Highlights
-
•
Alltest lateral flow immunoassay is reliable to diagnose SARS-CoV-2 infection.
-
•
This immunoassay showed a specificity of 100 % and a sensitivity of 64 %.
-
•
The sensitivity increased up to 88 % from 14 days after symptoms onset.
-
•
Alltest is useful in patients with suspected SARS-CoV-2 pneumonia but negative PCR.
-
•
The test was positive in 89 % of patients with pneumonia and negative PCR.
Keywords: SARS-CoV-2, COVID-19, Lateral flow immunoassay, Serologic rapid test, Diagnosis, Alltest
Abstract
Objectives
SARS-CoV-2 infection diagnosis is challenging in patients from 2 to 3 weeks after the onset of symptoms, due to the low positivity rate of the PCR. Serologic tests could be complementary to PCR in these situations. The aim of our study was to analyze the diagnostic performance of one serologic rapid test in COVID-19 patients.
Methods
We evaluated a lateral flow immunoassay (AllTest COVID-19 IgG/IgM) which detects IgG and IgM antibodies. We validated the serologic test using serum samples from 100 negative patients (group 1) and 90 patients with COVID-19 confirmed by PCR (group 2). Then, we prospectively evaluated the test in 61 patients with clinical diagnosis of pneumonia of unknown etiology that were negative for SARS-CoV-2 by PCR (group 3).
Results
All 100 patients from group 1 were negative for the serologic test (specificity = 100 %). Regarding group 2 (PCR-positive), the median time from their symptom onset until testing was 17 days. For these 90 group-2 patients, the test was positive for either IgM or IgG in 58 (overall sensitivity = 64.4 %), and in patients tested 14 days or more after the onset of symptoms, the sensitivity was 88.0 %. Regarding the 61 group-3 patients, median time after symptom onset was also 17 days, and the test was positive in 54 (88.5 % positivity).
Conclusions
Our study shows that Alltest lateral flow immunoassay is reliable as a complement of PCR to diagnose SARS-CoV-2 infection after 14 days from the onset of symptoms and in patients with pneumonia and negative PCR for SARS-CoV-2.
1. Introduction
The pandemic due to SARS-CoV-2 that started in Wuhan in December 2019 [1,2] has caused until May 16, 2020, a total of 4,425,485 cases and 302,059 deaths worldwide [3]. Spain is the country of the European region that has been most affected by the infection, accounting for 230,183 cases and 33,998 deaths by May 16 [3]. From the beginning of the pandemic, one of the main concerns was the complexity and excessive time to results of the diagnostic test, based on polymerase chain reaction (PCR) [4,5]. Few clinical microbiology laboratories were prepared at this time to process such a massive volume of samples that grew exponentially. In our hospital, which is a medium-sized center (490 beds), from March 5 to April 6, a total of 7,453 respiratory samples (the vast majority nasopharyngeal exudates) were processed for SARS-CoV-2 PCR, reaching a positivity rate between 20 and 40 %. Another problem was the low positivity rate of nasopharyngeal samples in patients presenting a clinical syndrome compatible with COVID-19 in the second and third week of infection [1,[6], [7], [8]], which is generally the period in which patients are admitted to the hospital [1]. Besides, most patients presented a non-productive cough [9], and this fact, together with the high risk of generating aerosols in bronchoscopies explains that most respiratory samples came from the upper respiratory tract, where the virus concentration is lower beyond the first week after the onset of symptoms [8,10]. As a consequence, the positivity rate of the PCR in these patients could be lower than expected and many of them were hospitalized with a provisional diagnosis of pneumonia of unknown etiology and possible COVID-19.
These limitations have led to development of different serologic microplate ELISA tests [11,12]. Recently published studies confirm the usefulness of combining PCR in nasopharyngeal exudates with the detection of IgM and IgG antibodies in the blood [13]. The combination of molecular and serologic techniques has allowed some authors to achieve a sensitivity of 97 % for diagnosis of SARS-CoV-2 infection [11]. However, those time-consuming tests based on ELISA are not as suitable for clinical use as rapid tests and, as a matter of fact, cannot be included in the management algorithms in emergency departments [11,[13], [14], [15]].
Since the beginning of the epidemic in Spain, information emerged about the availability of rapid serological diagnostic kits that detected IgG and IgM antibodies using immunochromatographic (ICT) tests also known as lateral flow immunoassays. However, there are very few published studies about the clinical application of these kits [15]. Our aim was to evaluate the diagnostic performance of one of these serologic rapid tests, first by a validation of the test in negative control patients and confirmed cases of COVID-19, and then by a prospective evaluation in patients with pneumonia of unknown etiology and a clinical diagnosis of COVID-19 with negative PCR for SARS-CoV-2.
