Highlights
-
•
Performance of the LightMix® E-gene kit for SARS-CoV-2 detection was evaluated.
-
•
The LightMix® kit had similar analytical and diagnostic sensitivity as the comparator in-house assays.
-
•
Cycle threshold values of the three assays showed excellent correlation.
-
•
The LightMix® kit performed well as a stand-alone test for SARS-CoV-2 detection.
Keywords: LightMix E-gene, SARS-CoV-2, COVID-19, Diagnostic, Evaluation, PCR
Abstract
Background
Rapid and sensitive diagnostic assays for SARS-CoV-2 detection are required for prompt patient management and infection control. The analytical and clinical performances of LightMix® Modular SARS and Wuhan CoV E-gene kit, a widely used commercial assay for SARS-CoV-2 detection, have not been well studied.
Objective
To evaluate the performance characteristics of the LightMix® E-gene kit in comparison with well-validated in-house developed COVID-19 RT-PCR assays.
Study design
Serial dilutions of SARS-CoV-2 culture isolate extracts were used for analytical sensitivity evaluation. A total of 289 clinical specimens from 186 patients with suspected COVID-19 and 8 proficiency testing (PT) samples were used to evaluate the diagnostic performance of the LightMix® E-gene kit against in-house developed COVID-19-RdRp/Hel and COVID-19-N RT-PCR assays.
Results
The LightMix® E-gene kit had a limit of detection of 1.8 × 10−1 TCID50/mL, which was one log10 lower than those of the two in-house RT-PCR assays. The LightMix® E-gene kit (149/289 [51.6%]) had similar sensitivity as the in-house assays (144/289 [49.8%] for RdRp/Hel and 146/289 [50.5%] for N). All three assays gave correct results for all the PT samples. Cycle threshold (Cp) values of the LightMix® E-gene kit and in-house assays showed excellent correlation. Reproducibility of the Cp values was satisfactory with intra- and inter-assay coefficient of variation values <5%. Importantly, the LightMix® E-gene kit, when used as a stand-alone assay, was equally sensitive as testing algorithms using multiple COVID-19 RT-PCR assays.
Conclusions
The LightMix® E-gene kit is a rapid and sensitive assay for SARS-CoV-2 detection. It has fewer verification requirements compared to laboratory-developed tests.
1. Background
In late December 2019, a novel coronavirus, now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in patients with pneumonia in Wuhan, China [1]. SARS-CoV-2 is efficiently transmitted from person to person and has rapidly disseminated globally [2,3]. The World Health Organization declared Coronavirus Disease 2019 (COVID-19) as a pandemic in early March 2020. As of 23 May 2020, over 4.9 million COVID-19 cases including more than 327,000 deaths attributable to SARS-CoV-2 have been reported globally (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Rapid, sensitive and specific diagnostic tests for COVID-19 are of paramount importance to facilitate early identification of cases, contact tracing, and isolation [4,5].
Reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard for laboratory diagnosis of COVID-19 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance). While a number of in-house and commercial RT-PCR assays for COVID-19 have been developed in the past few months, the clinical performance of some of these assays has not been well studied.
2. Objectives
In the present study, we evaluated the performance of a commercially available LightMix® Modular SARS and Wuhan CoV E-gene kit capable of detecting SARS-CoV-2 RNA against well-validated in-house developed RT-PCR assays targeting the RNA-dependent RNA polymerase (RdRp)/Helicase (Hel) and nucleocapsid (N) regions of SARS-CoV-2 using archived clinical specimens and proficiency testing samples [6].
