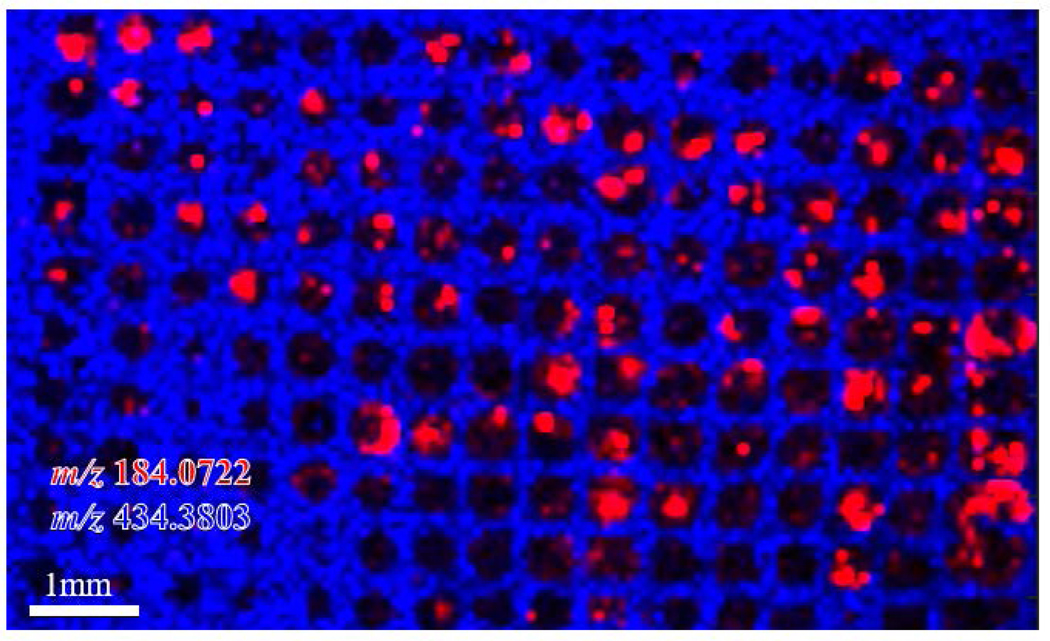
While 800μm microarrays are ideal for aligning the organoids on the same z-axis for sectioning, it is possible for multiple smaller organoids to cluster and appear as a single organoid. We next investigated whether the use of 200 μm microarrays could prevent this clustering. In this experiment, organoids from the same patient were isolated closer to the time of passaging, resulting in smaller sized organoids. The same workflow shown in Figure 2 was applied using 200μm well microarrays. The detected organoids are shown in 200μm wells in Figure 5, shown in red as m/z 184.0722. The microarray is shown as peak m/z 434.3803, which is localized to the microarray. This peak is specific to the gelatin background, but does not appear in the organoids. The minimum spatial resolution used on the MALDI Orbitrap is 75μm, while this method of using small microarrays for organoids (<200μm) would be best applied using instrumentation capable of handling much higher spatial resolutions (5–10μm) for improved visualization of the spatial heterogeneity within small organoids. Although this method does not necessarily ensure that only one organoid will be located in each microwell, the organization of small organoids into a microarray makes them easy to identify. This smaller microarray approach can be applied for smaller organoid sizes, as some tumor organoids are not capable of forming organoids as large as 800μm, even with longer incubation and maturation times.
Figure 5. 200μm Microwell Organoid Microarray:
m/z 184.0722, a phosphatidyl choline head group, is shown in red to demonstrate the location of the small pancreatic cancer organoids. m/z 434.3803, shown in blue, is a background signal from gelatin microarray only. This overlay analysis of the organoids and the microarray demonstrates that organoids are aligned on similar z-axis for sectioning and hundreds of organoids can be imaged in a microarray by MSI.
Conclusion
This gelatin microarray technology enables high-throughput alignment of tens to hundreds of organoids in a single section for MSI analysis. This technique eliminates background noise by centrifuging organoids out of Matrigel, which shows high background signal in MSI analysis and contains similar interfering peaks with the organoids. These improvements in organoid sample preparation can be easily implemented using commercially available microwells or customized microfluidic technologies for easy implementation in labs that routinely perform MSI. For applications of MSI analysis of organoids in pre-clinical drug discovery and personalized medicine, the improved throughput and reduced sample preparation time resulting from the methods described in this manuscript will be important to reach timely and informed, clinically relevant chemotherapy decisions using human tissue.
Supplementary Material
Acknowledgement:
We would like to thank the Translation Science BioCore BioBank and its staff at the University of Wisconsin Carbone Cancer Center for the collection of donated patient tissue. The authors would also like to acknowledge Joseph Vecchi for his recommendation of the commercially available 3D Petri Dish® microwell mold for culturing spheroids, available from Sigma Aldrich.
