Abstract
Microglia are the primary innate immune cells in the CNS. In the healthy brain, they exhibit a unique molecular homeostatic ‘signature’, consisting of a specific transcriptional profile and surface protein expression pattern, which differs from that of tissue macrophages. In recent years, there have been a number of important advances in our understanding of the molecular signatures of homeostatic microglia and disease-associated microglia that have provided insight into how these cells are regulated in health and disease and how they contribute to the maintenance of the neural environment.
Our understanding of the origin and functions of microglia has grown to such an extent that it is as if a new CNS cell has been described1–5. These advances have major implications for understanding normal CNS function and have opened up new avenues to understand the role of microglia in disease. Most importantly, they have created the opportunity to consider ways in which microglia may be imaged and targeted for the treatment of disease.
The origin of microglia has now been clearly defined and shown to relate to the early colonization of the CNS by mesodermal progenitors6 that arise from the yolk sac7,8 (Box 1). It is now recognized that monocytes and tissue macrophages are not, as had been previously proposed9, microglia progenitors in either health or disease10,11, and that, in adulthood, microglia are an independent self-renewing population12–14 (FIG. 1a). Human microglia turn over at a yearly median rate of 28% and live, on average, for 4.2 years. Thus, most of the microglial population is renewed several times over the course of a lifetime15. In support of the importance of microglial self-renewal, a recent study demonstrated that the repopulated microglia that rapidly replenish the adult brain’s microglial population after microglial depletion are solely derived from the proliferation of residual microglia and not from newly generated progenitors16. However, it has also been demonstrated that, in some circumstances, peripherally derived macrophages can replace depleted microglia with cells that maintain their own unique identity (distinct from that of microglia)17 and that these cells may play a distinct role in the progression or resolution of neurological diseases (FIG. 1b). In support of this idea, it was shown that abrogation of transforming growth factor-βl (TGFβ1) signalling in peripherally recruited myeloid cells, which enter the brain after microglial depletion, led to rapid onset of a progressive and fatal demyelinating motor disease18.
Box 1 |. Key features of mammalian microglial development.
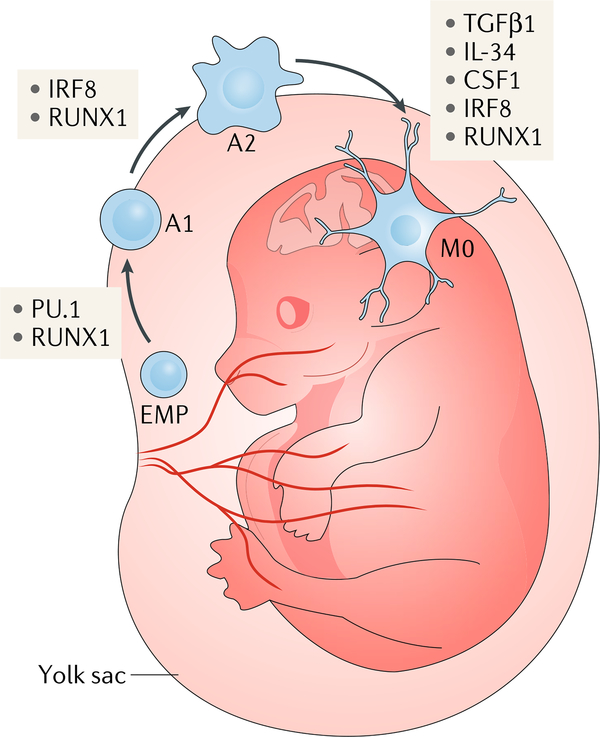
Fate mapping of runt-related transcription factor 1 (RuNX1)-expressing haematopoietic stem cell (HSC)-derived precursors in mice showed that microglia do not arise from these cells but instead originate between embryonic day (E)7 and E7.5 (REF.8) from early, uncommitted erythromyeloid progenitor (EmP)-derived yolk sac macrophages that express mast/stem cell growth factor receptor Kit (KIT)10. The EmP-derived yolk sac macrophages enter the embryonic brain and take up residence before the differentiation of other cell types. HSCs are detected in bone marrow only at E10.5, whereas microglial precursors expressing the markers adhesion G protein-coupled receptor E1 (ADGRE1) and αm integrin (ITGAm) begin to appear in the brain at E9.5 (REFS2,212,213). Thus, microglial precursors appear in the developing brain earlier than HSCs, indicating different origins of these two cell types213. murine induced pluripotent stem cell (iPSC)-derived yolk sac macrophage-like cells adopt a microglia-like morphology during co-culture with neurons and can engraft into the brain and differentiate into microglia-like cells214, showing that they have microglial potential that is determined by the brain milieu.
Microglial development is independent of the transcription factor MYB215, which controls HSC differentiation, but is dependent on the transcription factors RUNX1, PU.1 and interferon regulatory factor 8 (IRF8)8,10,216. During development, PU.1 and RUNX1 drive the transition of EMPs to the immature A1 state (cells that express CD45 and low levels of KIT but do not express CX3C chemokine receptor 1 (CX3CR1) or ADGRE1) at E8 in mice (see the figure). IRF8 and RUNX1 are required for the maturation of A1 cells to A2 cells, which start to express CX3CR1 and ADGRE1 and lose KIT expression at E9 (REF.10). The cytokines interleukin-34 (IL-34), colony-stimulating factor 1 (CSF1) and transforming growth factor-β1 (TGFβ1) are also essential for microglial development25,68. Human pluripotent stem cells can be differentiated into microglia-like cell subsets, including those resembling homeostatic (M0) microglia, via the induction of CSF1, IL-34 and TGFβ1 signalling37,38. Thus, brain environmental cues and transcriptional regulation define microglia as a unique population separate from myeloid cells.
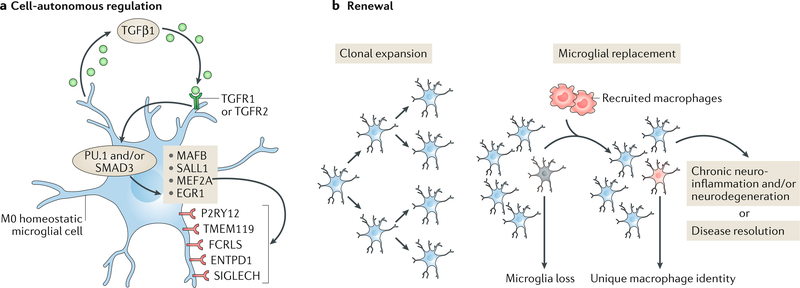
Once microglia establish themselves in the brain, they sustain the microglial pool via local clonal expansion throughout life12–14,217. However, peripherally derived macrophages can replace dying microglia (while maintaining their own unique identity)17,18 (FIG. 1).
Fig. 1 |. Regulation of microglia homeostasis.
a | Microglia are maintained in a cell-autonomous fashion via transforming growth factor-β1 (TGFβ1) signalling, which drives the transcriptional regulation (via the transcription factors PU.1 and/or mothers against decapentaplegic homologue 3 (SMAD3)) of genes that include those encoding the transcription factor MafB (MAFB), Sal-like protein 1 (SALL1), myocyte-specific enhancer factor 2A (MEF2A) and early growth response protein 1 (EGR1)25,75. Several unique surface receptors — P2Y purinoceptor 12 (P2RY12), transmembrane protein 119 (TMEM119), Fc receptor-like S, scavenger receptor (FCRLS), ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1; also known as CD39) and sialic acid binding Ig-like lectin H, isoform CRA_a (SIGLECH) — identify microglia. P2RY12 senses damaged tissue through the detection of extracellular nucleotides53, whereas ENTPD1 controls microglial process ramification218 and its loss is associated with handling-induced seizures in mice219. The role of the other receptors is as yet unknown. b | Microglial renewal occurs by local clonal expansion13. Furthermore, under certain conditions, lost microglial populations can be replaced with peripherally derived macrophages that are recruited to the CNS. These cells maintain a unique molecular identity and have distinct functional roles as compared with microglia17, although TMEM119 and other microglial markers may also be expressed by these macrophages220. Loss of TGFβ1 signalling in these macrophages has been shown to lead to a progressive and fatal demyelinating motor disease18, suggesting that the manipulation of engrafted myeloid cells has therapeutic potential in neurodegenerative disease. TGFR1 and TGFR2, TGFβ receptors type 1 and 2, respectively.
Monocytes.
Mononuclear cells that are derived from the bone marrow and circulate in the bloodstream. They pass into the body tissues, where they differentiate into various types of macrophages.
Tissue macrophages.
Tissue-resident macrophages that are distributed throughout the body, including microglia, liver Kupffer cells, lung alveolar cells and splenic and peritoneal macrophages. They are established before birth and maintain themselves during adulthood independent of replenishment by blood monocytes. They become mobile when stimulated by inflammation and migrate to affected areas.
Microglia exist in resting or activated states depending on the inflammatory milieu, which differs in the healthy CNS and in various disease states19–22. In addition, microglia exhibit both phenotypic and functional plasticity in healthy and diseased brains. In peripheral macrophages (the innate immune cells that are a homologue of brain microglia), cell phenotypes have been determined by the varied expression of cell surface receptors by these cells23. Similarly, microglial phenotypes were, in the past, characterized by the presence of particular cell surface molecules and the expression of specific sets of cytokines and were classified as either M1-like (exhibiting pro-inflammatory signalling and neurotoxicity) or M2-like (participating in the resolution of inflammation)24. However, it is now clear that this simplistic view of microglial phenotypes does not adequately describe the complex physiology of microglial cells5. The characterization of microglial diversity in health and disease has therefore been redefined in both mice25–29 and humans30–32 with the help of newly developed technologies, including RNA-sequencing, quantitative proteomics and epigenetic studies.
Cytokines.
Proteins secreted by a cell that act in a paracrine or autocrine fashion and affect the behaviour of other cells.
RNA-sequencing.
An unbiased whole transcriptome deep-sequencing technology that uses next-generation sequencing to detect novel or known features and to quantify RNA activity.
Epigenetic studies.
Investigations of inheritance by mechanisms other than those that involve changes to the underlying DNA sequence. Types of epigenetic mechanisms include DNA methylation, post-translational histone modifications and non-coding RNAs.
