Dear Editor,
We read with interest the work by Bermejo-Martin et al. [1] underlining the role of lymphopenia as a predictive marker of severe COVID-19 pneumonia. In an epidemic setting biomarkers can be useful at both patient level and for adequate resources allocation.
The gold standard for COVID-19 diagnosis relies on SARS-CoV-2 RNA detection by reverse transcription polymerase chain reaction (RT-PCR) through nasal and oropharyngeal swabs (despite suboptimal sensitivity, with the proportion of false negative results ranging from 56 to 83% [2]). Through this observational, cross-sectional study from a large Italian teaching hospital, we aimed to derive a diagnostic score to rapidly identify the possibility of being affected by COVID-19 at hospital admission, thus limiting the use of second-level diagnostic tests or a second swab.
Randomly selected, adult (≥18 years-old) patients coming to first aid with symptoms consistent with COVID-19 were considered for this analysis. The diagnosis relied on the results of RT-PCR on nasopharyngeal/oropharyngeal swabs, as recommended by International guidelines [3,4]. For most patients with initial negative swab, testing was repeated at least after 24 hours to definitely exclude the diagnosis.
The derivation cohort consisted in patients arrived at first aid between the 1st and the 15th of March 2020 (corresponding to the beginning of the epidemic in our hospital). The validation cohort included patients who came to first aid between the 21st and the 15th of April (i.e. at least after 14 days since national lockdown measures were declared by Italian government). Factors associated with a positive swab for SARS-CoV-2 (at a p-value≤0.05) were identified through a backward step-wise logistic regression. The multivariable regression model was then transformed into a point-based rule, as described by Sullivan et al.[5]. The discriminatory power and calibration of the prediction rule in the derivation and validation cohorts were assessed by the area under the receiver-operator characteristic curve (ROC AUC) and the Hosmer-Lemeshow test, respectively.
Data from 194 patients were analysed, 103 (53.4%) of whom in the derivation set and 91 (46.6%) in the validation set.
Fifty persons (48.5%) in the derivation set tested positive for SARS-CoV-2. Patients with negative swabs had other bacterial or viral infections (35 cases, 66.0%), cardiovascular and gastroenteric diseases (2 cases each, 7.6%), neoplasia (4, 7.6%), other/unspecified conditions (10, 18.8%). The derivation cohort was mainly composed of men (58.3%), with 53 years of median age (interquartile range, IQR, 38-70). Forty-one patients (39.8%) had at least one comorbidity among cardiovascular disease, hypertension, diabetes, active cancer and COPD. Fifty-two patients reported a possible contact with a case of COVID-19. At admission, the most frequent symptoms were represented by fever (88.3% of cases), cough (57.3%) and dyspnoea (35.9%). Twenty-three of 99 (23.2%) patients with a chest-X ray had interstitial pneumonia.
Five variables independently predicted COVID-19 diagnosis: the presence of an epidemiological link (aOR 10.35, p=0.001), total white blood cell count (per 100 cells/µL more, aOR 0.96, p=0.001), a CRP level<5 mL/min (versus ≥5 mL/min, aOR 0.07, p=0.002), the presence of the triad fever, cough and dyspnoea (versus the absence, aOR 10.02, p=0.012), time since symptoms onset (per 1 day more, aOR 1.33, p=0.001).
The validation group (90 persons) included 30 persons (33.3%) with a positive swab. Alternative diagnosis in patients without COVID-19 were: other viral or bacterial infections (30 of 60, 50.0%), cardiovascular disease (6, 10.0%), gastroenteric disease (8, 13.3%), neoplasia (9, 15.0%), other/unspecified conditions (7, 11.7%).
Overall, the validation group was mainly composed by men (57.8%), with 73 years of median age. Sixty-three patients (70.0%) had at least one comorbidity. Most of them also presented with fever (75.6%), cough (37.8%) and dyspnoea (55.6%). Compared with patients in the validation group, COVID-19 positive patients in derivation group were younger (60 versus 70 years of median age), had less frequently interstitial pneumonia at chest X-ray (38.0% versus 66.7%), more frequently fever (98.0% versus 86.7%) and cough (68.0% versus 40.0%) at admission, and lower median platelets count (169 versus 229 × 105 cells/µL).
Starting from regression coefficient, a score system was built in the derivation set (see table 1 ).
Table 1.
Estimated points corresponding to each risk factor category.