3. Study design
3.1. Viruses, clinical specimens and proficiency testing samples
SARS-CoV-2 was isolated from the nasopharyngeal aspirate specimen of a patient with COVID-19 in Hong Kong as previously described [7]. SARS-CoV-2 stock (1.8 × 107 TCID50/mL) was prepared by one additional passage in VeroE6 cells [8,9]. For analytical sensitivity evaluation, 10-fold serial dilutions of total nucleic acid (TNA) extracted from the SARS-CoV-2 isolate were used. For analytical specificity evaluation, TNA extracted from a clinical specimen positive for human coronavirus HKU1 (HCoV-HKU1) and 17 culture isolates of other human-pathogenic coronaviruses and respiratory viruses were used [6,10]. For clinical evaluation, 289 clinical specimens (236 respiratory tract and 53 non-respiratory tract) from 186 hospitalized patients (male : female = 90 : 96; median age: 37 years; range: 18–97 years) with suspected COVID-19 were selected for SARS-CoV-2 RNA detection. In addition to clinical specimens, 8 proficiency testing samples from Quality Control for Molecular Diagnostics (QCMD) with different concentrations of SARS-CoV-2 RNA or negative for SARS-CoV-2 RNA were also evaluated.
3.2. Nucleic acid extraction
TNA extraction was performed using NucliSENS easyMAG extraction system (BioMerieux, Marcy-l'Étoile, France) according to the manufacturer’s instructions and as previously described [[11], [12], [13]]. Briefly, 250 μL of each respiratory tract specimen, rectal swab and stool specimen were subjected to extraction with an elution volume of 55 μL; and 100 μL of each plasma specimen were subjected to extraction with an elution volume of 25 μL. The extracts were stored at −80 °C until use.
3.3. Real-time RT-PCR assays for SARS-CoV-2 RNA detection
LightMix® Modular SARS and Wuhan CoV E-gene kit (TIB Molbiol, Berlin, Germany) with LightCycler Multiplex RNA Virus Master (Roche, Basel, Switzerland) was used according to the manufacturer’s instructions. Briefly, each 20 μL reaction mixture contained 5.4 μL of water, 4 μL of Roche Master, 0.5 μL of reagent mix, 0.1 μL of RT Enzyme, and 10 μL of TNA as the template. RT-PCR was performed on a LightCycler 480 II Real-Time PCR System (Roche). The thermal cycling condition was 55 °C for 5 min, 95 °C for 5 min, followed by 45 cycles of 95 °C for 5 s, 60 °C for 15 s and 72 °C for 15 s.
In-house developed COVID-19-RdRp/Hel and COVID-19-N RT-PCR assays were performed using QuantiNova Probe RT-PCR Kit (QIAGEN, Hilden, Germany) on the LightCycler 480 II Real-Time PCR System (Roche) as previously described [6]. Each 20 μL reaction mixture contained 10 μL of 2x QuantiNova Probe RT-PCR Master Mix, 0.2 μL of QN Probe RT-Mix, 1.6 μL of each 10 μM forward and reverse primer, 0.4 μL of 10 μM probe, 1.2 μL of nuclease-free water and 5 μL of TNA as the template. The thermal cycling condition was 45 °C for 10 min, 95 °C for 5 min, followed by 45 cycles of 95 °C for 5 s and 55 °C for 30 s.
3.4. Statistical analysis
Fisher’s exact test was used to compare the performance of the assays. Spearman’s correlation was used to assess the relation between the Cp values of different assays. The Cp values obtained from the three assays were compared using ANOVA Friedman test with Dunn’s multiple comparisons test (a Cp value of 41 was assigned to specimens that tested negative in the real-time RT-PCR assay). Statistical analysis was performed using GraphPad Prism 8. P < 0.05 was considered statistically significant.
4. Results
4.1. Analytical sensitivity, analytical specificity and imprecision of the LightMix® E-gene assay
To determine the analytical sensitivity of the LightMix® E-gene assay, the limit of detection (LOD) was evaluated by using TNA extracted from the SARS-CoV-2 isolate. Serial 10-fold dilutions of SARS-CoV-2 TNA extracted from the viral culture isolate were prepared and tested in triplicate for each concentration in two independent runs. The LOD of the E-gene assay was 1.8 × 10−1 TCID50/mL (Table 1 ).
