Abstract
As the Coronavirus 2019 (COVID-19) pandemic evolves, the development of immunoassays to help determine exposure and potentially predict immunity has become a pressing priority. In this report we present the performance of the EUROIMMUN enzyme-linked immunosorbent assay (ELISA) for semi-quantitative detection of IgA and IgG antibodies in serum and plasma samples using recombinant S1 domain of the SARS-CoV-2 spike protein as antigen. Specimens from patients, with and without COVID-19 infection, were tested at the University of Chicago Clinical Microbiology and Immunology Laboratory. Of 86 samples from SARS-CoV-2 PCR-negative patients, including 28 samples positive for common human coronavirus strains, 76 tested negative and 10 tested positive for IgA (88.4% agreement, 95% CI: 79.9–93.6) while 84 tested negative and 2 tested positive for IgG (97.7% agreement, 95% CI: 91.9–99.6). Of 82 samples from SARS-CoV-2 PCR-positive patients, 14 tested negative and 68 tested positive for IgA (82.9% agreement, 95% CI: 73.4–89.5) while 27 tested negative and 55 tested positive for IgG (67.1% agreement, 95% CI: 56.3–76.3). Of samples collected ≥4 days after positive PCR, 38 of 42 (90.5% agreement, 95% CI: 77.9–96.2) were positive for IgA, and 42 of 42 (100% agreement, 95% CI: 91.6–100) were positive for IgG, respectively.
The EUROIMMUN Anti-SARS-CoV-2 ELISA Assay demonstrated good sensitivity for detection of IgA and excellent sensitivity for detection of IgG antibodies from samples collected ≥4 days, after COVID-19 diagnosis by PCR. This assay demonstrated good specificity for IgA and excellent specificity for IgG and demonstrated only borderline cross reaction in 2 of the 28 samples from patients with common human coronaviruses infection, types NL63 and OC43.
Keywords: Serology, COVID-19, SARS-CoV-2
1. Introduction
In December 2019 a novel coronavirus emerged as the cause of severe respiratory disease and quickly spread causing a worldwide pandemic. Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2) was determined to be the agent of coronavirus disease 2019 (COVID-19). The virus belongs to the Betacoronavirus genus of the Coronaviridae family, which also includes Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [1]. For diagnostic purposes many nucleic acid amplification assays were quickly developed and received Emergency Use Authorization (EUA) from the US Food and Drug Administration (FDA). Multiple manufacturers are offering serological assays, but few have received EUA; the EUROIMMUN IgG assay has received EUA from the FDA. Serological testing may be useful in conjunction with other laboratory tests and clinical findings of COVID-19 infection for epidemiological monitoring and outbreak control. Of the immunoassays currently available, choice of SARS-CoV-2 target antigens include the spike protein (S) or the nucelocapsid (N) [2,6,7]. IgA antibodies can show higher sensitivity, while IgG antibodies typically have longer duration, better specificity, and are better suited for serosurveillance studies [[3], [4], [5]].
2. Materials and methods
The EUROIMMUN Anti-SARS-CoV-2 Assay is an enzyme-linked immunosorbent assay (ELISA) that provides semi-quantitative in vitro determination of human antibodies of immunoglobulin classes IgA and IgG against SARS-CoV-2 in serum or EDTA plasma [6,7].
Each kit contains microplate strips with 8 break-off reagent wells coated with recombinant structural protein of SARS-CoV-2. In the first reaction step, diluted patient samples are incubated in the wells. In the case of positive samples, specific antibodies will bind to the antigens. To detect the bound antibodies, a second incubation is carried out using an enzyme-labelled antihuman IgA or IgG (enzyme conjugate) catalyzing a color reaction.
Results are evaluated semi-quantitatively by calculation of a ratio of the extinction of the control or patient sample over the extinction of the calibrator. This ratio is interpreted as follows: < 0.8 negative; ≥ 0.8 to <1.0 borderline; ≥ 1.1 positive. Borderline results were considered positive for analysis.