In this Review, we discuss the features that define homeostatic microglia, including their transcriptional and surface protein signatures, and describe the factors that regulate these signatures. We relate the physiological functions of microglia, including their roles in synaptogenesis, trophic support, chemotaxis and neurogenesis, to their phenotypic diversity. Finally, we consider what microglial signatures can reveal about the role of these cells in ageing and disease as well as the potential impact of understanding these signatures in the development of therapeutic strategies to target microglia for the treatment of CNS disease.
Homeostatic microglia
Microglial transcriptional signature
Until recently, the molecular and functional characteristics of homeostatic microglia had not been identified, which had impeded the field as there was no basis to understand how microglia function in health and how they change and react to disease. A further complication was the lack of understanding of whether (and how) microglia fundamentally differ from peripheral myeloid cells. Taking advantage of new technological advances, including RNA-sequencing, quantitative proteomics, epigenetics and bioinformatics, several groups simultaneously identified a unique transcriptional signature for homeostatic microglia in adult mice25,27–29. Specifically, microglia — but not peripheral myeloid cells — were shown to express P2Y purinoceptor 12 (P2ry12), transmembrane protein 119 (Tmem119), sialic acid binding Ig-like lectin H (Siglech), probable G protein coupled receptor 34 (Gpr34), suppressor of cytokine signalling 3 (Socs3), β-hexosaminidase subunit β (Hexb), olfactomedin-like protein 3 (Olfml3) and Fc receptor-like S, scavenger receptor (Fcrls). Several of these microglial genes were also observed in human microglia, including P2RY12 and TMEM119 (REFs25,33–36). Identification of this homeostatic microglial transcriptional signature has enabled the development of robust tools, including microglia-specific antibodies and transgenic mice in which microglia are specifically targeted, the generation of microglia from induced pluripotent stem cells (iPSCs)37–39 and the investigation of microglial biology in health and disease40.
Peripheral myeloid cells.
Cells originating from haematopoietic cells, as opposed to tissue-resident myeloid cells, such as microglia.
Induced pluripotent stem cells.
(iPsCs). stem cells derived from skin or blood that have been reprogrammed into an embryonic-like pluripotent state.
Microglial surface protein signature
Microglia express macrophage markers, including adhesion G protein-coupled receptor E1 (ADGRE; also known as cell surface glycoprotein F4/80), crystallizable fragment (Fc) receptors, and αM integrin (ITGAM; also known as CD11b), in mice41 and in humans42. Furthermore, murine microglia express a number of other macrophage markers, such as the macrophage colony-stimulating factor 1 receptor (CSF1R; also known as CD115), the inhibitory immune receptor cell surface glycoprotein CD200 receptor 1 (CD200R1), the surface enzyme tyrosine-protein phosphatase non-receptor type substrate 1 (SIRPA; also known as CD172a), the fractalkine receptor CX3C-chemokine receptor 1 (CX3CR1) and the calcium-binding protein allograft inflammatory factor 1 (AIF1; also known as IBA1)43.
Although many of these proteins are expressed by all macrophages, the levels of expression of some protein markers can be used to distinguish microglia from related cell types. For example, microglia express lower levels of receptor-type tyrosine-protein phosphatase C (PTPRC; also known as CD45) than monocytes, allowing these two cell types to be discriminated, whereas the expression of scavenger receptor cysteine-rich type 1 protein M130 (CD163) by microglia enables their distinction from perivascular macrophages35,44–47. Finally, sialoadhesin (SIGLEC1; also known as CD169) is a marker for mature dendritic cells and macrophages in both mice and humans48 and has been proposed as a marker for invading monocyte-derived macrophages in the CNS as it is not expressed in microglia in health or disease49–52. However, this method of distinguishing between cell types relies on relative surface marker expression as assessed by flow cytometry, which can have some limitations. For example, CD45 surface expression is known to be upregulated on microglia associated with amyloid-β (Aβ) plaques in the APP/PS1 mouse model of Alzheimer disease (AD)52.
Microglia can also be distinguished from related cell types by their binding to specific antibodies. Vaccination of rats with adult microglia yielded two microglial-specific antibodies — an ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1; also known as CD39) antibody and 4D4, an antibody for which the protein specificity has not yet been identified — the binding of which could be used to distinguish between non-overlapping populations of monocytes and resident microglia in mice49. Furthermore, on the basis of identification of the unique transcriptomic signature of homeostatic microglia (see above)25,27–29, several additional antibodies that label microglia have been developed and characterized, including those recognizing receptors involved in microglial maintenance, such as FCRLS25, P2RY12 (REFs25,53), TMEM119 (REFs33,35) and SIGLECH54 (FIG. 1a).
Microglial heterogeneity and plasticity
Despite the existence of the molecular signatures described above, the presence of microglial heterogeneity has been recognized for many years. Initial descriptions noted microglia with different morphologies55 and the differential responses of microglia to various stimuli22 and brain irradiation56. Of note, in a number of different CNS areas, it has been observed that not all microglia express triggering receptor expressed on myeloid cells 2 (TREM2)57, an immune receptor that is found in brain microglia and, when mutated, is a risk factor for non-familial AD58. A study in which a genome-wide analysis of gene expression in microglia from different brain regions across the adult lifespan of the mouse was performed found that there are region-specific transcriptional profiles and age-dependent regional variability in gene expression59. Interestingly, in the young-adult brain, microglia in different brain regions exhibited differences in the expression of genes in bioenergetic and immunoregulatory pathways59. The findings further suggested that cerebellar and hippocampal microglia exist in a more ‘immune-vigilant’ state than microglia in forebrain regions. Cerebellar and hippocampal microglia were found to undergo a region-specific response to age-related neurodegeneration59 that was associated with reduced expression of genes associated with TGFβ signalling52. Indeed, another study confirmed that microglia in the white matter of the cerebellum undergo substantial changes during ageing, including alterations in morphology and gene expression60. However, a study that employed high-dimensional single-cell cytometry to characterize myeloid subsets in the brain in health, ageing and in mouse models of AD and multiple sclerosis (MS) found that all microglia are homogenously affected in neuroinflammatory diseases61.
The diversity of microglial phenotype and function enables them to adapt and face challenges. For example, during inflammation, microglia change from a ‘bushy’ shape with extended processes to become amoeboid and produce inflammatory factors62,63. Microglial plasticity and its regulation are crucial features of their biology because these cells have several functions in the brain, including both normal brain maintenance and response to injury (see below)64–67. However, microglial plasticity in inflammatory conditions is a more complex phenomenon than was assumed by initial hypotheses that were based on macrophage morphoplasticity alone23. It is now known that the regulation of microglia involves several different classes of transcription factors that interact to select and activate various regulatory elements.
Mechanisms of homeostatic microglia regulation
In order to understand how microglial plasticity comes about, we must first understand the mechanisms that establish and maintain the phenotype of homeostatic microglia. For example, it has been shown that the cytokine CSF1, acting via its receptor CSF1R, is important for microglial development8. Consistent with this, it has been demonstrated that interleukin-34 (IL-34), which is an additional ligand for CSF1R, promotes the tissue-specific signalling pathway that is required for the differentiation of myeloid cells that populate the skin and microglia in the CNS68. In addition, it has been shown that tonic activity of potassium channel subfamily K member 13 (KCNK13; also known as THIK1) promotes microglial ramification and surveillance of the brain parenchyma69. Given that THIK1 is also required for the release of IL-1β by microglia, this finding may have therapeutic implications: these may include stimulation of THIK1 signalling to induce microglial motility and eliminate Aβ plaque deposition in AD or inhibition of THIK1 signalling to reduce microglial-mediated damage to bystander neurons in other diseases.
Studies of factors that regulate microglia have revealed a number of molecular mechanisms that regulate microglial transcription programmes. The transcription factor PU.1 serves as a master regulator of myeloid cells70. PU.1 plays a crucial role in determining macrophage lineages and microglial genesis and is a major factor in selecting the set of enhancers expressed by microglia. Of note, PU.1-deficient mice have several haematopoietic abnormalities, including the loss of B cell and macrophage populations71. PU.1 deficiency is also associated with loss of microglia in mice and in zebrafish72. PU.1 binds to the CSF1R promoter to drive gene transcription73 and to most enhancers in both mouse and human microglia. PU.1 has an important role as an essential lineage-determining transcription factor (LDTF)74. Motif analysis of PU.1 binding sites has shown enrichment for consensus sequences for mothers against decapentaplegic homologue 3 (SMAD3), myocyte-specific enhancer factor 2A (MEF2A) and transcription factor MafB (MAFB), suggesting that these transcription factors cooperate with PU.1 to establish microglial-specific enhancer profiles75. Consistent with these findings, PU.1 binds microglial-specific enhancer regions of genes encoding the MEF2 family of transcription factors, which are expressed in microglia but not in other peripheral immune cells76. These factors partner with PU.1 to establish the microglial-specific molecular signature that is described above.
Enhancers.
Short regions of DNA, typically occupied by multiple transcription factors, that are sufficient to drive expression of a gene with temporal and/or cell-type specificity.
Lineage-determining transcription factor.
(LDTF). Transcription factors that create regions of open chromatin, which enables the recruitment of secondary transcription factors required for cell-type-specific gene expression.
In addition to establishing microglial transcriptional signatures, these transcription factors have a number of additional roles in the maintenance of the homeostatic microglial phenotype. In one study, it was reported that the transcription factor MEF2C acts in homeostatic microglia to suppress the inflammatory response and that its expression declines during ageing in an interferon (IFN)-1-dependent manner77. This study showed that loss of microglial MEF2C was linked to downregulation of CX3CR1, a molecule that is crucial for restraining immune responses in microglia78,79. On the other hand, MAFB is part of a family of transcription factors that binds to the transcription factor Maf (MAF) recognition element and forms heterodimers with the transcription factors MAF, AP-1 (JUN) and proto-oncogene c-Fos (FOS). In macrophages, MAFB promotes an anti-inflammatory phenotype, and its loss exacerbates inflammatory conditions80 and controls self-renewal81. Expression of MAFB increases during development, and its deletion in transgenic mice disrupts gene expression in adult microglia to a greater extent than it does in immature microglia82. MAFB also antagonizes granulocyte-macrophage colony-stimulating factor (GM-CSF), which represses microglial differentiation83. Furthermore, its expression is downregulated in the superoxide dismutase 1 (SOD1) mouse model of amyotrophic lateral sclerosis (ALS), in which inflammation is increased52,84. In primary microglia isolated from the newborn mouse brain, MAFB was shown to regulate cell division by dampening the response to CSF1 (REF.83). Thus, MAFB acts to maintain a healthy microglial phenotype by inhibiting both inflammatory and proliferative responses.