| Risk factor | Categories | Reference value (Wij) | βi | Points= βi(Wij − WiREF)/B |
|---|---|---|---|---|
| Epidemiological link (presence vs absence) |
Absent Present |
0 = WiREF 1 |
2.34 |
0 3 |
|
Total white blood cell count (per cells/µL increase) |
0-4,000 >4,000-6,000 >6,000-8,000 >8,000-10,000 >10,000-12,000 >12,000-14,000 >14,000-16,000 >16,000 |
3,000 5,000 7,000= WiREF 9,000 11,000 13,000 15,000 17,000 |
-0,00038 | 2 1 0 -1 -2 -3 -4 -4 |
| C-reactive protein |
≥5 mL/L <5 mL/L |
0 = WiREF 1 |
-2.62 |
0 -3 |
| Fever, cough and dyspnoea at admission |
Absence Presence |
0 = WiREF 1 |
2.31 |
0 3 |
| Days since symptoms onset | 0-3 >3-6 >6-9 >9-12 >12-15 |
1.5 4.5 = Ref 7.5 10.5 13.5 |
0.29 | -1 0 1 2 3 |
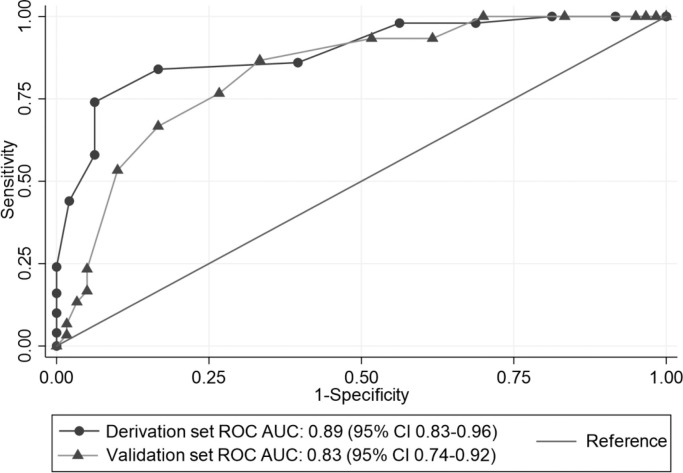
In the derivation group the ROC AUC for this model was 0.89 (95% CI 0.83-0.96) indicating excellent discriminatory power. Result of the Hosmer-Lemeshow chi-squared testing (χ2 13.50, p=0.334) also indicated good calibration. A cut-off of ≥-3 showed the highest negative predictive value (100%), whereas a cut-off of ≥5 exhibited the highest positive predictive value (100%).
When applied to the validation set, the predictive score demonstrated good predictive power, with a ROC AUC of 0.83 (95% CI 0.74-0.92), and a good calibration (Hosmer-Lemeshow χ2 7.58, p=0.476). A score cut-off of ≥-3 exhibited the highest negative predictive value (100%) and a cut-off of ≥3 had the highest positive predictive value (72.7%). Fig. 1 compares the score ROC AUC in both derivation and validation sets.
Fig. 1.
Receiver-operator characteristic curves (ROC AUC) for the scoring system in the derivation and validation set.
As noticed by Bermejo-Martin et al. [1], biomarkers are urgently requested for correct categorization of patients with COVID-19. Several evidences showed that lymphopenia could have a prognostic value in COVID-19 [1], in addition to its diagnostic value, already recognized in Chinese guidelines [6]. Interestingly, in our work total white cells count was more associated with a diagnosis of COVID-19 than lymphocytes count. This could be related with an overall low severity spectrum of disease shown by our patients (only a half of COVID-19 patients had interstitial pneumonia at admission). CRP, epidemiological link, clinical symptoms and time since symptoms onset were also associated with the pre-test probability of COVID-19. Particularly, the derived prediction-rule showed higher utility in ruling-out COVID-19 diagnosis: in the derivation and validation groups only 1 of 21 (4.8%) and 2 of 29 (6.9%) patients with a score of less than -1 tested positive for SARS-CoV-2. Despite its intrinsic limitations (retrospective data collection, derivation and validation cohorts belonging to the same hospital), our works represents to our knowledge the first attempt to measure the impact of an easily-available score for stratifying the risk of COVID-19. This could represent an important tool for assisting clinicians as well for driving hospital policies of infection control, particularly in resource-limited settings.
Funding
This study was performed as part of our routine work.
Transparency declarations
The authors report no conflicts of interest relevant to this work.
Declaration of Competing Interest
None.
Acknowledgements
Residents against COVID-Unit Elite (ResCUE)-Team: F. Agostini, S. Bibbo, A. D'Angelillo, A. Dusina, D. Farinacci, D. Feliciani, M. Garcovich, F.Landi, F. Mangiola, D. Moschese, C. Picarelli, V. Popolla, L. Salvatore, M. Sanguinetti, F. Santopaolo, R. Talerico, A. Tosoni.
References
- 1.Bermejo-Martin JF, Almansa R, Menéndez R, Mendez R, Kelvin DJ, Torres A. Lymphopenic community acquired pneumonia as signature of severe COVID-19 infection. J Infect. 2020;80(5):e23–e24. doi: 10.1016/j.jinf.2020.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kokkinakis I, Selby K, Favrat B, Genton B, Cornuz J. Performance du frottis nasopharyngé-PCR pour le diagnostic du Covid-19 - Recommandations pratiques sur la base des premières données scientifiques [Covid-19 diagnosis: clinical recommendations and performance of nasopharyngeal swab-PCR] Rev Med Suisse. 2020;16(689):699–701. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons Under Investigation (PUIs) for Coronavirus Disease 2019 (COVID-19) 2020;14 https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html February. (Accessed on March 15, 2020) [Google Scholar]
- 4.Interim guidance. WHO; 2020. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases.https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 19 MarchAvailable at. [Google Scholar]
- 5.Sullivan LM1, Massaro JM, D'Agostino RB., Sr Presentation of multivariate data for clinical use: The Framingham Study risk score functions. Stat Med. 2004;23(10):1631–1660. doi: 10.1002/sim.1742. May 30. [DOI] [PubMed] [Google Scholar]
- 6.National Health Commission of the People's Republic of China, The Diagnosis and Treatment Plan for 2019-nCoV (The Seventh Trial Edition).