Abstract
In this paper, we have trained several deep convolutional networks with introduced training techniques for classifying X-ray images into three classes: normal, pneumonia, and COVID-19, based on two open-source datasets. Our data contains 180 X-ray images that belong to persons infected with COVID-19, and we attempted to apply methods to achieve the best possible results. In this research, we introduce some training techniques that help the network learn better when we have an unbalanced dataset (fewer cases of COVID-19 along with more cases from other classes). We also propose a neural network that is a concatenation of the Xception and ResNet50V2 networks. This network achieved the best accuracy by utilizing multiple features extracted by two robust networks. For evaluating our network, we have tested it on 11302 images to report the actual accuracy achievable in real circumstances. The average accuracy of the proposed network for detecting COVID-19 cases is 99.50%, and the overall average accuracy for all classes is 91.4%.
Keywords: Deep learning, Convolutional neural networks, COVID-19, Coronavirus, Transfer learning, Deep feature extraction, Chest X-ray images
Highlights
-
•
We introduce a deep convolution network based on the concatenation of Xception and ReNet50V2 to improve the accuracy.
-
•
We propose a training technique for dealing with unbalanced datasets.
-
•
We evaluate our networks on 11302 chest X-ray images.
-
•
We have evaluated ResNet50V2 and Xception on our dataset and compared our proposed network with them. .
1. Introduction
The pervasive spread of the coronavirus around the world has quarantined many people and crippled many industries, which has had a devastating effect on human life quality, due to the high transmissibility of coronavirus, the detection of this disease (COVID-19) plays an important role in controlling it and planning preventative measures.
On the other hand, demographic conditions such as age and sex of individuals and many urban parameters such as temperature and humidity affect the prevalence of this disease in different parts of the world, which is more effective in spreading this disease [1,2].
The lack of detective tools and the limitations in their production has slowed disease detection; as a result, it increases the number of patients and casualties. The incidence of other diseases and the prevalence and number of casualties due to COVID-19 disease will decrease if it is detected quickly.
The first step is detection, recognize the symptoms of the disease, and use distinctive signs to detect the coronavirus accurately. Depending on the type of coronavirus, symptoms can range from those of the common cold to fever, cough, shortness of breath, and acute respiratory problems. The patient may also have a few days of cough for no apparent reason [3].
Unlike SARS, coronavirus affects not only the respiratory system but also other vital organs, such as the kidneys and liver [4]. Symptoms of a new coronavirus leading to COVID-19 usually begin a few days after the person becomes infected, where, in some people, the symptoms may appear a little later. According to Ref. [5]; WHO [6], respiratory problems are one of the main symptoms of COVID-19, which can be detected by X-ray imaging of the chest. CT scans of the chest can also show the disease when symptoms are mild, so analyzing these images can well detect the presence of the disease in suspicious people and even without initial symptoms [7]. Using these data can also overcome the limitations of other tools, such as lack of diagnostic kits and limitations of their production. The advantage of using CT scans and X-ray images is the availability of CT scan devices and x-ray imaging systems in most hospitals and laboratories, and the ease of access to the data needed to train the network and thus detect the disease. In the absence of common symptoms such as fever, the use of CT scans and X-ray images of the chest has a relatively good ability to detect the disease [8].
The use of specialists to diagnose the disease is a common method of detecting COVID-19 in laboratories. In this method, the specialist uses the symptoms and injuries in the chest radiology image to detect COVID-19 disease from a healthy person or person that is suffering from other diseases. This procedure has significant cost [5,9].
In recent years, computer vision and Deep Learning have been used to detect many different diseases and lesions in the body automatically [10]. Some examples are: Detection of tumor types and volume in lungs, breast, head and brain [11,12]; state-of-the-art bone suppression in x-rays, diabetic retinopathy classification, prostate segmentation, nodule classification [10]; skin lesion classification, analysis of the myocardium in coronary CT angiography [13]; sperm detection and tracking 14; etc.
Given that chest CT scan or X-ray images analysis is one of the methods of diagnosing COVID-19, the use of computer vision and Deep Learning can play a beneficial role in diagnosing this disease. Since the disease became widespread, many researchers have used machine vision and Deep Learning methods and obtained good results.
Due to the sensitivity of the Covid-19 diagnosis, the diagnostic accuracy is one of the main challenges we face in our research. On the other hand, our focus is on increasing the detection efficiency due to the limited open-source data available.
In this article, we try to improve COVID-19 detection and diminish false COVID-19 detections. This is done by combining two robust deep convolutional neural networks and optimizing the training parameters. Be- sides, we also propose a method for training the network when the dataset is imbalanced.
In [8,15]; statistical analysis of CT scans was performed by several specialists and diagnosticians, who classified the suspects into several classes for diagnosis and treatment.
Because of the superiority of computer vision and Deep Learning in the analysis of medical images, after the reliability of CT scans of the chest for COVID-19 detection, the researchers used these tools to diagnose COVID-19. Immediately, artificial intelligence became useful to detect the disease and measure the rate of infection and damage to the lungs using CT scans and the course of the disease, with promising results [16].
In [17]; they have used an innovative CNN to classify and predict COVID-19 using lung CT scans. [16] has used Deep Learning to detect COVID-19 and segment the lung masses caused by the coronavirus using 2D and 3D images. COVID-Net uses a lightweight residual projection-expansion- projection-extension (PEPX) design pattern to investigate quantitative analysis and qualitative analysis [18]. In another research study, pre-trained ResNet50, InceptionV3, and Inception ResNetV2 models have been used with transfer learning techniques to classify Chest X-ray images normal and COVID-19 classes [19]. In Ref. [20]; they present COVNet to predict COVID-19 from CT scans that have been segmented using U-net [21].