Table 1.
Test results for determining the limit of detection of the LightMix® Modular SARS and Wuhan CoV E-gene assay with genomic RNA extracted from a SARS-CoV-2 culture isolate.
| Virus titer (TCID50/mL) | Cp (Intra-run) |
Cp (Inter-run) |
||||
|---|---|---|---|---|---|---|
| Test 1 | Test 2 | Test 3 | Test 1 | Test 2 | Test 3 | |
| 1.8 × 101 | 30.04 | 30.05 | 29.91 | 30.23 | 30.42 | 30.44 |
| 1.8 × 10° | 33.24 | 33.61 | 33.64 | 33.57 | 34.01 | 33.77 |
| 1.8 × 10−1 | 36.50 | 36.03 | 36.64 | 37.25 | 37.61 | 37.05 |
| 1.8 × 10−2 | 40.00 | 40.00 | – | – | – | 38.16 |
| 1.8 × 10−3 | 37.87 | – | – | – | – | – |
Abbreviations: -, negative; Cp, cycle number at detection threshold.
To investigate whether the LightMix® E-gene assay would non-specifically amplify other human-pathogenic coronaviruses and respiratory viruses, we tested TNA extracted from the clinical respiratory specimen with HCoV-HKU1, and TNAs extracted from the 17 culture isolates of SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E, influenza A ((H1N1)pdm09 and H3N2) viruses, influenza B virus, influenza C virus, parainfluenza virus types 1–4, respiratory syncytial virus, human metapneumovirus, human rhinovirus and human adenovirus. The LightMix® E-gene assay did not cross react with these respiratory viruses, except SARS-CoV.
Different concentrations of TNA extracted from the SARS-CoV-2 isolate were used to evaluate intra- and inter-assay variations by the LightMix® E-gene assay. Each concentration was tested in triplicate in two independent runs. The total imprecision (% CV) values ranged from 0.72% to 1.54% (Table 2 ).
Table 2.
Imprecision of the LightMix® Modular SARS and Wuhan CoV E-gene assay using the SARS-CoV-2 culture isolate extracts.
| Intra-assay | Inter-assay | ||
|---|---|---|---|
| Virus titer (TCID50/mL) | Number of positive replicates | Mean Cp ± SD (% coefficient of variation) |
Mean Cp ± SD (% coefficient of variation) |
| 1.8 × 101 | 3 | 30.00 ± 0.08 (0.26) | 30.18 ± 0.22 (0.72) |
| 1.8 × 10° | 3 | 33.50 ± 0.22 (0.67) | 33.64 ± 0.25 (0.75) |
| 1.8 × 10−1 | 3 | 36.39 ± 0.32 (0.88) | 36.85 ± 0.57 (1.54) |
4.2. Comparative performance of the LightMix® E-gene assay and the in-house COVID-19-RdRp/Hel and COVID-19-N assays for the detection of SARS-CoV-2 RNA in clinical specimens and proficiency testing samples
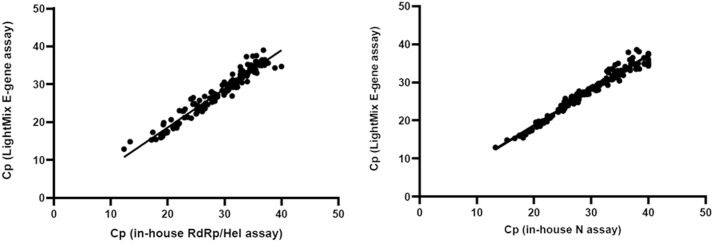
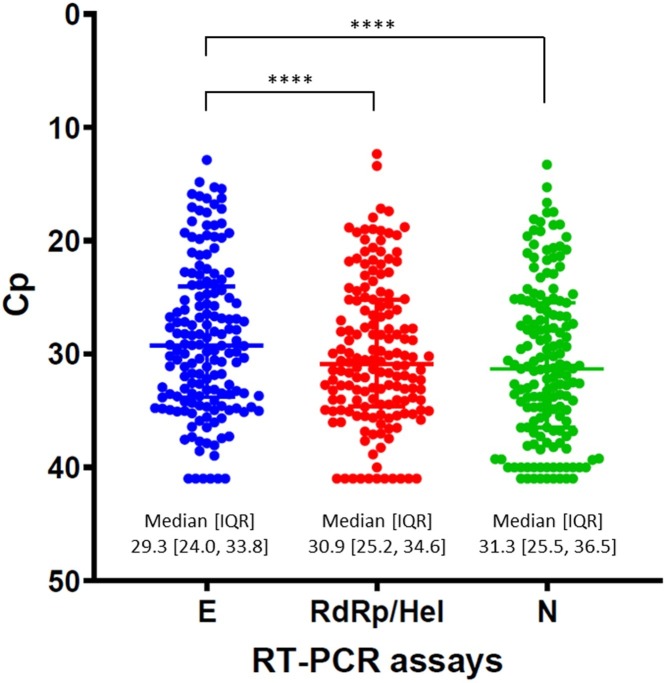