The University of Chicago Medicine uses two different RT-PCR assays allowed by the FDA under EUA. The Roche cobas 6800 SARS-CoV-2 assay relies on amplification of the SARS-CoV-2 specific ORF1 gene as well as a portion of the E-gene conserved across the sarbecoviruses, a subgenus of coronaviruses which includes SARS-CoV-2. The Cepheid Xpert Xpress SARS-CoV-2 assay also detects the pan-sarbecovirus E-gene but uses the SARS-CoV-2 specific N-gene rather than ORF1 as its primary target. Samples tested include nasopharyngeal and nasal mid-turbinate swabs transported in viral transport or liquid Amies media.
The BioFire FilmArray Respiratory Panel 2 (RP2) is a multiplex in vitro molecular diagnostic test for the simultaneous and rapid detection of 21 pathogens, including 4 common human coronavirus strains, directly from nasopharyngeal swab (NPS) samples.
Stored residual serum and plasma samples submitted to the University of Chicago Medicine Clinical Laboratories for routine testing were recovered for this evaluation.
Percent agreement was determined and the hybrid Wilson/Brown method was used to calculate 95% confidence intervals of proportions (95% CI). All statistical analyses were performed using GraphPad Prism version 8.4.1.
3. Results
Eighty six blood samples were tested from 84 patients thought to be negative for exposure to SARS-CoV-2 (70 samples collected from ambulatory patients at the University of Chicago with negative results for SARS-CoV-2 by PCR, from March to May 2020 and 16 collected in early 2019, prior to the current pandemic, stored at −20 °C). Twenty-eight unique samples were from patients who had tested positive by the BioFire FilmArray RP2 respiratory viral panel for common coronavirus strains (6 samples positive for HKU1, 10 positive for NL63, 9 positive for OC43, 2 positive for 229E, and one positive for both OC43 and 229E).
Of these 86 samples, 76 tested negative and 10 tested positive for IgA (88.4% agreement, 95% CI: 79.9–93.6) while 84 tested negative and 2tested positive for IgG (97.7% agreement, 95% CI: 91.9–99.6). Four borderline results for IgA and one borderline result for IgG were included in the positive results.
One sample was highly positive for both IgA and IgG. This positive serological result and negative PCR could represent prior infection with SARS-CoV-2 with clearance of the virus.
Borderline cross reactivity was detected in one patient with NL63 infection for IgG and in one patient with coinfection OC43 and rhinovirus for IgA. For the other eight samples that tested either borderline or low positive for IgA and negative for IgG, it is unclear if positivity is due to patient prior exposure to SARS-CoV-2, beginning of infection with false negativity with SARS-CoV-2 PCR or cross reactivity with viruses not detectable on the respiratory viral panel.
Eighty two blood samples collected from 64 patients from March to May 2020 with PCR-positive SARS-CoV-2 were tested. Of these samples, 68 tested positive and 14 tested negative for IgA (82.9% agreement, 95% CI: 73.4–89.5) while 55 tested positive and 27 tested negative for IgG (67.1% agreement, 95% CI: 56.3–76.3). Six borderline results for IgA and two borderline results for IgG were included in the positive results.