Financial Support: This work was supported in part by Stand Up to Cancer (SU2C-AACR-IG-08-16, SU2CAACR-PS-18) awarded to MCS, and the University of Wisconsin Carbone Cancer Center (233-AAC9675), the UWCCC Pancreatic Cancer Taskforce, and NIH R56MH110215 for Funding. The MALDI Orbitrap instrument was purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531). LL acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
References
- 1.Clinton J; McWilliams-Koeppen P, Initiation, Expansion, and Cryopreservation of Human Primary Tissue-Derived Normal and Diseased Organoids in Embedded Three-Dimensional Culture. Curr Protoc Cell Biol 2019, 82 (1), e66. [DOI] [PubMed] [Google Scholar]
- 2.Drost J; Clevers H, Organoids in cancer research. Nature Reviews Cancer 2018, 18 (7), 407–418. [DOI] [PubMed] [Google Scholar]
- 3.Romero-Calvo I; Weber CR; Ray M; Brown M; Kirby K; Nandi RK; Long TM; Sparrow SM; Ugolkov A; Qiang W; Zhang Y; Brunetti T; Kindler H; Segal JP; Rzhetsky A; Mazar AP; Buschmann MM; Weichselbaum R; Roggin K; White KP, Human Organoids Share Structural and Genetic Features with Primary Pancreatic Adenocarcinoma Tumors. Mol Cancer Res 2019, 17 (1), 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai S; McOlash L; Palen K; Johnson B; Duris C; Yang Q; Dwinell MB; Hunt B; Evans DB; Gershan J; James MA, Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018, 18 (1), 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beshiri ML; Tice CM; Tran C; Nguyen HM; Sowalsky AG; Agarwal S; Jansson KH; Yang Q; McGowen KM; Yin J; Alilin AN; Karzai FH; Dahut WL; Corey E; Kelly K, A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin Cancer Res 2018, 24 (17), 4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weeber F; van de Wetering M; Hoogstraat M; Dijkstra KK; Krijgsman O; Kuilman T; Gadellaa-van Hooijdonk CG; van der Velden DL; Peeper DS; Cuppen EP; Vries RG; Clevers H; Voest EE, Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A 2015, 112 (43), 13308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X; Flinders C; Mumenthaler SM; Hummon AB, MALDI Mass Spectrometry Imaging for Evaluation of Therapeutics in Colorectal Tumor Organoids. J Am Soc Mass Spectrom 2018, 29 (3), 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii M; Shimokawa M; Date S; Takano A; Matano M; Nanki K; Ohta Y; Toshimitsu K; Nakazato Y; Kawasaki K; Uraoka T; Watanabe T; Kanai T; Sato T, A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18 (6), 827–38. [DOI] [PubMed] [Google Scholar]
- 9.Salahudeen AA; Kuo CJ, Toward recreating colon cancer in human organoids. Nat Med 2015, 21 (3), 215–6. [DOI] [PubMed] [Google Scholar]
- 10.Moreira L; Bakir B; Chatterji P; Dantes Z; Reichert M; Rustgi AK, Pancreas 3D Organoids: Current and Future Aspects as a Research Platform for Personalized Medicine in Pancreatic Cancer. Cell Mol Gastroenterol Hepatol 2018, 5 (3), 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boj SF; Hwang CI; Baker LA; Chio II; Engle DD; Corbo V; Jager M; Ponz-Sarvise M; Tiriac H; Spector MS; Gracanin A; Oni T; Yu KH; van Boxtel R; Huch M; Rivera KD; Wilson JP; Feigin ME; Öhlund D; Handly-Santana A; Ardito-Abraham CM; Ludwig M; Elyada E; Alagesan B; Biffi G; Yordanov GN; Delcuze B; Creighton B; Wright K; Park Y; Morsink FH; Molenaar IQ; Borel Rinkes IH; Cuppen E; Hao Y; Jin Y; Nijman IJ; Iacobuzio-Donahue C; Leach SD; Pappin DJ; Hammell M; Klimstra DS; Basturk O; Hruban RH; Offerhaus GJ; Vries RG; Clevers H; Tuveson DA, Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160 (1–2), 324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SH; Hu W; Matulay JT; Silva MV; Owczarek TB; Kim K; Chua CW; Barlow LJ; Kandoth C; Williams AB; Bergren SK; Pietzak EJ; Anderson CB; Benson MC; Coleman JA; Taylor BS; Abate-Shen C; McKiernan JM; Al-Ahmadie H; Solit DB; Shen MM, Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173 (2), 515–528.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roelofs C; Hollande F; Redvers R; Anderson RL; Merino D, Breast tumour organoids: promising models for the genomic and functional characterisation of breast cancer. Biochem Soc Trans 2019, 47 (1), 109–117. [DOI] [PubMed] [Google Scholar]