Overall, 289 clinical specimens from 186 patients were evaluated in this study. The LightMix® E-gene kit detected 149/289 [51.6%] specimens, and had similar sensitivity as the in-house assays (144/289 [49.8%] for RdRp/Hel, P = 0.739; and 146/289 [50.5%] for N, P = 0.868). Of these 289 specimens, 195 were initial specimens obtained from the 186 patients with suspected COVID-19. Among these 186 patients, 72 patients were positive for SARS-CoV-2 RNA by at least two of the assays and 114 patients were negative by all three assays in their initial specimens. For the initial specimens of these patients, the positive detection rate of the LightMix® E-gene assay was 71/72 (98.6%), while those of the in-house COVID-19-RdRp/Hel and COVID-19-N assays were 71/72 (98.6%) and 70/72 (97.2%), respectively. The remaining specimens were follow-up specimens of the confirmed cases. Among a total of 94 follow-up specimens obtained from the laboratory-confirmed COVID-19 patients, 11 were negative by all three assays. Seventy-eight (83.0%) were positive by the LightMix® E-gene assay, while 73 (77.7%) and 76 (80.9%) were positive by the in-house COVID-19-RdRp/Hel and COVID-19-N assays, respectively. We then compared the sensitivity of the LightMix® E-gene assay for the follow-up clinical specimens against a testing algorithm involving combination of two of the three or all three assays. We found that adding the in-house assays to the LightMix® E-gene assay did not result in a significant increase in sensitivity (Table 3 ). There was no significant difference in the detection rate between the LightMix® E-gene assay and our in-house assays. The sensitivity of these assays did not differ significantly for both respiratory and non-respiratory tract specimens (Table 4 ). For the specimens with discordant results, their mean cycle threshold (Cp) value was 36.7, which represented very low viral RNA load. Among the 8 proficiency testing samples from QCMD, all three assays provided 100% correct results. A good agreement in the performance of the LightMix® E-gene assay compared to the in-house assays was evidenced by a strong correlation (Spearman’s ρ > 0.97; P < 0.0001) (Fig. 1 ). The Cp values obtained from the 3 different assays were also examined. The median Cp value of the LightMix® E-gene assay (29.3) was significantly lower than those of the COVID-19-RdRp/Hel (30.9; P < 0.0001) and COVID-19-N (31.3; P < 0.0001) assays (Fig. 2 ).
Table 3.