In addition to the factors described above, there are a number of secondary transcription factors involved in microglial homeostatic regulation. These include the SMAD proteins, which are TGFβ1-dependent and contribute to establishing transcription of specific microglial genes75. Furthermore, the development of microglia is dependent on CSF1R, which transmits signals upon activation by its ligands CSF1 and IL-34 and serves as a survival factor68. It has also been shown that the development of microglia relies on the secondary transcription factor IFN regulatory factor 8 (IRF8), which is essential for the development of the mature microglia precursors known as A2 cells10 (Box. 1). Another secondary transcription factor, Sal-like protein 1 (SALL1), regulates cortical neurogenesis and is expressed in human and mouse microglia25. Mutations in SALL1 result in neural or behavioural abnormalities in humans85 and abnormal microglial morphology and downregulation of the microglial homeostatic signature in mice86, suggesting that SALL1 inhibits reactive microglia and promotes a physiological surveillant phenotype. At this time, mechanisms connecting extrinsic signals to transcriptional responses remain poorly defined.
Finally, it has been shown that the absence of TGFβ1, a factor known to be required for the in vitro development of the molecular signature characteristic of adult microglia, in the CNS results in a paralytic neurologic disease in mice25. Importantly, TGFβ1 signalling has also been shown to be inhibited in microglia that are associated with neurodegeneration, in which the homeostatic signature is also suppressed52, and an additional study has further highlighted the critical function of TGFβ1 in preventing microglia and/or macrophage-mediated CNS pathology18. Similarly, CNS-conditional Tgfb1−/− mice exhibit motor deficits, a loss of homeostatic microglia and the appearance of peripherally derived monocytes in the CNS25. Mice deficient in negative regulator of reactive oxygen species (NRROS), which is preferentially expressed in myeloid cells (including macrophages, neutrophils, microglia and perivascular macrophages) and regulates the production of reactive oxygen species, lack microglia and instead are populated by perivascular macrophage-like cells87. The neurological abnormalities observed in TGFβ1-deficient mice were also observed in NRROS-deficient mice and were associated with the loss of the homeostatic microglial signature and a defect in TGFβ and SALL1 signalling. The lack of microglia in mice lacking NRROS shows that this factor is important for the programme that generates the homeostatic microglial phenotype.
Microglial physiological functions
The features of homeostatic microglia discussed above make them well suited to their physiological functions in the CNS.
Synaptic plasticity and synaptogenesis
Microglia play a crucial role in synaptic and axonal pruning during development by phagocytizing inappropriate synaptic connections and axons, thereby shaping neural circuits in the developing brain41,88–90. Similarly, it has been shown that microglia shape postnatal neural circuits in an activity-dependent and complement-dependent manner that is dependent on neural activity91. In follow-up studies, the contribution of complement and microglia to synaptic loss in mouse models of AD has been investigated92,93. It is known that synapse loss correlates with cognitive decline in AD and that microglia may play an important role in neural inflammation. Inhibition of complement reduces the number of phagocytic microglia as well as the extent of early synapse loss. Furthermore, the complement component C1q is needed for the toxic effects of Aβ oligomers92. These findings suggest that, in AD, the ability of microglia to prune excess synapses in development is inappropriately activated and participates in synapse loss.
Trophic support
Microglia secrete neurotrophic factors, such as nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT3), fibroblast growth factor (FGF) and insulin-like growth factor 1 (IGF1), that control the activity of receptor tyrosine kinases and are associated with signalling through phosphoinositide 3-kinase (PI3K)94. It has been suggested that, at prenatal stages, the secretion of these factors mediates the effects of microglia on synaptogenesis95, and it was shown that microglia promote learning-dependent synapse formation through the secretion of BDNF in mice96. In addition to secreting neurotrophins, microglia also respond to these factors with increased proliferation97 or phagocytic activity98.
Microglia also respond to injury by expressing specific growth factors. Following striatal injury, activated microglia and macrophages induce the sprouting of dopaminergic neurons in the injured striatum and express both BDNF and glia cell line-derived neurotrophic factor (GDNF)99. In another study, it was demonstrated that BDNF secreted by microglia caused a shift in the neuronal anion gradient underlying neuropathic pain100. The secretion of BDNF by microglia is suppressed by IL-1β101. Related to this, the suppression of synaptic plasticity by Aβ, which also disrupts the homeostatic microglial signature (see below), is also sensitive to IL-1β: excessive activation of IL-1β disrupts the formation of dendritic spines and thus interferes with memory consolidation101. Thus, the microglial molecular signature ultimately finds its expression in microglial function.
Chemotaxis and immune cell recruitment
Microglia attract monocytes to the brain in a CC-chemokine ligand 2 (CCL2)-dependent fashion. These monocytes play an important role in neuroinflammation but do not contribute to the resident microglial pool102. For example, in the SOD1 mouse model of ALS, splenic monocytes expressing CC-chemokine receptor 2 (CCR2) and high levels of lymphocyte antigen 6C (LY6C) are recruited to the CNS by spinal cord microglia49. The recruitment of CCR2-expressing monocytes also plays an important role in mouse models of AD, in which genetic deletion of CCR2 in mice impairs monocyte accumulation and accelerates disease progression103. It has also been shown that infiltrating monocytes promote brain inflammation and exacerbate neuronal damage after status epilepticus104. Levels of CCL2 and other chemokines are expressed at the highest levels when microglia switch from homeostatic to disease-associated phenotypes. Thus, one of the functions of disease-associated microglia is to facilitate recruitment of monocytes to the CNS.
Neurogenesis
Microglia contribute to neurogenesis in two ways: through phagocytosis of excessive newborn cells and by promoting neurogenesis through the secretion of cytokines. Most newborn cells in the developing nervous system undergo apoptotic death as they transition from neural progenitors to neuroblasts and are rapidly cleared by microglia-mediated phagocytosis105. In addition, microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis105. Phagocytosis is linked to the homeostatic microglial phenotype and is specifically dependent on the expression of receptor tyrosine kinase MerTK (MERTK), which is responsible for routine non-inflammatory clearance of dead brain cells in the neurogenic niche106.
In adult mice, hippocampal neurogenesis may be induced by exposure to an enriched environment, a process that is associated with the recruitment of T cells and the activation of microglia107. Similarly, activated microglia accumulate in the early postnatal subventricular zone, where they enhance neurogenesis via release of cytokines108. On the other hand, microglial activation is suppressed by the interaction of fractalkine, expressed by neurons, with microglial CX3CR1. This may repress neurogenesis and is thus a target for the therapeutic modulation of microglia109.
It is known that microglia are also involved in oligodendrocyte development. A unique molecular signature of the microglia involved in regulating the development and homeostasis of oligodendrocytes and their progenitors has been identified and shown to be essential for these functions, as confirmed by microglial depletion experiments in mice110. Interestingly, this signature included genes such as C-type lectin domain containing 7A (Clec7a), secreted phosphoprotein 1 (Spp1), Igf1, αX integrin (Itgax) and transmembrane glycoprotein NMB (Gpnmb), which have been described in ageing microglia and in disease models (see below)52,111. In addition, this ITGAX-expressing microglial subset has been shown to have a key role in myelinogenesis in the developing brain112.
Inflammation also plays a role in regulating the influence of microglia on neurogenesis and oligodendrogenesis. Microglia activated by IL-4 or IFNγ induced oligodendrogenesis or neurogenesis, respectively, from adult stem cell progenitor cells both in vivo and in vitro113. High, but not low, levels of IFNγ induced a microglial phenotype that impeded oligodendrogenesis. IL-4 reduced this impediment and overcame the blockage of IGF1 production that was caused by IFNγ113. Consistent with these observations, an ITGAX-expressing microglial subset was identified as the source of IGF1 in the developing brain112.
A better understanding of how microglia in different states of activation regulate adult neurogenesis under physiological and pathological conditions will provide the foundation for using modulation of microglial activation as a therapeutic strategy.
Microglia in disease and ageing
It is now known that, during the course of many diseases, microglia lose their homeostatic molecular signature and functions26,84,114, such as their roles in synaptic plasticity115, and become chronically inflammatory116. At the present time, a common neurodegenerative microglial signature has been identified for diseases such as ALS, AD and MS. Investigations are beginning to probe the unique microglial signatures related to specific diseases and to specific stages of disease; however, these have yet to be defined.
Amyotrophic lateral sclerosis
One of the first descriptions of a disease-associated microglial signature was provided by studies that examined the SOD1 mouse model of ALS28. Tau, presenilin 2 and apolipoprotein E (ApoE) pathways were found to be activated in microglia in the SOD1 mouse, with Apoe being one of the transcripts that was the most significantly induced during disease progression. This upregulation was not seen in response to lipopolysaccharide (LPS) activation or in M1 or M2 macrophages. In concordance with these results, a later study also reported increased ApoE signalling in microglia in both SOD1 mice and human ALS84. Moreover, this study also revealed loss of the homeostatic microglial signature described above as early as 2 months before measurable disease onset in the SOD1 mouse.
Studies have also provided information on the mechanisms driving the changes in microglial signature in ALS. ALS is associated with neuronal death and the activation of TREM2-ApoE signalling in microglia by apoptotic neurons52. This signalling leads to the induction of the expression of miR-155, which has been shown to be a pro-inflammatory signal117. Indeed, genetic deletion of Apoe abrogated induction of miR-155 in microglia52. Importantly, genetic ablation of miR-155 itself restored the homeostatic microglial signature and extended survival in SOD1 mice84. Many of the microglial changes observed in ALS were also observed in an AD mouse model (see below), suggesting a common microglial response to different disease processes.