2. Methods
2.1. Population and study period
We included three groups of patients in our study:
2.1.1. Group 1 (negative controls)
A randomly selected group of 100 patients who had a serum sample taken for other serologic studies, from September 1 to November 30, 2019 (before the first cases of COVID-19 were reported).
2.1.2. Group 2 (confirmed cases of SARS-CoV-2 infection)
90 patients admitted to the Emergency department between March 1 and April 6, 2020, with suspicion of COVID-19. The PCR was positive for SARS-CoV-2 for all of them.
2.1.3. Group 3 (pneumonia of unknown etiology)
61 patients admitted for at least 5 days between February 9 and April 2, 2020, with a clinical and radiological diagnosis of pneumonia of unknown etiology, in which the PCR for SARS-CoV-2 was negative. They were prospectively studied after the validation of the serologic test.
2.2. Diagnostic methods
2.2.1. Molecular techniques
Two automatic extractors were used to obtain viral RNA from clinical samples: MagCore HF16 (RBC bioscience, Taipei, Taiwan) and Hamilton Microlab Starlet (Hamilton Company, Bonaduz, Switzerland). RNA amplification was made using two real-time PCR platforms: VIASURE SARS-CoV-2 Real Time PCR Detection Kit (Certest Biotech, Zaragoza, Spain) and Allplex 2019-nCoV assay (Seegene, Seoul, South Korea). All equipments were used according to the manufacturer's instructions for both the handling and the interpretation of the results.
2.2.2. Serology
We applied the AllTest COV-19 IgG/IgM kit (AllTest Biotech, Hangzhou, China) for the serological diagnosis. This test is a qualitative membrane-based immunoassay (immunochromatography or lateral flow immunoassay, LFA) for the detection of IgG and IgM antibodies against SARS-CoV-2 in whole blood, serum or plasma samples. We used 10 μL of serum for the performance of the test. For the negative control group (group 1), cryopreserved archive samples were obtained, which were previously defrosted and tempered to room temperature before analysis. The performance of the test and the interpretation of the results were done according to the manufacturer's instructions.
2.3. Clinical data
Demographic and clinical variables of the study population were obtained from the medical records (age, sex, hospital and ICU admission, outcome and disease severity). Severity of infection was classified according to WHO criteria. Briefly, patient infections were classified as: mild disease, pneumonia, severe pneumonia, acute respiratory distress syndrome (ARDS), sepsis and septic shock [16]. The time from the onset of symptoms was calculated in groups 2 and 3 from the day of onset of symptoms to the day of the extraction of the serum sample.
2.4. Serologic test validation
The serologic test was evaluated on clinical samples from groups 1 and 2 in order to assess the sensitivity and specificity of the test:
2.4.1. Group 1 (negative controls)
They were used to evaluate the specificity of the serological test. 100 aliquots of cryopreserved sera, corresponding to 100 different controls, were recovered from the serum archive.
2.4.2. Group 2: (patients with positive PCR for SARS-CoV-2)
They were used to evaluate the sensitivity of the serological test, using PCR as a gold standard. A total of 90 confirmed cases of SARS-CoV-2 infection were included, and cryopreserved aliquots of serum of those patients were used. Those aliquots were previously obtained from samples sent to the laboratory to carry out other serologies.
2.5. Diagnostic performance of the serologic test
The assessment was performed on patients from group 3 (pneumonia of unknown etiology with negative PCR for SARS-CoV-2). Fresh serum samples from these 61 patients were studied.
2.6. Statistical analysis
We considered a positive result for samples in which IgG, IgM or both of them were detected. Continuous variables were expressed as median and interquartile range (IQR) and categorical variables as proportions. Comparisons between continuous variables were made using the t test or Mann–Whitney test, depending on the normality of the distribution. For these comparisons, a p value less than or equal to 0.05 was considered significant. Specificity and sensitivity were calculated for the serologic tests using the results from group 1 and group 2 patients, respectively. Statistical analysis was performed using Stata/IC 13.1 (StataCorp, Texas, USA).