- 14.Sachs N; de Ligt J; Kopper O; Gogola E; Bounova G; Weeber F; Balgobind AV; Wind K; Gracanin A; Begthel H; Korving J; van Boxtel R; Duarte AA; Lelieveld D; van Hoeck A; Ernst RF; Blokzijl F; Nijman IJ; Hoogstraat M; van de Ven M; Egan DA; Zinzalla V; Moll J; Boj SF; Voest EE; Wessels L; van Diest PJ; Rottenberg S; Vries RGJ; Cuppen E; Clevers H, A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172 (1–2), 373–386.e10. [DOI] [PubMed] [Google Scholar]
- 15.Yan HHN; Siu HC; Law S; Ho SL; Yue SSK; Tsui WY; Chan D; Chan AS; Ma S; Lam KO; Bartfeld S; Man AHY; Lee BCH; Chan ASY; Wong JWH; Cheng PSW; Chan AKW; Zhang J; Shi J; Fan X; Kwong DLW; Mak TW; Yuen ST; Clevers H; Leung SY, A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23 (6), 882–897.e11. [DOI] [PubMed] [Google Scholar]
- 16.Junttila MR; de Sauvage FJ, Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501 (7467), 346–54. [DOI] [PubMed] [Google Scholar]
- 17.Tiriac H; Belleau P; Engle DD; Plenker D; Deschênes A; Somerville TDD; Froeling FEM; Burkhart RA; Denroche RE; Jang GH; Miyabayashi K; Young CM; Patel H; Ma M; LaComb JF; Palmaira RLD; Javed AA; Huynh JC; Johnson M; Arora K; Robine N; Shah M; Sanghvi R; Goetz AB; Lowder CY; Martello L; Driehuis E; LeComte N; Askan G; Iacobuzio-Donahue CA; Clevers H; Wood LD; Hruban RH; Thompson E; Aguirre AJ; Wolpin BM; Sasson A; Kim J; Wu M; Bucobo JC; Allen P; Sejpal DV; Nealon W; Sullivan JD; Winter JM; Gimotty PA; Grem JL; DiMaio DJ; Buscaglia JM; Grandgenett PM; Brody JR; Hollingsworth MA; O’Kane GM; Notta F; Kim E; Crawford JM; Devoe C; Ocean A; Wolfgang CL; Yu KH; Li E; Vakoc CR; Hubert B; Fischer SE; Wilson JM; Moffitt R; Knox J; Krasnitz A; Gallinger S; Tuveson DA, Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 2018, 8 (9), 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Öhlund D; Handly-Santana A; Biffi G; Elyada E; Almeida AS; Ponz-Sarvise M; Corbo V; Oni TE; Hearn SA; Lee EJ; Chio II; Hwang CI; Tiriac H; Baker LA; Engle DD; Feig C; Kultti A; Egeblad M; Fearon DT; Crawford JM; Clevers H; Park Y; Tuveson DA, Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017, 214 (3), 579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh AJ; Castellanos JA; Nagathihalli NS; Merchant NB; Skala MC, Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response. Pancreas 2016, 45 (6), 863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchberger AR; DeLaney K; Johnson J; Li L, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal Chem 2018, 90 (1), 240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chughtai K; Heeren RM, Mass spectrometric imaging for biomedical tissue analysis. Chem Rev 2010, 110 (5), 3237–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaysse PM; Heeren RMA; Porta T; Balluff B, Mass spectrometry imaging for clinical research - latest developments, applications, and current limitations. Analyst 2017, 142 (15), 2690–2712. [DOI] [PubMed] [Google Scholar]
- 23.Bergmann S; Lawler SE; Qu Y; Fadzen CM; Wolfe JM; Regan MS; Pentelute BL; Agar NYR; Cho CF, Blood-brain-barrier organoids for investigating the permeability of CNS therapeutics. Nat Protoc 2018, 13 (12), 2827–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lietz CB; Gemperline E; Li L, Qualitative and quantitative mass spectrometry imaging of drugs and metabolites. Adv Drug Deliv Rev 2013, 65 (8), 1074–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo C; Lewis EEL; Flint L; Clench MR, Mass Spectrometry Imaging of 3D Tissue Models. Proteomics 2018, 18 (14), e1700462. [DOI] [PubMed] [Google Scholar]
- 26.Haeberle L; Steiger K; Schlitter AM; Safi SA; Knoefel WT; Erkan M; Esposito I, Stromal heterogeneity in pancreatic cancer and chronic pancreatitis. Pancreatology 2018. [DOI] [PubMed] [Google Scholar]
- 27.Kleinman H; McGarvey M; Hassell J; Star V; Cannon F; Laurie G; Martin G, Basement membrane complexes with biological activity. Biochemistry, 1986; Vol. 2, pp 312–8. [DOI] [PubMed] [Google Scholar]
- 28.Kleinman HK; Martin GR, Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol 2005, 15 (5), 378–86. [DOI] [PubMed] [Google Scholar]