Ageing
Aged microglia display a dystrophic phenotype118–120. It is also known that ageing, similar to neurodegeneration, induces a hypersensitive, pro-inflammatory microglial phenotype that some have referred to as ‘primed microglia’26. In ageing mice, most of the ‘sensome’ genes (which encode proteins involved in sensing endogenous ligands and microorganisms) that are expressed by homeostatic microglia are downregulated27. These cells also express pro-inflammatory cell surface markers such as galectin 3 (LGALS3), tyrosine-protein kinase receptor UFO (AXL), CLEC7A, major histocompatibility complex (MHC) class II molecules and CXC-chemokine receptor 4 (CXCR4)26 and appear immunologically activated but not polarized to a pro- or anti-inflammatory phenotype25. Microglia from transgenic mice that lack multifunctional protein 2 (MFP2) and thus model the peroxisomal β-oxidation deficiency common to ageing express inflammatory surface markers and downregulate the homeostatic purinergic receptor P2RY12 (REF.121) in the same manner as aged microglia27. In MFP2-deficient mice TGFβ receptor 1 (Tgfbr1) is downregulated, suggesting that the loss of homeostatic properties in ageing microglia is also related to the downregulation of this receptor121.
Alzheimer disease
Reactive gliosis and neural inflammation are hallmarks of AD4,122–125. In the past, microglial changes had been considered to be secondary events; however, it is now clear that, although not being the primary initiator of AD, microglia play an important cell-autonomous role in the disease. Immunohistochemical analysis has shown that microglia surround amyloid plaques in both human AD and in animal models of the disease and may be either beneficial or detrimental in halting disease progress4. Genetic studies show that a number of genes that have been linked to early-onset AD risk are expressed primarily in microglia. These include those encoding TREM2 (REFs126,127), membrane-spanning 4-domains subfamily A (MS4A) members128, ATP-binding cassette subfamily A member 7 (ABCA7)128, the ε4 allele of ApoE (ApoE4)129–131, myeloid cell surface antigen CD33 (REFs132–134), PU.1 (REF.135) and Myc box-dependent-interacting protein 1 (BIN1)116,136. In addition, microglial expression of the complement receptor CR1 has been linked to late-onset AD137,138 and may reflect the microglial phagocytosis of synapses that has been proposed to contribute to AD pathogenesis.
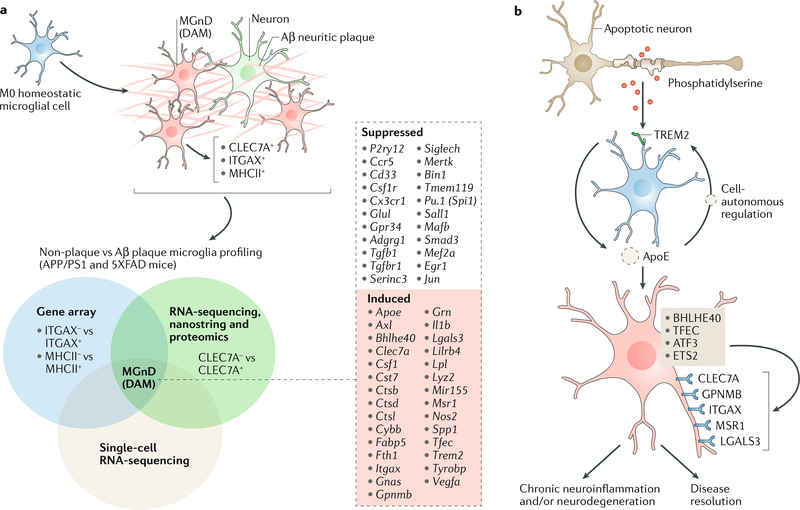
Transcriptomic analysis of microglia in an AD mouse model and in human AD has revealed a proinflammatory signature present in Aβ-plaque-associated microglia52,111,139,140 (FIG. 2a). This phenotype was not observed in microglia in non-plaque areas, suggesting that the neuroinflammation associated with AD predominantly occurs in plaque-associated microglia. The upregulated genes in Aβ-plaque-associated microglia, also known as disease-associated microglia (‘DAM microglia’), included Axl, Apoe, Clec7a, Itgax, Lgals3 and Cst7, which are now known to be part of a common neurodegenerative52 and disease-associated signature that has been observed by several groups26,111,141,142 (see below; FIG. 2a). These DAM microglia were shown to be generated through a two-step activation process in which homeostatic microglia first transition to an intermediate stage (known as stage 1 DAM) in a TREM2-independent manner, followed by a second TREM2-dependent transition to stage 2 DAM111. These studies emphasize that cellular reprogramming of microglia occurs in response to neurodegeneration and that this process is related to unique underlying transcriptional programmes. DAM microglia have been observed in other conditions including ageing, ALS and frontotemporal dementia (FTD) and in an inducible CK-p25 mouse model of severe neurodegeneration, and it appears that the DAM expression phenotype is nonspecifically triggered by altered brain homeostasis111. In a mouse AD model, microglia typified by the expression of modules of co-regulated type 1 and type 2 IFN-responsive genes and their proliferation were shown to be present as an early response to neuronal injury142. Moreover, two distinct reactive microglial phenotypes were identified at later stages of neurodegeneration142. Independent of the specific disease, it appears that, over time, microglia may transform into an activated state and that this change is related to signals from the periphery that occur with ageing or disease143.
Fig. 2 |. Regulation of the neurodegenerative microglial phenotype.
a | Several laboratories profiled microglia gene and protein expression through microglial isolation using αX integrin (ITGAX, also known as CD11c)139 major histocompatibility complex class II (MHCII) antibodies140, C-type lectin domain containing 7A (CLEC7A) antibodies52 and single-cell RNA-sequencing111 (see Venn diagram). Comparison of these findings shows that microglia associated with amyloid-β (Aβ) plaques acquire a specific phenotype: these have been defined as MGnD (microglial neurodegenerative phenotype)52 or disease-associated microglia (DAM microglia)111. This phenotype is associated with expression of CLEC7A, ITGAX and MHCII molecules52,111. The middle box lists the top suppressed and induced genes that are common to both MGnD and DAM microglia. b | The schematic depicts triggering receptor expressed on myeloid cells 2 (TREM2)-apolipoprotein E (ApoE) signalling, which induces a microglial phenotype switch from homeostatic to the MGnD phenotype. TREM2 present on M0 homeostatic microglia senses phosphatidylserine on apoptotic or damaged cells and activates ApoE signalling, which induces an MGnD neurodegenerative phenotype in a cell-autonomous manner. This phenotype is associated with the suppression of the M0 homeostatic signature and induction of molecules associated with inflammation, including CC-chemokine ligand 2 (CCL2), CLEC7A, macrophage colony-stimulating factor 1 (CSF1), secreted phosphoprotein 1 (SPP1), miR-155 and nitric oxide synthase, inducible (NOS2). This phenotype may have dual roles. ATF3, cAMP-dependent transcription factor ATF3; BHLHE40, class E basic helix-loop-helix protein 40; ETS2, protein C-ets-2; GPNMB, transmembrane glycoprotein NMB; LGALS3, galectin 3; MSR1, macrophage scavenger receptor types I and II; TFEC, transcription factor EC.
Although it is clear that the DAM microglia identified in these studies are associated with Aβ plaques, it is not clear whether they have a protective or disease-inducing function (FIG. 2b). It is known that innate immune cells can protect against AD by reducing the accumulation of Aβ, which occurs during early stages of disease. For example, in mice, peripheral monocytes can enter the brain, clear Aβ and affect disease pathology144, an effect that is lost in Ccr2−/− animals103. It may be that as microglia age they lose their ability to phagocytose Aβ in the brain27,145. Furthermore, with the deposition of Aβ, the phenotype of microglial cells becomes more neurotoxic118,146–148: they express several inflammatory cytokines, including IL-1β, IL-6, tumour necrosis factor (TNF) and CCL3 (also known as MIP1α)146. The neurotoxicity of microglial cells may also be related to their expression of tau, which is triggered by the deposition of Aβ in the brain and is known to be neurotoxic149.
In summary, microglia are thought to have multiple functions in AD. The identification of the factors associated with both their beneficial and detrimental functions and the development of strategies to restore the homeostatic microglial signature or to induce the DAM microglial signature could improve our ability to target microglia and treat AD. It is important to note that there are likely to be multiple microglial phenotypes associated with disease and that DAM microglia as defined may have either beneficial or detrimental function. These functions will need to be defined in more detail in order to target microglia therapeutically.
Multiple sclerosis
MS is an inflammatory disease of the CNS in which microglia play an important role. In the experimental autoimmune encephalomyelitis (EAE) mouse model of MS, microglia have a transcriptional phenotype that is similar to that observed in ALS and AD models (see below)52. Aspects of this transcriptional profile may contribute to disease. For example, microglia have been shown to express chemokines (such as CCL2) that facilitate recruitment of inflammatory CCR2+ monocytes from the periphery. Microglia are the only cells in the brain to express CX3CR1, which serves an immunomodulatory role in the homeostatic state and the genetic ablation of which results in neurotoxicity78. Moreover, suppression of microglial Cx3cr1 expression is associated with disease progression in EAE52. The disease-associated microglial transcriptional phenotype in EAE also contributes to the inflammatory cascade by activating astrocytes150. In human MS, microglia lose the expression of the P2RY12 homeostatic microglial marker and adopt a pro-inflammatory phenotype that includes the expression of phagocytic-related markers (macrosialin (also known as CD68)), antigen presentation markers (MHC class I and II molecules and T lymphocyte activation antigen CD86) and proteins involved in the production of reactive oxygen species (cytochrome b-245 light chain (CYBA))36. During the progressive stages of MS, microglia have been shown to accumulate iron151,152. Minocycline has been shown to specifically affect microglia153–155, and a trial in MS showed positive trends156, although minocycline worsened patients with ALS157. These differences may be related to how minocycline differentially affects the microglial signature.
Parkinson disease
Parkinson disease (PD) is characterized by the loss of dopamine neurons in the substantia nigra and the presence of intracellular proteinaceous inclusions (Lewy bodies) in neurons composed of α-synuclein that activate microglia158,159. Although molecular profiling of microglia in animal models of PD has yet to be attempted, activated microglia are known to be prominent in mouse models and in human PD160. This activation includes the expression of AXL in disease associated microglia. Microglial activation appears to act via TAM (TYRO3, AXL and MERTK)-dependent mechanisms to have a detrimental role in a mouse model of PD106. Microglial activation in human PD has been demonstrated by positron emission tomography161, and it has been reported that α-synuclein-dependent activation of microglia may be linked to changes in the microbiome in both animal models and patients162.