3. Results
A total of 251 patients were studied. Median age was 61 years (IQR: 46–74) and 152 (60.6 %) were males. The overall serologic results from the three groups of patients are summarized in Table 1 . Demographic and clinical characteristics of group 2 (PCR positive) and group 3 patients (pneumonia of unknown etiology and negative PCR) are summarized in Supplementary Table 1. Briefly, regarding group 2 patients (n = 90), 14 (15.6 %) of them were discharged from emergency department. Remaining 76 (84.4 %) patients were admitted to our hospital and 11 (14.5 %) required ICU admission. Regarding the severity of the infection, 17 patients (18.9 %) presented mild disease, there were 47 (52.2 %) cases of non-severe pneumonia, 20 (22.2 %) patients with severe pneumonia, 3 (3.3 %) patients of ARDS and another 3 (3.3 %) patients with septic shock. The overall mortality in group 2 was 17.1 % (13 patients).
Table 1.
Overall serologic results from the three groups of patients.
| Group of patients | Group 1 (negative controls) | Group 2 (PCR positive) | Group 3 (pneumonia of unknown etiology and negative PCR) |
|---|---|---|---|
| No. patients | 100 | 90 | 61 |
| Age (years) | 50 (33−65) | 64 (55−79) | 67 (57−73) |
| Sex (male) | 55 (55.0 %) | 52 (57.8 %) | 45 (73.8 %) |
| Time from onset of symptoms (days) | Not applicable | 17 (9−25) | 17 (15−20) |
| IgG positive | 0 (0%) | 54 (60.0 %) | 54 (88.5 %) |
| IgM positive | 0 (0%) | 25 (28.9 %) | 23 (37.7 %) |
| Positive result | 0 (0%) | 58 (64.4 %) | 54 (88.5 %) |
Statistics: values are expressed as median (interquartile range) and absolute count (percentage). A positive serologic result was defined for samples that resulted positive for either IgM or IgG antibodies.
Regarding group 3 patients (n = 61), most of them presented a non-severe pneumonia (40 patients, 65.6 %), followed by severe pneumonia (20 patients, 32.8 %) and one case of ARDS (1.6 %). Mortality rate in group 3 was 4.9 % (3 patients).
3.1. Serologic test validation
All patients included in group 1 (negative controls) showed negative results for serological tests. Thus, the serological test presented a specificity of 100 % (95 % Confidence Interval [95 %CI]: 96.5–100.0%). The overall sensitivity of the test was 64.4 % (95 %CI: 53.7–74.3%) compared to PCR (Table 1). The sensitivity increased within the first 2 weeks both for IgM and IgG (Table 2 ), reaching a sensitivity of 88.0 % (95 %CI: 75.7–95.5%) after 14 days from the onset of symptoms.
Table 2.
Serologic results in group 2 patients (PCR positive) according to the time from the onset of symptoms.
| Time from onset of symptoms | Global | ≤ 7 days | 8−14 days | 15−21 days | 22−28 days | > 28 days |
|---|---|---|---|---|---|---|
| No. patients | 90 (100.0 %) |
19 (21.1 %) |
21 (23.3 %) |
15 (16.7 %) |
20 (22.2 %) |
15 (16.7 %) |
| IgG positive | 54 (60.0 %) |
4 (21.1 %) |
7 (33.3 %) |
13 (86.7 %) |
16 (80.0 %) |
14 (93.3 %) |
| IgM positive | 26 (28.9 %) |
4 (21.1 %) |
4 (19.0 %) |
9 (60.0 %) |
6 (30.0 %) |
3 (20.0 %) |
| Positive result | 58 (64.4 %) |
5 (26.3 %) |
9 (42.9 %) |
14 (93.3 %) |
16 (80.0 %) |
14 (93.3 %) |
Statistics: values are expressed as absolute count (percentage). A positive serologic result was defined for samples that resulted positive for either IgM or IgG antibodies.
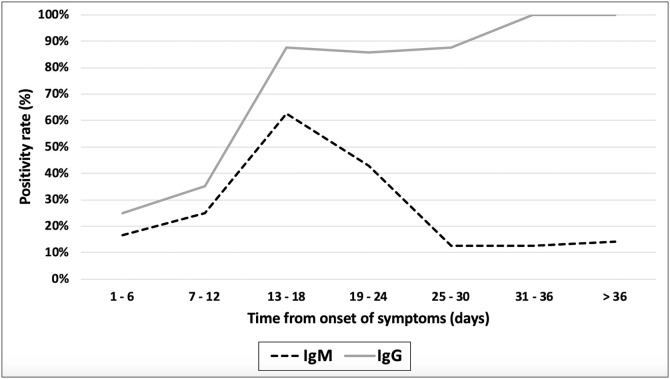
Fig. 1 shows the evolution in the positivity rates of the test in group 2: for IgM antibodies, the positivity increased to a maximum level of 62.5 % that was reached approximately 13–18 days after the onset of symptoms and then began to decrease until reaching its minimum at 25–30 days. IgG positivity rates increased up to 100 % at 31–36 days after the onset of symptoms.