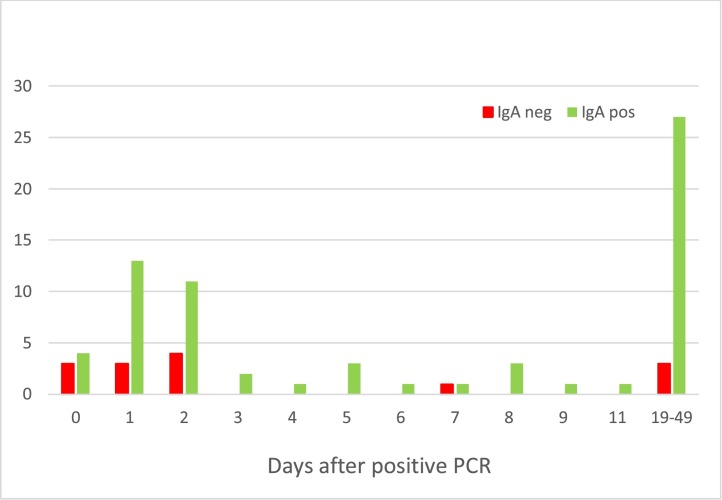
These 82 samples from PCR positive patients were collected 0–49 days after PCR testing. The 11 IgA-negative samples and the 27 IgG-negative samples were collected within 7 days of PCR testing. Since antibody development occurs after viremia and requires time to reach a detectable concentration, it is likely that these samples were collected too early in the course of disease to expect antibody production. Of samples collected ≥4 days after positive PCR, 38 of 42 (90.5% agreement, 95% CI: 77.9–96.2) were positive for IgA Fig. 1 , and 42 of 42 (100% agreement, 95% CI: 91.6–100) were positive for IgG Fig. 2 , respectively.
Fig. 1.
Timeline of IgA results from PCR positive patients.
Fig. 2.
Timeline of IgG results from PCR positive patients.
4. Cross-reactivity
A total of 28 unique samples that tested positive by BioFire FilmArray RP2 panel for common human coronaviruses including types 229E, NL63, HKU1 and OC43 were tested for cross reactivity. This assay demonstrated only borderline cross reaction in 2 of the 28 samples from patients with common human coronaviruses (types NL63 and OC43).
5. Discussion and conclusions
A positive or negative test for the SARS-CoV2 antibody is difficult to interpret at this time. It is not yet known if antibodies serve as an indication of the presence or absence of protective or sustained immunity. Negative SARS-CoV-2 antibody results do not rule out SARS-CoV-2 infection, particularly in those who have been in contact with the virus. Results from antibody testing should not be used as the sole basis to diagnose or exclude SARS-CoV-2 infection or to inform infection status. The EUROIMMUN Anti-SARS-CoV-2 ELISA Assay demonstrated good sensitivity for detection of IgA and excellent sensitivity for detection of IgG antibodies from samples collected ≥4 days after COVID-19 diagnosis by PCR. The EUROIMMUN Anti-SARS-CoV-2 ELISA Assay demonstrated good specificity for IgA and excellent specificity for IgG and did not demonstrate cross reaction with common human coronaviruses, except borderline cross reactivity in 2 of the 28 samples.
Limitations of this study include a small sample size and a lack of specimens collected more than 49 days following a positive PCR.
Declaration of competing interest
All authors declared no conflict of interest. Study was internally funded.
Acknowledgement
The authors thank Caroline Guenette and Caroline Hokl from Occupational Medicine at the University of Chicago.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang A.T., Garcia-Carreras B., Hitchings MDT Yang B., Katzelnick L., Rattigan S.M., Borgert B., Moreno C., Solomon B.D., Rodriguez-Barraquer I., Lessler J., Salje H., Burke D.S., Wesolowski A., Cummings DAT . 2020. A Systematic Review of Antibody Mediated Immunity to Coronaviruses: Antibody Kinetics, Correlates Of Protection, And Association of Antibody Responses with Severity of Disease. medRxiv 04.14.20065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C.Y., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. 2020. Evaluation of Nine Commercial SARS-CoV-2 Immunoassays. medRxiv 04.09.20056325. [DOI] [Google Scholar]
- 5.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamer M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2 − specific antibody responses in coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EUROIMMUN . 2020. Anti-SARS-CoV-2 ELISA IgG, Package Insert. EI_2606G_A_US_C02.docx Version: 2020-05-04. [Google Scholar]
- 7.EUROIMMUN . 2020. Anti-SARS-CoV-2 ELISA IgA, Package Insert. EI_2606A_A_US_C01.docx Version: 2020-03-24. [Google Scholar]