Comparative performance of the three RT-PCR assays in follow-up specimens of confirmed COVID-19 patients.
| Combination of different assays | No. of specimens missed by the assay(s) / total no. of positive follow-up specimens (%) | P-value* |
|---|---|---|
| LightMix® E-gene | 5/83 (6.0) | N/A |
| COVID-19-RdRp/Hel | 10/83 (12.0) | 0.279 |
| COVID-19-N | 7/83 (8.4) | 0.766 |
| LightMix® E-gene + COVID-19-RdRp/Hel | 0/83 (0) | 0.059 |
| LightMix® E-gene + COVID-19-N | 3/83 (3.6) | 0.720 |
| COVID-19-RdRp/Hel + COVID-19-N | 3/83 (3.6) | 0.720 |
| All 3 assays | 0/83 (0) | 0.059 |
* P-value for comparison between LightMix® E-gene assay and other assay combinations. N/A: not applicable.
Table 4.
Comparative performance of the three RT-PCR assays in respiratory and non-respiratory tract specimens.
| No. of positive test results/no. of specimens (%) |
|||||
|---|---|---|---|---|---|
| Specimen typea | LightMix® Modular E-gene | COVID-19-RdRp/Hel | COVID-19-N | P valueb | P valuec |
| Respiratory tract | 117/236 (49.6) | 113/236 (47.9) | 115/236 (48.7) | 0.782 | 0.927 |
| NPA/NPS/TS | 94/202 (46.5) | 87/202 (43.1) | 90/202 (44.6) | 0.548 | 0.764 |
| Saliva | 23/34 (67.6) | 26/34 (76.5) | 25/34 (73.5) | 0.590 | 0.791 |
| Non-respiratory | 32/53 (60.4) | 31/53 (58.5) | 31/53 (58.5) | 1 | 1 |
| Stool/rectal swabs | 23/33 (69.7) | 24/33 (72.7) | 22/33 (66.7) | 1 | 1 |
| Plasma | 9/20 (45.0) | 7/20 (35.0) | 9/20 (45.0) | 0.748 | 1 |
| Total | 149/289 (51.6) | 144/289 (49.8) | 146/289 (50.5) | 0.739 | 0.868 |
aAbbreviations: NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab; TS, throat swab bP value for LightMix® E-gene and COVID-19-RdRp/Hel assays cP value for LightMix® E-gene and COVID-19-N assays.
Fig. 1.
Correlation of the Cp values of the specimens tested positive for SARS-CoV-2 RNA by the assays. (A) LightMix® E-gene assay vs in-house COVID-19-RdRp/Hel assay and (B) LightMix® E-gene assay vs in-house COVID-19-N assay.
Fig. 2.
Comparison of the Cp values of the three RT-PCR assays in this study. **** indicates P < 0.0001.
5. Discussion
An increasing number of in-house and commercial COVID-19 RT-PCR assays have been described in the past 5 months [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]]. The commercially available LightMix® Modular SARS and Wuhan CoV E-gene kit is widely used in clinical laboratories, but its performance has not been thoroughly evaluated with clinical specimens. In the present study, we compared the performance of the LightMix® E-gene kit with two previously established and validated in-house COVID-19 RT-PCR assays using a variety of clinical specimens and proficiency testing samples [6].
According to the manufacturer instructions, the LightMix® E-gene assay can detect not only SARS-CoV-2, but other sarbecoviruses including SARS-CoV and bat SARS-related coronaviruses. In our analytical specificity evaluation, the LightMix® E-gene assay detected SARS-CoV but not other common human-pathogenic coronaviruses and respiratory viruses, while our in-house COVID-19-RdRp/Hel and COVID-19-N assays were specific for SARS-CoV-2 without cross-reactivity with SARS-CoV. The LightMix® E-gene assay was likely intentionally designed to cross-react with SARS-CoV because of the scarce information on the genetic diversity of SARS-CoV-2 in human and animals in late December 2019 when it was developed. To avoid under-diagnosis, the primers and probe targeting the viral E gene were designed to detect not only SARS-CoV-2 but also other sarbecoviruses. This strategy was similarly used for designing other RT-PCR assays in the earlier phase of the COVID-19 pandemic [14,15]. It would therefore be a reasonable strategy to use the sensitive LightMix® E-gene assay as the first-line screening assay for suspected COVID-19 cases, followed by confirmation by sequencing or another RT-PCR assay specific to SARS-CoV-2 (https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117).