Fig. 1.
Temporal evolution of the positivity rate for IgM and IgG antibodies in group 2 patients (PCR positive).
3.2. Diagnostic performance of the serologic test
We assessed the serologic test in the group 3 patients (patients with pneumonia of unknown etiology and negative PCR). There were no patients with less than 7 days from onset of symptoms in this group. The median time from onset of symptoms was 17 days (IQR: 15–20), showing no significant differences compared to group 2 (median time of 17 days, IQR: 9–25; p = 0.635). Antibodies against SARS-CoV-2 were detected in 54 out of 61 patients (88.5 %), being all of them positive for IgG antibodies. Twenty-three patients (37.7 %) were also positive for IgM antibodies (Table 1). The positivity rate was 86.7 % within the second week (8–14 days) and increased to 89.1 % after 14 days (Table 3 ).
Table 3.
Serologic results in group 3 patients (pneumonia of unknown etiology and negative PCR) according to the time from the onset of symptoms.
| Time from onset of symptoms | Overall | < 7 days | 8−14 days | 15−21 days | 22−28 days | > 28 days |
|---|---|---|---|---|---|---|
| No. patients | 61 (100.0 %) |
0 (0.0 %) |
15 (24.6 %) |
31 (50.8 %) |
14 (23.3 %) |
1 (1.7 %) |
| IgG positive | 54 (88.5 %) |
– | 13 (86.7 %) |
30 (96.8 %) |
10 (71.4 %) |
1 (100.0 %) |
| IgM positive | 23 (37.7 %) |
– | 5 (33.3 %) |
14 (45.2 %) |
4 (28.6 %) |
0 (0.0 %) |
| Positive result | 54 (88.5 %) |
– | 13 (86.7 %) |
30 (96.8 %) |
10 (71.4 %) |
1 (100.0 %) |
Statistics: values are expressed as absolute count (percentage). A positive serologic result was defined for samples that resulted positive for either IgM or IgG antibodies.
4. Discussion
Our study shows that Alltest COVID-19 IgG/IgM lateral flow immunoassay is a reliable tool to diagnose SARS-CoV-2 infection from 14 days of onset of symptoms, being especially useful in hospitalized patients with pneumonia of unknown etiology with 14 or more days from the onset of symptoms and in whom the PCR has been negative.
The current situation of the COVID-19 pandemic requires an urgent and coordinated answer to the inherent problems of the PCR-based diagnosis: on the one hand the low capacity to carry out PCR techniques in some laboratories and also the low sensitivity of PCR test in nasopharyngeal samples, specially from the second week of infection [2,6]. This study shows that the AllTest COVID-19 IgG/IgM rapid test for the detection of IgG and IgM is very specific (100 %) and reaches a sensitivity of 88.0 % from day 14 of onset of symptoms in patients with previous positive PCR in a nasopharyngeal exudate. According to our data, the vast majority of patients seroconvert from day 14 and this is a key aspect in the management of health care personnel [17] and in population immunity studies related to pandemic control [18]. There is increasing evidence on the usefulness of serology for diagnosis of SARS-CoV-2 infection, but most of these studies were based on microplate ELISA tests to detect IgA, IgM and IgG antibodies [[11], [12], [13]]. These techniques have shown high sensitivity and specificity, but they also require special equipment, trained personnel and take several hours to perform. Due to this, there is an increasing interest about the usefulness of serologic rapid tests, but there is scarce information about their diagnostic performance. In a recently published study, Li et al. [15] performed a multicenter evaluation of a serologic rapid test that the authors had developed. In their study, the overall sensitivity was 88.7 % and the specificity was 90.6 %. However, these authors did not present any data about the time after the onset of symptoms except from 58 out of 525 patients enrolled in the study. Moreover, for this subgroup of patients they only described that the time from the onset of symptoms was 8–33 days. As this is a serological test, this kind of information is key in order to interpret properly the sensitivity and specificity results. Other authors such as Montesinos et al. [19] and Imai et al. [20] evaluated different lateral flow immunoassays taking into account the time from the onset of symptoms, finding the same results as in the present work, with a significant increase in sensitivity from 14 days after the onset of symptoms [19,20].