Autism spectrum disorder
Autism spectrum disorder (ASD) is a neurodevelopmental disorder that includes abnormalities in social interaction, difficulties with language and repetitive behaviour. Although considered primarily a neuronal disease, the role of microglia in sculpting and regulating neuronal synapses suggests that microglial dysfunction contributes to ASD. In humans, post-mortem genome-wide transcriptome analysis has revealed age-related changes in the trajectory of microglial and synaptic dysfunction that may reflect an early developmental alteration in microglial function163. Consistent with this, mice lacking CX3CR1 exhibit a transient reduction in the numbers of microglia present in the brain during the early postnatal period and a subsequent deficit in synaptic pruning associated with impaired social behaviour164.
Regulation of disease-associated microglial signatures
As described above, microglia change during ageing and in disease to acquire different phenotypes and activate specific transcriptional programmes as a result of processes that are complex and result in both protective and disease-associated microglia. To determine whether there are common patterns in the changes observed in different diseases, microglial transcriptomes in ageing and in mouse models of ALS, AD and MS have been analysed52. Across disease models, the induction of genes related to ApoE signalling and the suppression of TGFβ signalling in microglia were associated with neurodegeneration, resulting in a phenotype that was termed ‘MGnD’ (microglia neurodegenerative phenotype). The MGnD microglial phenotype overlaps to some extent with that of the DAM microglia found in models of AD (see above). In disease, several core homeostatic disease genes were suppressed, including those encoding the transcriptional regulators Mef2a, Mafb, Sall1 and early growth response protein 1 (Egr1). At the same time, there was an increase in the expression of genes encoding transcription factors such as class E basic helix-loop-helix protein 40 (Bhlhe40), transcription factor EC (Tfec), protein C-ets-2 (Ets2) and cAMP-dependent transcription factor ATF3 (Atf3). Another study analysed 18 data sets from transcriptional profiles of CNS myeloid cells from diverse mouse models of neurodegeneration and identified a set of co-regulated genes associated with neurodegenerative disease in mice that were also observed in human AD brains165. Many of these genes overlap with the MGnD microglial phenotype. Interestingly, some of the inflammatory genes that were observed in AD brains, were not observed in mouse models of AD.
A major question is how microglial signatures are regulated in disease. Phagocytosis of apoptotic neurons that were injected into the brain of naive mice suppressed the microglial homeostatic signature and resulted in a transcriptional profile that mimicked the molecular signature of MGnD microglia. This microglial phenotype switch was shown to be regulated by activation of TREM2-ApoE signalling via phosphatidylserine exposed on the apoptotic neurons52. Indeed, TREM2 is an important microglial molecule25,27,28 thought to be involved in the regulation of microglial signatures in disease1,58. TREM2 is a receptor that detects damage-associated lipids, allowing microglia to sense Aβ accumulation, and its loss has been shown to augment Aβ accumulation in a mouse model of AD166. It has been reported that TREM2 promotes microglial survival by activating the WNT-β-catenin signalling pathway167. Consistent with this, TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury in mice168. A rare mutation of TREM2 (R47H) is associated with a substantial increase in the risk of developing AD126.
Aβ-induced microglial activation is impaired in humanized transgenic mice expressing human R47H TREM2 (REF.169), and the R47H mutation impairs TREM2 binding to anionic lipids166. Furthermore, an elevation in the levels of soluble TREM2 in the cerebrospinal fluid in AD is coincident with markers of neural damage and the onset of clinical dementia170. It has been proposed that TREM2 signalling is anti-inflammatory for microglia and that agonizing TREM2 signalling would suppress neuroinflammation58,171. Indeed, in EAE it has been shown that TREM2 is upregulated in spinal cord microglia and that an anti-TREM2 antibody exacerbates disease172. Thus, stimulating TREM2 represents a potential therapeutic strategy in neurodegenerative disease. However, others have reported that Trem2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in AD mouse models173, which is seemingly inconsistent with the link between TREM2 loss-of-function mutations and an increased risk of AD. One possible explanation for this discrepancy is that TREM2 is highly expressed on inflammatory monocytes, which are recruited to the plaques, and that genetic ablation of Trem2 eliminates their recruitment. However, in parabiosis experiments it was found that amyloid-associated myeloid cells are brain-resident microglia rather than recruited peripheral monocytes174. In the absence of TREM2, it has been reported that microglia do not fully surround Aβ plaques and that Aβ plaques are more diffuse and associated with greater neuritic damage174. These findings are consistent with the interpretation that TREM2 protects from AD by enabling microglia to surround and alter Aβ plaque structure, thereby limiting neuritic damage.
To further explore these differences, the effects of Trem2 deficiency at early and late stages of disease in AD mouse models have been explored. Trem2 deficiency ameliorated amyloid pathology at early stages but exacerbated it at late stages, suggesting that TREM2 plays distinct roles at different stages of disease175. Given that TREM2 mutations are associated with increased risk of AD in humans126,176,177, a protective role for this receptor appears to be the predominant biologic effect. Consistent with this, it was found that TREM2 also affects microglial metabolic fitness: patients with AD carrying TREM2 risk variants and TREM2-deficient mice have abundant autophagic vesicles in their microglia178. Although it is not entirely clear how TREM2 regulates microglial signature phenotypes, TREM2 is known to recognize phosphatidylserine on the surface of apoptotic or damaged cells and to initiate ApoE signalling52. ApoE is a ligand for TREM2 in microglia179–181. It binds to apoptotic neurons and increases TREM2-mediated microglial phagocytosis of neurons179. ApoE also suppresses the expression of major transcription factors of homeostatic microglia52. Moreover, genetic ablation of Apoe in MGnD microglia restored their homeostatic properties in a cell-autonomous fashion. Genetic targeting of Trem2 suppressed the ApoE pathway and restored homeostatic microglia in both AD and SOD1 mice52. In the tau model of neurodegeneration, genetic deletion of either Apoe or Trem2 also arrested neurodegeneration, demonstrating that this pathway negatively regulates the homeostatic signature182,183.
As outlined above, it is still a matter of debate whether MGnD or DAM microglia63 are beneficial or detrimental in neurodegenerative disease. One report showed that microglia-derived apoptosis-associated speck-like protein containing a CARD (PYCARD) facilitates spreading of Aβ pathology in an AD mouse model184. MGnD microglia that are associated with Aβ significantly upregulate their expression of Pycard. Thus, Aβ-plaque-associated microglia may play a detrimental role in AD by facilitating Aβ plaque seeding rather than restricting Aβ plaque formation63. Consistent with a potential detrimental role of microglia, it was shown that ablation of microglia by a compound (PLX3397) that inhibits CSF1R did not reduce plaque burden, although it did protect synapses185. In addition, PLX3397 treatment was reported to reduce accumulation of intraneuronal Aβ, neuritic plaque deposition and pre-fibrillar oligomers in the same mouse model of AD186.
Therapeutic targeting of microglia
As described above, normal microglial functions include phagocytosis, synaptic pruning, chemotaxis and regulation of neurogenesis and CNS immunity. Because microglia have such a fundamental role in brain function, the potential therapeutic landscape in targeting microglia is extraordinarily broad, including developmental diseases such as autism, acquired diseases such as MS and genetic and degenerative diseases such as AD, PD and ALS. As outlined above, it is now recognized that microglia are resident cells of the nervous system that are independent from myeloid cells that migrate into the brain from the periphery. Thus, therapeutic targeting of microglia themselves must target them in the CNS.
Broadly speaking, one would argue that the goal of microglial-targeted therapy is to maintain homeostatic microglial function and to restrain or inhibit inflammatory or disease-promoting microglia, such as those displaying the MGnD transcriptional profile described above. Thus, one would want to agonize molecular targets that enhance the homeostatic signature and antagonize those that drive MGnD. On the basis of our understanding of homeostatic and MGnD microglial signatures, a number of specific microglial molecular targets are beginning to emerge. Targets for which the goal would be to boost function include miR-124, TGFβ, MERTK, CX3CR1, P2RY12, MEF2C, SALL1 and MAFB. Targets for antagonistic therapeutics include TREM2, ApoE4, AXL, CSF1R, complement C3, miR-155 and SPP1. As discussed above, some molecules, such as TREM2, may be either beneficial or detrimental depending on disease stage, a characteristic that becomes important when therapeutics targeting these molecules are applied to humans.
Recent studies that have used an anti-CSF1R antibody187 and PLX3397 to block CSF1R suggest that inhibition of CSF1R causes the depletion of microglia188. Treatment with PLX5622, which also targets CSF1R, prevented the association of microglia with plaques and improved cognition in the 3xTg-AD mouse model of AD189. Similarly, an oral tyrosine kinase inhibitor (GW2580) inhibited CSF1R in the APP/PS1 mouse model of AD, which reduced microglial proliferation and the microglial inflammatory profile190. Pharmacological targeting of CSF1R in this study also improved cognition and prevented synaptic degeneration190. These studies therefore support the potential of CSF1R inhibitory strategies in the treatment of AD.
TAM receptors are involved in multiple aspects of microglial physiology106. As discussed above, microglia are damage sensors of the CNS and act as phagocytes for the routine, non-inflammatory clearance of dead brain cells. These functions are regulated by the TAM receptor tyrosine kinases MER and AXL. Animals deficient in Mer and Axl accumulate apoptotic cells106. Microglial expression of Axl is upregulated and Mer is downregulated in microglia exhibiting the MGnD phenotype52. Thus, TAM receptors may control microglial physiology and are potential targets for therapeutic intervention.
Other well-studied inhibitors of inflammation include glucocorticoids, minocycline, vitamin E, vitamin D, endocannabinoids, TGFβ1, rapamycin and other synthetic drugs191. However, although some anti-inflammatory drugs have been shown to diminish neuroinflammation, few have direct functional effects on microglial activity192. On the basis of our current understanding of microglial physiology, these treatments would be expected to mediate their effects by modulating the microglial homeostatic signature.