The LightMix® E-gene assay was highly sensitive for SARS-CoV-2 RNA detection, with LOD of 1.8 × 10−1 TCID50/mL, which is one log10 TCID50/mL lower than our previously described COVID-19-RdRp/Hel and COVID-19-N assays (1.8 TCID50/mL) [6]. The median Cp value of the LightMix® E-gene assay was also significantly lower than the in-house assays. This might be due to the higher volume of specimen template used in the LightMix® E-gene assay than the in-house assays. Another possibility is that the LightMix® E-gene assay and our in-house assays were performed using different PCR reagents and thermocycling conditions. Nevertheless, no significant difference in the sensitivity was noted among these three assays for both respiratory and non-respiratory tract specimens. Reproducibility of the Cp values was satisfactory with the intra- and inter-assay coefficient of variation values of <5% [[27], [28], [29]]. The Cp values of the LightMix® E-gene/COVID-19-RdRp/Hel and E-gene/COVID-19-N assays showed excellent correlation. All three assays performed well in the proficiency testing samples from QCMD. These findings suggested that the LightMix® E-gene assay and our in-house assays showed excellent diagnostic performance for SARS-CoV-2 RNA detection.
Healthcare facilities including our hospital use RT-PCR negativity as a criterion for hospital discharge. However, the false-negative rate of RT-PCR assays may rise during the convalescent phase of illness as the patient’s viral RNA load decreases. Thus, it remains controversial as to how many RT-PCR assays targeting different gene regions should be used to test convalescent phase patients. Our results showed that the LightMix® E-gene assay performed well as a stand-alone test with similar sensitivity as other testing algorithms using multiple tests for follow-up clinical specimens. This feature is reassuring and obviates the need for testing follow-up specimens with multiple assays, especially in areas where diagnostic kits are limited.
In addition to the analytical and clinical performance, the turnaround time and cost are also essential factors affecting the choice of diagnostic assays, especially when there is a large number of clinical specimens from patients with suspected COVID-19 during this pandemic. The sample-to-extract time was the same among the three assays because the same extraction method was used, while the PCR running time of the LightMix® E-gene assay (66 min) was slightly shorter than our in-house COVID-19-RdRp/Hel and COVID-19-N assays (72 min). For the reagent cost including the PCR reagents and primers/probes, our in-house assays (US$2 per reaction) were much lower than the LightMix® E-gene assay (US$10 per reaction). For clinical laboratories without the necessary expertise in the development of in-house assays, the LightMix® E-gene kit may be an alternative commercially available diagnostic option.
In conclusion, the LightMix® Modular SARS and Wuhan CoV E-gene kit is a rapid and highly sensitive assay for screening suspected cases of COVID-19. Further confirmation can be achieved by performing another assay specific to SARS-CoV-2, such as our in-house COVID-19-RdRp/Hel and COVID-19-N assays with lower cost.
Ethics approval and consent to participate
This study was approved by Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. Data records were de-identified and completely anonymous, so informed consent was waived.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Cyril Chik-Yan Yip: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing - original draft. Siddharth Sridhar: Formal analysis, Writing - original draft, Writing - review & editing. Andrew Kim-Wai Cheng: Validation, Investigation, Data curation. Kit-Hang Leung: Validation, Investigation. Garnet Kwan-Yue Choi: Methodology. Jonathan Hon-Kwan Chen: Investigation. Rosana Wing-Shan Poon: Investigation. Kwok-Hung Chan: Investigation. Alan Ka-Lun Wu: Investigation. Helen Shuk-Ying Chan: Investigation. Sandy Ka-Yee Chau: Investigation. Tom Wai-Hin Chung: Investigation. Kelvin Kai-Wang To: Investigation. Owen Tak-Yin Tsang: Investigation. Ivan Fan-Ngai Hung: Investigation, Resources. Vincent Chi-Chung Cheng: Resources. Kwok- Yung Yuen: Writing - review & editing, Supervision. Jasper Fuk-Woo Chan: Conceptualization, Formal analysis, Writing - original draft, Writing - review & editing, Supervision.