- 29.Vukicevic S; Kleinman HK; Luyten FP; Roberts AB; Roche NS; Reddi AH, Identification of multiple active growth factors in basement membrane Matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res 1992, 202 (1), 1–8. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y; Lukacova V; Reindl K; Balaz S, Quantitative characterization of binding of small molecules to extracellular matrix. J Biochem Biophys Methods 2006, 67 (2–3), 107–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serban MA; Prestwich GD, Modular extracellular matrices: solutions for the puzzle. Methods 2008, 45 (1), 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinman HK; McGarvey ML; Hassell JR; Star VL; Cannon FB; Laurie GW; Martin GR, Basement membrane complexes with biological activity. Biochemistry 1986, 25 (2), 312–8. [DOI] [PubMed] [Google Scholar]
- 33.Lee MY; Kumar RA; Sukumaran SM; Hogg MG; Clark DS; Dordick JS, Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci U S A 2008, 105 (1), 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Achilli TM; McCalla S; Meyer J; Tripathi A; Morgan JR, Multilayer spheroids to quantify drug uptake and diffusion in 3D. Mol Pharm 2014, 11 (7), 2071–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabadi PK; Vantangoli MM; Rodd AL; Leary E; Madnick SJ; Morgan JR; Kane A; Boekelheide K, Into the depths: Techniques for in vitro three-dimensional microtissue visualization. Biotechniques 2015, 59 (5), 279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napolitano AP; Dean DM; Man AJ; Youssef J; Ho DN; Rago AP; Lech MP; Morgan JR, Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. Biotechniques 2007, 43 (4), 494, 496–500. [DOI] [PubMed] [Google Scholar]
- 37.Shibata T; Ebata T; Fujita K; Shimokata T; Maeda O; Mitsuma A; Sasaki Y; Nagino M; Ando Y, Optimal dose of gemcitabine for the treatment of biliary tract or pancreatic cancer in patients with liver dysfunction. Cancer Sci 2016, 107 (2), 168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bokhart MT; Nazari M; Garrard KP; Muddiman DC, MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J Am Soc Mass Spectrom 2018, 29 (1), 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robichaud G; Garrard KP; Barry JA; Muddiman DC, MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 2013, 24 (5), 718–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guijas C; Montenegro-Burke JR; Domingo-Almenara X; Palermo A; Warth B; Hermann G; Koellensperger G; Huan T; Uritboonthai W; Aisporna AE; Wolan DW; Spilker ME; Benton HP; Siuzdak G, METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal Chem 2018, 90 (5), 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gill EL; Yost RA; Vedam-Mai V; Garrett TJ, Precast Gelatin-Based Molds for Tissue Embedding Compatible with Mass Spectrometry Imaging. Anal Chem 2017, 89 (1), 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter CL; Jones JW; Farese AM; MacVittie TJ; Kane MA, Inflation-Fixation Method for Lipidomic Mapping of Lung Biopsies by Matrix Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging. Anal Chem 2016, 88 (9), 4788–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon HY; Kim IK; Kang J; Sohn SK; Lee KY, In Vitro Adenosine Triphosphate-Based Chemotherapy Response Assay as a Predictor of Clinical Response to Fluorouracil-Based Adjuvant Chemotherapy in Stage II Colorectal Cancer. Cancer Res Treat 2016, 48 (3), 970–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JS; Kim JK; Yoon DS, Correlation of Early Recurrence With In Vitro Adenosine Triphosphate Based Chemotherapy Response Assay in Pancreas Cancer With Postoperative Gemcitabine Chemotherapy. J Clin Lab Anal 2016, 30 (6), 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalski CW; Erkan M; Sauliunaite D; Giese T; Stratmann R; Sartori C; Giese NA; Friess H; Kleeff J, Ex vivo chemosensitivity testing and gene expression profiling predict response towards adjuvant gemcitabine treatment in pancreatic cancer. Br J Cancer 2008, 99 (5), 760–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharick JT; Walsh CM; Sprackling CM; Pasch CA; Parikh AA; Matkowskyj KA; Deming DA; Skala MC, Optical Metabolic Imaging of Heterogeneous Drug Response in Pancreatic Cancer Patient Organoids. bioRxiv 2019, 542167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.