What then are the modalities by which one may target microglia? The most obvious and easiest to develop are monoclonal antibodies specific for microglial targets. Virtually any surface structure on microglia could serve as an antibody target. Investigators are currently developing an antibody against ApoE4 (REF.193) as well as antibodies against TREM2 (REF.172). As shown in the trial of the anti-Aβ monoclonal antibody aducanumab for AD, it appears that enough of a systemically administered monoclonal antibody reaches the CNS to have an effect194. Another approach involves microRNAs. miR-155 has been associated with inflammatory microglia and is a major driver of inflammation in innate immunity. Targeting miR-155 in microglia has shown to be a benefit in models of ALS84,195 and is currently being pursued as a possible therapeutic strategy. Other microRNAs have anti-inflammatory functions themselves. For example, it has been shown that agonizing miR-124 is beneficial in models of MS196. Thus, it appears that microRNAs may regulate microglial transcriptional signatures. Consistent with this, in MGnD microglia from Apoe−/− mice, miR-155 expression was suppressed52.
In addition to classical pharmacological approaches, there are other physiological methods to drive changes in microglia. One intriguing example arises from a report that environmental enrichment reduces Aβ levels and amyloid deposition in a mouse model of AD197. Moreover, environmental enrichment prevents the microglial-dependent neural inflammation that is driven by human Aβ protein polymers198. Thus, environmental or behavioural modification could have positive effects on brain function by affecting microglia.
Environmental enrichment.
Creation of complex environmental stimulation through the use of toys, ladders, tunnels and a running wheel in the environment in which animals are housed.
Finally, the microbiome plays a central role in health and disease, and the host microbiome controls maturation and function of microglia199. Animals raised in a germ-free environment or mice treated with broad-spectrum antibiotics develop dysfunctional microglia, an effect that can be ameliorated with short-chain fatty acids. In addition, it has been shown that the microbiome influences prenatal and adult microglia in a sex-dependent and time-dependent manner200. A gut-brain axis exists involving both neural and metabolic communication that could affect microglia199. Thus, it is possible that modulation of the microbiome could treat neurodegenerative diseases through its effects on microglia. Indeed, studies of the microbiome in neurodegenerative diseases such as PD162, AD201 and MS202 are in progress.
Central to our understanding of microglial function in disease and our ability to target microglia is a requirement to be able to measure microglial function via biomarkers. The most widely used biomarkers in neurodegenerative disease involve imaging. Developing treatment for diseases such as MS has required that therapy affects the imaging measures of disease203. In AD, measurements of Aβ and tau are used as measures of therapeutic efficacy204. Methods to image microglia exist and are being used to test the effect of treatments on microglia205–210. With the identification of new microglial markers that are linked to homeostatic or disease-associated states, novel imaging ligands may provide a clearer window to assess microglia in disease and the effect of therapy. Other biomarkers to measure microglial function could be present in the cerebrospinal fluid. For example, soluble TREM2 has been shown to increase with time with the development of AD211.
In summary, the expanding molecular characterization of homeostatic and disease associated microglia provides an avenue for understanding and treating human nervous system disease.
Conclusions
As described in this article, microglia have now been redefined as having either a homeostatic or a disease-associated phenotype on the basis of their molecular signature, and many of the pathways that regulate these signatures have been elucidated. Nonetheless, the distinction between homeostatic and disease-associated signatures is not absolute, particularly in terms of their impact on disease: some microglial disease signatures that are not homeostatic may in fact have beneficial roles in halting the disease process, as illustrated by the paradoxical findings with TREM2. Future directions in this field will relate to the development of strategies to manipulate microglia to treat disease states. The microglial signatures that have been identified to date will provide a basis to identify new therapeutic targets and to measure the effect of microglial-directed therapies.
Acknowledgements
O.B. is supported by the US National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (R01 NS088137, R21 NS104609 and R21 NS101673), National Institute on Aging (NIA; R01 AG051812 and R01 AG054672) and National Eye Institute (R01 EY027921); the National Multiple Sclerosis Society (5092A1); a Nancy Davis Foundation Faculty Award; a Cure Alzheimer’s Fund Award; the Amyotrophic Lateral Sclerosis Association; and Sanofi. H.L.W. is supported by the US Department of Defense (AL120029), the Thome Foundation and the NIH NIA (R01AG043975 and R01AG040092).
Footnotes
Competing interests
O.B. and H.L.W. hold patent applications entitled ‘Targeting Apolipoprotein E (APOE) in Neurologic Disease’ (PCT/US2015/056492), ‘Micrornas in Neurodegenerative Disorders’ (PCT/US2012/059671) and ‘Mir-155 Inhibitors for Treating Amyotrophic Lateral Sclerosis (ALS)’ (Application #20180161357), which have exclusive licensing rights from the Brigham and Women’s Hospital. O.B. is an adviser and collaborator for Sanofi and Nanostring.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Neuroscience thanks B. Eggen, M. Prinz and M.-E. Tremblay for their contribution to the peer review of this work.
References
- 1.Colonna M & Butovsky O Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginhoux F, Lim S, Hoeffel G, Low D & Huber T Origin and differentiation of microglia. Front. Cell. Neurosci. 7, 45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q & Barres BA Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. (2017). [DOI] [PubMed] [Google Scholar]
- 4.Sarlus H & Heneka MT Microglia in Alzheimer’s disease. J. Clin. Invest. 127, 3240–3249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ransohoff RM A polarizing question: do M1 and M2 microglia exist? Nat. Neurosci. 19, 987–991 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kaur C, Hao AJ, Wu CH & Ling EA Origin of microglia. Microsc. Res. Tech. 54, 2–9 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Alliot F, Godin I & Pessac B Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain Res. 117, 145–152 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Ginhoux F et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).References 7 and 8 identify microglia as originating from yolk-sac-derived primitive macrophages.
- 9.van Furth R & Cohn ZA The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128, 415–435 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kierdorf K et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).This study shows that microglia derived from erythromyeloid precursors develop into CD45+ c-kitlo CX3CRI− immature (A1) cells and mature into CD45+c-kit−CX3CR1+ (A2) cells. Both PU.1 and IRF8 transcription factors are vital for the development of A2 microglia.
- 11.Neumann H & Wekerle H Brain microglia: watchdogs with pedigree. Nat. Neurosci. 16, 253–255 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Bruttger J et al. Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity 43, 92–106 (2015).This study reports that microglia have the potential to self-renew without the need for a contribution of peripheral myeloid cells.
- 13.Tay TL et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Askew K et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 18, 391–405 (2017).References 13 and 14 show that microglia self-renew stochastically in the healthy brain and expand clonally during disease. The resulting excess in microglia is resolved by cell egress and programmed cell death.
- 15.Reu P et al. The lifespan and turnover of microglia in the human brain. Cell Rep. 20, 779–784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci. 21, 530–540 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Cronk JC et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 10.1084/jem.20180247 (2018).This study shows that microglial populations can be replaced with peripherally derived macrophages that maintain a unique identity and distinct functional role in the CNS compared with microglia.
- 18.Lund H et al. Fatal demyelinating disease is induced by monocyte-derived macrophages in the absence of TGF-beta signaling. Nat. Immunol. 19, 1–7 (2018).This study demonstrates that peripherally derived macrophages engraft the brain after microglia depletion and that TGFβ plays a critical role in preventing microglia-and/or macrophage-mediated CNS pathology.
- 19.Ransohoff RM & Cardona AE The myeloid cells of the central nervous system parenchyma. Nature 468, 253–262 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Kettenmann H, Hanisch UK, Noda M & Verkhratsky A Physiology of microglia. Physiol. Rev. 91, 461–553 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Hanisch UK & Kettenmann H Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 10, 1387–1394 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Hanisch UK Functional diversity of microglia - how heterogeneous are they to begin with? Front. Cell. Neurosci. 7, 65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantovani A, Sica A & Locati M Macrophage polarization comes of age. Immunity 23, 344–346 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Martinez FO & Gordon S The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6, 13 (2014).This review discusses the limitations of using the M1 and M2 paradigm, which does not represent the broader functional repertoire of macrophage biology.
- 25.Butovsky O et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).This study identifies the molecular and functional signature of homeostatic microglia, which depends on TGFβ signalling.
- 26.Holtman IR et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol. Commun. 3, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman SE et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu IM et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier EL et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosselin D et al. An environment-dependent transcriptional network specifies human microglia identity. Science 10.1126/science.aal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galatro TF et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20, 1162–1171 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Olah M et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoh J et al. TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Zhu C et al. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 5, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett ML et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zrzavy T et al. Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain 140, 1900–1913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muffat J et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 22, 1358–1367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abud EM et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douvaras P et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 8, 1516–1524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crotti A & Ransohoff RM Microglial physiology and pathophysiology: insights from genome-wide transcriptional profiling. Immunity 44, 505–515 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Perry VH, Hume DA & Gordon S Immunohistochemical localization of macrophages and microglia in the adult and developing mouse brain. Neuroscience 15, 313–326 (1985). [DOI] [PubMed] [Google Scholar]
- 42.Akiyama H & McGeer PL Brain microglia constitutively express beta-2 integrins. J. Neuroimmunol. 30, 81–93 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Ginhoux F & Prinz M Origin of microglia: current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 7, a020537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vainchtein ID et al. In acute experimental autoimmune encephalomyelitis, infiltrating macrophages are immune activated, whereas microglia remain immune suppressed. Glia 62, 1724–1735 (2014). [DOI] [PubMed] [Google Scholar]
- 45.van den Berg TK, Puklavec MJ, Barclay AN & Dijkstra CD Monoclonal antibodies against rat leukocyte surface antigens. Immunol. Rev. 184, 109–116 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Kim WK et al. CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. Am. J. Pathol. 168, 822–834 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sousa C, Biber K & Michelucci A Cellular and molecular characterization of microglia: a unique immune cell population. Front. Immunol. 8, 198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies LC, Jenkins SJ, Allen JE & Taylor PR Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butovsky O et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122, 3063–3087 (2012).This study shows the relevance of innate inflammation for ALS pathogenesis and identifies miR-155 as a potential therapeutic target.