To the best of our knowledge, our study is the first evaluation of this serologic rapid test (AllTest COVID-19 IgG/IgM) which also includes data about its prospective implementation in patients with pneumonia of unknown etiology. In our experience, the use of these tests allowed diagnosis of COVID-19 infection in 88.5 % of a group of 61 patients admitted with a clinical diagnosis of COVID-19 pneumonia and negative PCR in nasopharyngeal exudate. However, the majority of patients included in group 3 were in the second and third week from the onset of symptoms. Thus, the decrease on the positivity rate observed between 22 and 28 days could be explained by the smaller sample size in this subgroup that could have underestimated it.
Our study is subject to some limitations. First, it has been conducted in a single hospital. Further multicenter studies are necessary to reinforce our findings. Second, group 3 patients were selected according to the diagnostic needs of our hospital. Consequently, they were all patients with negative PCR with clinical and radiological criteria of pneumonia and because of that, our results could not be generalized to other COVID-19 patients with other clinical syndromes and further studies including all kinds of clinical presentations are needed in order to reinforce our conclusions. Finally, we have analyzed the results of only one the commercial lateral flow immunoassays (AllTest COVID-19 IgG/IgM). Nowadays there are many commercial LFAs, but their usefulness is questionable due to a lack of official performance validations [21]. Because of this, it is recommended to perform a validation of each diagnostic test before its clinical implementation and consequently, our results should not be extrapolated to other available commercial LFAs.
The question about the reliability of serologic rapid tests is still under debate [22,23] and more research is needed on this topic. We think that our study may help to point out the usefulness of this rapid test.
5. Conclusion
Alltest COVID-19 IgG/IgM lateral flow immunoassay is reliable to diagnose SARS-CoV-2 infection as a complement of PCR from 14 days after the onset of symptoms. This test was especially useful in hospitalized patients with pneumonia and negative PCR for SARS-CoV-2.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Informed consent
Since the present study is retrospective, informed consent was not required.
Ethical approval
The study was conducted according to the ethical requirements established by the Declaration of Helsinki. The Ethics Committee of Hospital Universitario Príncipe de Asturias (Madrid) approved the study.
Author contributions
Study concept and design: FPG, RPT and JCG
Patients’ selection and clinical data acquisition: FPG, RPT, JR, TA, PGH and JCG
Sample processing: FPG, JR, TA and PGH
Statistical analysis and interpretation of data: FPG and RPT
Writing of the manuscript: FPG, RPT, JR and JCG
Critical revision of the manuscript for relevant intellectual content: JR, JCG
Supervision and visualization: JCG
All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We want to thank Carolyn Brimley Norris, from the University of Helsinki Language Services for her help with the preparation of the manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104473.
Contributor Information
Felipe Pérez-García, Email: felipe.perez.garcia.87@gmail.com.
Ramón Pérez-Tanoira, Email: ramontanoira@hotmail.com.
Juan Romanyk, Email: jromanyk@yahoo.es.
Teresa Arroyo, Email: tarroyo26@hotmail.com.
Peña Gómez-Herruz, Email: pgherruz@gmail.com.
Juan Cuadros-González, Email: juan.cuadros@uah.es.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omer S.B., Malani P., Del Rio C. The COVID-19 pandemic in the US: a clinical update. JAMA. 2020 doi: 10.1001/jama.2020.5788. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . 2020. Novel Coronavirus (2019-nCoV) Situation Reports. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed May 16. [Google Scholar]
- 4.Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vashist S.K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics Basel (Basel) 2020;10(4) doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W., Cao Q., Qin L., Wang X., Cheng Z., Pan A. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):a multi-center study in Wenzhou city, Zhejiang, China. J. Infect. 2020;80(4):388–393. doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of Nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L., Ren L., Yang S., Xiao M., Chang Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020;(94):49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization . 2020. Clinical Management of Severe Acute Respiratory Infection (SARI) When COVID-19 Disease Is Suspected.https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected Available at: [Google Scholar]
- 17.Xiang Y.T., Jin Y., Wang Y., Zhang Q., Zhang L., Cheung T. Tribute to health workers in China: a group of respectable population during the outbreak of the COVID-19. Int. J. Biol. Sci. 2020;16(10):1739–1740. doi: 10.7150/ijbs.45135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9(3) doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai K., Tabata S., Ikeda M., Noguchi S., Kitagawa Y., Matuoka M. Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020 doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization . 2020. Advice on the Use of Point-of-care Immunodiagnostic Tests for COVID-19. Available at: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19. Accesed Apr 11. [Google Scholar]
- 23.Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020 doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.