Declaration of Competing Interest
J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited, and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. S.S. has received speaker’s honoraria from Abbott Laboratories Limited. The other authors declared no conflict of interests. The funding sources had no role in study design, data collection, analysis or interpretation or writing of the report.
Acknowledgements
This study was partly supported by the donations of Lo Ying Shek Chi Wai Foundation, Richard Yu and Carol Yu, May Tam Mak Mei Yin, the Shaw Foundation Hong Kong, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, and Kai Chong Tong; and the National Program on Key Research Project of China (grant no. 2020YFA0707500 and 2020YFA0707504); the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government; and the Theme-Based Research Scheme (T11/707/15) of the Research Grants Council, Hong Kong Special Administrative Region. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Contributor Information
Kwok- Yung Yuen, Email: kyyuen@hku.hk.
Jasper Fuk-Woo Chan, Email: jfwchan@hku.hk.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng V.C., Wong S.C., Chuang V.W., So S.Y., Chen J.H., Sridhar S. The role of community-wide wearing of face mask for control of coronavirus disease 2019 (COVID-19) epidemic due to SARS-CoV-2. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng V.C.C., Wong S.C., Chen J.H.K., Yip C.C.Y., Chuang V.W.M., Tsang O.T.Y. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020;41:493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse Transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV: implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yip C.C., Ho C.C., Chan J.F., To K.K., Chan H.S., Wong S.C. Development of a novel, genome subtraction-derived, SARS-CoV-2-Specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yip C.C., Chan W.M., Ip J.D., Seng C.W., Leung K.H., Poon R.W. Nanopore Sequencing Reveals Novel Targets for Detection and Surveillance of Human and Avian Influenza A Viruses. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02127-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N. Engl. J. Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L. Comparative performance of SARS-CoV-2 detection assays using seven different Primer/Probe sets and one assay kit. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamaoto K., Shirato K., Nao N., Saito S., Kageyama T., Hasegawa H. An assessment of real-time RT-PCR kits for SARS-CoV-2 detection. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.108. [DOI] [PubMed] [Google Scholar]
- 20.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.J.I., Kim B.T. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2. J. Mol. Diagn. 2020 doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfefferle S., Reucher S., Norz D., Lutgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman H., Carter I., Basile K., Donovan L., Kumar S., Tran T. Interpret with caution: An evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan C., Cui J., Huang L., Du B., Chen L., Xue G. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong S.S.Y., Yip C.C.Y., Sridhar S., Leung K.H., Cheng A.K.W., Fung A.M.Y. Comparative evaluation of a laboratory-developed real-time PCR assay and RealStar(R) Adenovirus PCR Kit for quantitative detection of human adenovirus. Virol. J. 2018;15:149. doi: 10.1186/s12985-018-1059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip C.C.Y., Sridhar S., Cheng A.K.W., Fung A.M.Y., Cheng VCC Chan K.H. Comparative evaluation of a laboratory developed real-time PCR assay and the RealStar® HHV-6 PCR Kit for quantitative detection of human herpesvirus 6. J. Virol. Methods. 2017;246:112–116. doi: 10.1016/j.jviromet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Yip C.C.Y., Sridhar S., Leung K.H., Cheng A.K.W., Chan K.H., Chan J.F.W. Evaluation of RealStar® Alpha Herpesvirus PCR Kit for Detection of HSV-1, HSV-2, and VZV in Clinical Specimens. Biomed Res. Int. 2019;2019 doi: 10.1155/2019/5715180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.