- 50.Gao L et al. Infiltration of circulating myeloid cells through CD95L contributes to neurodegeneration in mice. J. Exp. Med. 212, 469–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zondler L et al. Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 132, 391–411 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Krasemann S et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017).This study identifies a major microglial neurodegenerative phenotype in rodent and humans regulated by TREM2-ApoE signalling.
- 53.Haynes SE et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Konishi H et al. Siglec-H is a microglia-specific marker that discriminates microglia from CNS-associated macrophages and CNS-infiltrating monocytes. Glia 65, 1927–1943 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Lawson LJ, Perry VH, Dri P & Gordon S Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Hua K, Schindler MK, McQuail JA, Forbes ME & Riddle DR Regionally distinct responses of microglia and glial progenitor cells to whole brain irradiation in adult and aging rats. PLOS ONE 7, e52728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid CD et al. Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J. Neurochem. 83, 1309–1320 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colonna M & Wang Y TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 17, 201–207 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Grabert K et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19, 504–516 (2016).This study describes the age-dependent and region-dependent transcriptional heterogeneity of microglia.
- 60.Raj D et al. Increased white matter inflammation in aging- and alzheimer’s disease brain. Front. Mol. Neurosci. 10, 206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mrdjen D et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Ladeby R et al. Microglial cell population dynamics in the injured adult central nervous system. Brain Res. Rev. 48, 196–206 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Ziebell JM, Adelson PD & Lifshitz J Microglia: dismantling and rebuilding circuits after acute neurological injury. Metab. Brain Dis. 30, 393–400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tremblay ME et al. The role of microglia in the healthy brain. J. Neurosci. 31, 16064–16069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomez-Nicola D & Perry VH Microglial dynamics and role in the healthy and diseased brain: a paradigm of functional plasticity. Neuroscientist 21, 169–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michell-Robinson MA et al. Roles of microglia in brain development, tissue maintenance and repair. Brain 138, 1138–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schafer DP & Stevens B Microglia function in central nervous system development and plasticity. Cold Spring Harb. Perspect. Biol. 7, a020545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madry C et al. Microglial ramification, surveillance, and interleukin-ip release are regulated by the two-pore domain K(+) channel THIK-1. Neuron 97, 299–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedman AD Transcriptional control of granulocyte and monocyte development. Oncogene 26, 6816–6828 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Anderson KL et al. Myeloid development is selectively disrupted in PU.1 null mice. Blood 91, 3702–3710 (1998).This study demonstrates that PU.1 gene disruption affects a number of developmentally regulated haematopoietic processes and myeloid development.
- 72.Herbomel P, Thisse B & Thisse C Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Satoh J, Asahina N, Kitano S & Kino YA Comprehensive profile of ChIP-Seq-based PU.1/Spi1 target genes in microglia. Gene Regul. Syst. Bio. 8, 127–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gosselin D et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lavin Y et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).References 75 and 76 provide evidence that the tissue environment is a major determinant of resident macrophage gene expression and that PU.1 binds SMAD3 to establish a microglial-specific enhancer profile.
- 77.Deczkowska A et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 8, 717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cardona AE et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat. Neurosci. 9, 917–924 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Limatola C & Ransohoff RM Modulating neurotoxicity through CX3CL1/CX3CR1 signaling. Front. Cell. Neurosci. 8, 229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuevas VD et al. MAFB determines human macrophage anti-inflammatory polarization: relevance for the pathogenic mechanisms operating in multicentric carpotarsal osteolysis. J. Immunol. 198, 2070–2081 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Soucie EL et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351, aad5510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matcovitch-Natan O et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016).This study uses single-cell RNA-sequencing and bioinformatics to describe microglial transcriptome profiles and temporal stages of development in early, pre-adult microglia, which are regulated by distinct regulatory circuits.
- 83.Koshida R, Oishi H, Hamada M & Takahashi S MafB antagonizes phenotypic alteration induced by GM-CSF in microglia. Biochem. Biophys. Res. Commun. 463, 109–115 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Butovsky O et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 77, 75–99 (2015).This study validates miR-155 as a therapeutic target to modulate microglia in a mouse model and in human ALS.
- 85.Harrison SJ, Nishinakamura R, Jones KR & Monaghan AP Sall1 regulates cortical neurogenesis and laminar fate specification in mice: implications for neural abnormalities in Townes-Brocks syndrome. Dis. Model. Mech. 5, 351–365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buttgereit A et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397–1406 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Wong K et al. Mice deficient in NRROS show abnormal microglial development and neurological disorders. Nat. Immunol. 18, 633–641 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Bialas AR & Stevens B TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat. Neurosci. 16, 1773–1782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Schafer DP & Stevens B Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr. Opin. Neurobiol. 23, 1034–1040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paolicelli RC et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Stevens B et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Hong S et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi Q et al. Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci. Transl Med 10.1126/scitranslmed.aaf6295 (2017).References 91–93 provide evidence for the role of complement in synaptic elimination by microglia.
- 94.Shi SH, Jan LY & Jan YN Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112, 63–75 (2003). [DOI] [PubMed] [Google Scholar]
- 95.Kettenmann H, Kirchhoff F & Verkhratsky A Microglia: new roles for the synaptic stripper. Neuron 77, 10–18 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Parkhurst CN et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang J et al. Neurotrophins regulate proliferation and survival of two microglial cell lines in vitro. Exp. Neurol. 183, 469–481 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Elkabes S, DiCicco-Bloom EM & Black IB Brain microglia/macrophages express neurotrophins that selectively regulate microglial proliferation and function. J. Neurosci. 16, 2508–2521 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Batchelor PE et al. Activated macrophages and microglia induce dopaminergic sprouting in the injured striatum and express brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor. J. Neurosci. 19, 1708–1716 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coull JA et al. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature 438, 1017–1021 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Tong L et al. Brain-derived neurotrophic factor-dependent synaptic plasticity is suppressed by interleukin-1beta via p38 mitogen-activated protein kinase. J. Neurosci. 32, 17714–17724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ajami B, Bennett JL, Krieger C, McNagny KM & Rossi FM Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14, 1142–1149 (2011).Using a parabiosis paradigm, this study shows that recruited monocytes do not contribute to the resident microglial pool.
- 103.El Khoury J et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 (2007). [DOI] [PubMed] [Google Scholar]
- 104.Varvel NH et al. Infiltrating monocytes promote brain inflammation and exacerbate neuronal damage after status epilepticus. Proc. Natl Acad. Sci. USA 113, E5665–E5674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sierra A et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fourgeaud L et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016).This study shows that microglial MERTK is essential for elimination of apoptotic cells in neurogenic regions of the CNS.
- 107.Ziv Y et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat. Neurosci. 9, 268–275 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y & Sato K Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34, 2231–2243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bachstetter AD et al. Fractalkine and CX 3 CR1 regulate hippocampal neurogenesis in adult and aged rats. Neurobiol. Aging 32, 2030–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hagemeyer N et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keren-Shaul H et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).This study utilizes unbiased single-cell RNA-sequencing analysis to define DAM microglia.
- 112.Wlodarczyk A et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Butovsky O et al. Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol. Cell. Neurosciences 31, 149–160 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Moore CS et al. P2Y12 expression and function in alternatively activated human microglia. Neurol. Neuroimmunol. Neuroinflamm. 2, e80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Riazi K et al. Microglia-dependent alteration of glutamatergic synaptic transmission and plasticity in the hippocampus during peripheral inflammation. J. Neurosci. 35, 4942–4952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naj AC et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 71, 1394–1404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Connell RM et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Streit WJ, Sammons NW, Kuhns AJ & Sparks DL Dystrophic microglia in the aging human brain. Glia 45, 208–212 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Streit WJ Microglial senescence: does the brain’s immune system have an expiration date? Trends Neurosciences 29, 506–510 (2006).References 118 and 119 propose that microglial senescence is important for age-related decline in cognitive function.
- 120.Flanary BE, Sammons NW, Nguyen C, Walker D & Streit WJ Evidence that aging and amyloid promote microglial cell senescence. Rejuven. Res. 10, 61–74 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Verheijden S et al. Identification of a chronic non-neurodegenerative microglia activation state in a mouse model of peroxisomal beta-oxidation deficiency. Glia 63, 1606–1620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McGeer PL, Itagaki S, Tago H & McGeer EG Reactive microglia in patients with senile dementia of the Alzheimer type are positive for the histocompatibility glycoprotein HLA-DR. Neurosci. Lett. 79, 195–200 (1987). [DOI] [PubMed] [Google Scholar]
- 123.Griffin WS et al. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc. Natl Acad. Sci. USA 86, 7611–7615 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Streit WJ Microglia and Alzheimer’s disease pathogenesis. J. Neurosci. Res. 77, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- 125.Streit WJ, Mrak RE & Griffin WS Microglia and neuroinflammation: a pathological perspective. J. Neuroinflamm. 1, 14 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jonsson T et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guerreiro R et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368, 117–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hollingworth P et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bennett C et al. Evidence that the APOE locus influences rate of disease progression in late onset familial Alzheimer’s disease but is not causative. Am. J. Med. Genet. 60, 1–6 (1995). [DOI] [PubMed] [Google Scholar]
- 130.Slooter AJ et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam study. Arch. Neurol. 55, 964–968 (1998). [DOI] [PubMed] [Google Scholar]
- 131.Blacker D et al. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology 48, 139–147 (1997). [DOI] [PubMed] [Google Scholar]
- 132.Naj AC et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griciuc A et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bradshaw EM et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16, 848–850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang KL et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052–1061 (2017).This study implicates innate immunity in AD through alteration of PU.1 core transcriptional regulation of microglia and blood monocytes.
- 136.Chapuis J et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thambisetty M et al. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol. Psychiatry 73, 422–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lambert JC et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009). [DOI] [PubMed] [Google Scholar]
- 139.Kamphuis W, Kooijman L, Schetters S, Orre M & Hol EM Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta 1862, 1847–1860 (2016). [DOI] [PubMed] [Google Scholar]
- 140.Yin Z et al. Immune hyperreactivity of Aβ plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging 55, 115–122 (2017). [DOI] [PubMed] [Google Scholar]
- 141.Srinivasan K et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun 7, 11295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mathys H et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hoeijmakers L, Heinen Y, van Dam AM, Lucassen PJ & Korosi A Microglial priming and Alzheimer’s disease: a possible role for (early) immune challenges and epigenetics? Front. Hum. Neurosci. 10, 398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Simard AR, Soulet D, Gowing G, Julien JP & Rivest S Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer’s disease. Neuron 49, 489–502 (2006). [DOI] [PubMed] [Google Scholar]
- 145.Streit WJ, Braak H, Xue QS & Bechmann I Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 118, 475–485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sastre M, Klockgether T & Heneka MT Contribution of inflammatory processes to Alzheimer’s disease: molecular mechanisms. Int. J. Dev. Neurosci. 24, 167–176 (2006). [DOI] [PubMed] [Google Scholar]
- 147.Perry VH, Cunningham C & Holmes C Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 7, 161–167 (2007). [DOI] [PubMed] [Google Scholar]
- 148.Rojo LE, Fernandez JA, Maccioni AA, Jimenez JM & Maccioni RB Neuroinflammation: implications for the pathogenesis and molecular diagnosis of Alzheimer’s disease. Arch. Med. Res. 39, 1–16 (2008). [DOI] [PubMed] [Google Scholar]
- 149.Heneka MT et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liddelow SA et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).This study shows that classically activated neuroinflammatory microglia induce toxic astrocytes, which contribute to the death of neurons and oligodendrocytes in neurodegenerative disorders.
- 151.Hametner S et al. Iron and neurodegeneration in the multiple sclerosis brain. Ann. Neurol. 74, 848–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dal-Bianco A et al. Slow expansion of multiple sclerosis iron rim lesions: pathology and 7 T magnetic resonance imaging. Acta Neuropathol. 133, 25–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kobayashi K et al. Minocycline selectively inhibits M1 polarization of microglia. Cell Death Dis. 4, e525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sriram K, Miller DB & O’Callaghan JP Minocycline attenuates microglial activation but fails to mitigate striatal dopaminergic neurotoxicity: role of tumor necrosis factor-alpha. J. Neurochem. 96, 706–718 (2006). [DOI] [PubMed] [Google Scholar]
- 155.Ledeboer A et al. Minocycline attenuates mechanical allodynia and proinflammatory cytokine expression in rat models of pain facilitation. Pain 115, 71–83 (2005). [DOI] [PubMed] [Google Scholar]
- 156.Metz LM & Eliasziw M Trial of minocycline in clinically isolated syndrome of multiple sclerosis. N. Engl. J. Med. 377, 789 (2017). [DOI] [PubMed] [Google Scholar]
- 157.Gordon PH et al. Efficacy of minocycline in patients with amyotrophic lateral sclerosis: a phase III randomised trial. Lancet Neurol. 6, 1045–1053 (2007). [DOI] [PubMed] [Google Scholar]
- 158.Kim C et al. Neuron-released oligomeric alpha-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat. Commun. 4, 1562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sanchez-Guajardo V, Annibali A, Jensen PH & Romero-Ramos M alpha-Synuclein vaccination prevents the accumulation of parkinson disease-like pathologic inclusions in striatum in association with regulatory T cell recruitment in a rat model. J. Neuropathol. Exp. Neurol. 72, 624–645 (2013). [DOI] [PubMed] [Google Scholar]
- 160.Joers V, Tansey MG, Mulas G & Carta AR Microglial phenotypes in Parkinson’s disease and animal models of the disease. Prog. Neurobiol. 155, 57–75 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koshimori Y et al. Imaging striatal microglial activation in patients with Parkinson’s disease. PLOS ONE 10, e0138721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sampson TR et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhan Y et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 17, 400–406 (2014). [DOI] [PubMed] [Google Scholar]
- 165.Friedman BA et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 166.Wang Y et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015).This study demonstrates that microglial TREM2 is a sensor for lipids associated with fibrillar Aβ and its loss affects microglia and augments Aβ accumulation.
- 167.Zheng H et al. TREM2 Promotes Microglial Survival by Activating Wnt/beta-Catenin Pathway. J. Neurosci. 37, 1772–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mazaheri F et al. TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 18, 1186–1198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Song WM et al. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215, 745–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Piccio L et al. Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol. 131, 925–933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Klesney-Tait J, Turnbull IR & Colonna M The TREM receptor family and signal integration. Nat. Immunol. 7, 1266–1273 (2006). [DOI] [PubMed] [Google Scholar]
- 172.Piccio L et al. Blockade of TREM-2 exacerbates experimental autoimmune encephalomyelitis. Eur. J. Immunol. 37, 1290–1301 (2007). [DOI] [PubMed] [Google Scholar]
- 173.Jay TR et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212, 287–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Y et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jay TR et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J. Neurosci. 37, 637–647 (2017).References 173–175 show time-dependent differential effects of TREM2-dependent microglial function in AD mouse models.
- 176.Abduljaleel Z et al. Evidence of trem2 variant associated with triple risk of Alzheimer’s disease. PLOS ONE 9, e92648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jin SC et al. TREM2 is associated with increased risk for Alzheimer’s disease in African Americans. Mol. Neurodegener. 10, 19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ulland TK et al. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Atagi Y et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem. 290, 26043–26050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yeh FL, Wang Y, Tom I, Gonzalez LC & Sheng M TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91, 328–340 (2016).References 179 and 180 find that ApoE binds to TREM2 and may thus explain the functional relevance of two AD-associated risk factors.
- 181.Bailey CC, DeVaux LB & Farzan M The triggering receptor expressed on myeloid cells 2 binds apolipoprotein. J. Biol. Chem. 290, 26033–26042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shi Y et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Leyns CEG et al. TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl Acad. Sci. USA 114, 11524–11529 (2017).References 182 and 183 connect TREM2 and ApoE signalling with exacerbation of neuroinflammation and tauopathy.
- 184.Venegas C et al. Microglia-derived ASC specks cross-seed amyloid-beta in Alzheimer’s disease. Nature 552, 355–361 (2017). [DOI] [PubMed] [Google Scholar]
- 185.Spangenberg EE et al. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-beta pathology. Brain 139, 1265–1281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sosna J et al. Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol. Neurodegener. 13, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gomez-Nicola D, Fransen NL, Suzzi S & Perry VH Regulation of microglial proliferation during chronic neurodegeneration. J. Neurosci. 33, 2481–2493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Elmore MR et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).This report shows rapid repopulation of microglia following pharmacological depletion.
- 189.Dagher NN et al. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflamm 12, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Olmos-Alonso A et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain 139, 891–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dheen ST, Kaur C & Ling EA Microglial activation and its implications in the brain diseases. Curr. Med. Chem. 14, 1189–1197 (2007). [DOI] [PubMed] [Google Scholar]
- 192.Lleo A, Galea E & Sastre M Molecular targets of non-steroidal anti-inflammatory drugs in neurodegenerative diseases. Cell. Mol. Life Sci. 64, 1403–1418 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liao F et al. Anti-ApoE antibody given after plaque onset decreases Abeta accumulation and improves brain function in a mouse model of Abeta amyloidosis. J. Neurosci. 34, 7281–7292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sevigny J et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016). [DOI] [PubMed] [Google Scholar]
- 195.Koval ED et al. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum. Mol. Genet. 22, 4127–4135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM & Weiner HL MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat. Med. 17, 64–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lazarov O et al. Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell 120, 701–713 (2005). [DOI] [PubMed] [Google Scholar]
- 198.Xu H et al. Environmental enrichment potently prevents microglia-mediated neuroinflammation by human amyloid β-protein oligomers. J. Neurosci. 36, 9041–9056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Erny D et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Thion MS et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vogt NM et al. Gut microbiome alterations in Alzheimer’s disease. Scientif. Rep. 7, 13537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jangi S et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bakshi R et al. MRI in multiple sclerosis: current status and future prospects. Lancet. Neurol. 7, 615–625 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Johnson KA, Fox NC, Sperling RA & Klunk WE Brain imaging in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nimmerjahn A Two-photon imaging of microglia in the mouse cortex in vivo. Cold Spring Harb. Protoc 10.1101/pdb.prot069294 (2012). [DOI] [PubMed] [Google Scholar]
- 206.Dupont AC et al. Translocator protein-18 kDa (TSPO) positron emission tomography (PET) imaging and its clinical impact in neurodegenerative diseases. Int. J. Mol. Sci. 10.3390/ijms18040785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ghadery C et al. Microglial activation in Parkinson’s disease using [18F]-FEPPA. J. Neuroinflamm. 14, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cerami C, Iaccarino L & Perani D Molecular imaging of neuroinflammation in neurodegenerative dementias: the role of in vivo PET imaging. Int. J. Mol. Sci. FF (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hamelin L et al. Early and protective microglial activation in Alzheimer’s disease: a prospective study using 18F-DPA-714 PET imaging. Brain 139, 1252–1264 (2016). [DOI] [PubMed] [Google Scholar]
- 210.Politis M, Su P & Piccini P Imaging of microglia in patients with neurodegenerative disorders. Front. Pharmacol. 3, 96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Brendel M et al. Increase of TREM2 during aging of an Alzheimer’s disease mouse model is paralleled by microglial activation and amyloidosis. Front. Aging Neurosci 9, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Prinz M, Erny D & Hagemeyer N Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 18, 385–392 (2017). [DOI] [PubMed] [Google Scholar]
- 213.Prinz M & Priller J Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300–312 (2014). [DOI] [PubMed] [Google Scholar]
- 214.Takata K et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47, 183–198 (2017). [DOI] [PubMed] [Google Scholar]
- 215.Schulz C et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 216.Kierdorf K & Prinz M Factors regulating microglia activation. Front. Cell. Neurosci. 7, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Fuger P et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 20, 1371–1376 (2017). [DOI] [PubMed] [Google Scholar]
- 218.Matyash M, Zabiegalov O, Wendt S, Matyash V & Kettenmann H The adenosine generating enzymes CD39/CD73 control microglial processes ramification in the mouse brain. PLOS ONE 12, e0175012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lanser AJ et al. Disruption of the ATP/adenosine balance in CD39(−/−) mice is associated with handling-induced seizures. Immunology 152, 589–601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bennett FC et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183 (2018).This study shows that transplanted macrophages from multiple tissues can express microglial genes in the brain; however, only those of yolk sac origin fully attain microglial identity.