Another research study has combined the Human-In-The-Loop (HITL) strategy that involved a group of chest radiologists with deep learning-based methods to segment and measure infection in CT scans [22]. In Ref. [23]; they have tried to detect COVID-19 and Influenza-A-viral-pneumonia from their data; they have used classical ResNet-18 network structure to extract the features, and another Innovative CNN network uses these features by creating the location-attention oriented model to classify the data.
The remainder of the paper is organized as follows: In Section 2, we describe the proposed neural network, the dataset, and training techniques. In Section 3, we have presented the experimental results, and then the paper is discussed in Section 4. In Section 5, we concluded our paper, and in the next, we presented the trained networks and the codes used in this research.
2. Methodology
2.1. Neural networks
Deep convolutional neural networks are useful in machine vision tasks. These have created advances in many field like Agriculture 24; medical disease diagnosis [25,26]; and industry [27]. The superiority of these networks comes from the robust and valuable semantic features they generate from input data. Here the main focus of deep networks is detecting infection in X-ray images, so classifying the X-ray images into normal, pneumonia or COVID-19. Some of the powerful and most used deep convolutional networks are VGG [28]; ResNet [29]; DenseNet [30]; Inception [31]; Xception [32].
Xception is a deep convolutional neural network that introduced new inception layers. These inception layers are constructed from depthwise convolution layers, followed by a point-wise convolution layer. Xception achieved the third-best results on the ImageNet dataset [33] after InceptionresnetV2 [34] and NasNet Large [35]. ResNet50V2 [36] is a modified version of ResNet50 that performs better than ResNet50 and ResNet101 on the ImageNet dataset. In ResNet50V2, a modification was made in the propagation formulation of the connections between blocks. ResNet50V2 also achieves a good result on the ImageNet dataset.
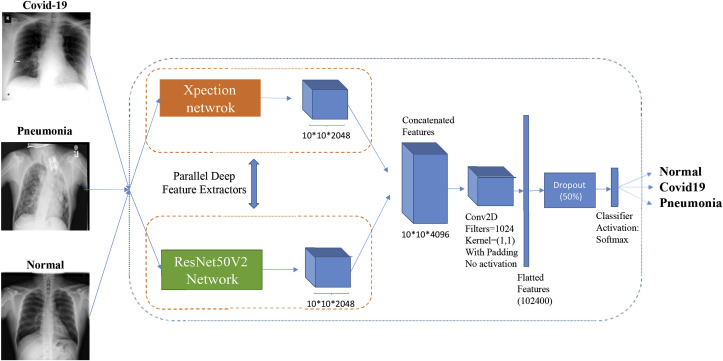
The pre-processed input images of our dataset are 300 × 300 pixels. Xception generates a 10 × 10 × 2048 feature map on its last feature extractor layer from the input image, and ResNet50V2 also produces the same size of feature map on its final layer. As both networks generate the same size of feature maps, we concatenated their features so that by using both of the inception-based layers and residual-based layers, the quality of the generated semantic features would be enhanced.
A concatenated neural network is designed by concatenating the extracted features of Xception and ResNet50V2 and then connecting the concatenated features to a convolutional layer that is connected to the classifier. The kernel size of the convolutional layer that was added after the concatenated features was 1 × 1 with 1024 filters and no activation function. This layer was added to extract a more valuable semantic feature out of the features of a spatial point between all channels, with each channel being a feature map. This convolutional layer helps the network learn better from the concatenated features extracted from Xception and ResNet50V2. The architecture of the concatenated network is depicted in Fig. 1 .
Fig. 1.
The architecture of the concatenated network.
2.2. Dataset
We have used two open-source datasets in our work. The covid chestxray dataset is taken from this GitHub repository (https://github.com/ieee8023/covid-chestxray-dataset), which has been prepared by Refs. [37]. This dataset consists of X-ray and CT scan images of patients infected to COVID-19, SARS, Streptococcus, ARDS, Pneumocystis, and other types of pneumonia from different patients.
In this dataset, we only considered the X-ray images, and in total, there were 180 images from 118 cases with COVID-19 and 42 images from 25 cases with Pneumocystis, Streptococcus, and SARS that were considered as pneumonia. The second dataset was taken from (https://www.kaggle.com/c/rsna-pneumonia-detection-challenge), which contains 6012 cases with pneumonia and 8851 normal cases. We combined these two datasets, and the details are listed in Table 1 .
Table 1.
Composition of the number of allocated images to training and validation set in both datasets.
| Dataset | COVID-19 | Pneumonia | Normal |
|---|---|---|---|
| covid chestxray dataset | 180 | 42 | 0 |
| rsna pneumonia detection challenge | 0 | 6012 | 8851 |
| Total | 180 | 6054 | 8851 |
| Training Set | 149 | 1634 | 2000 |
| Validation Set | 31 | 4420 | 6851 |
As stated, we only had 180 cases infected with COVID-19, which is few data for a class as compared to other classes. If we had combined lots many images from normal or pneumonia classes with few COVID-19 images for training, the network would become able to detect pneumonia and normal classes very well, but not the COVID-19 class because of the unbalanced dataset. In that case, although the network cannot identify COVID-19 properly, as there are many more images of pneumonia and normal classes than the COVID-19 class, the general accuracy would become very high, but not the COVID-19 detection accuracy. This condition is not our goal because the main purpose here is to achieve good results in detecting COVID-19 cases and not to identify wrong COVID-19 cases.
The best way to solve this problem is to make the dataset balanced and provide the network almost equal data of each class when training, so that the network will learn to identify all classes. Here because we do not access more open-source datasets of COVID-19 to increase this class data, we chose the number of pneumonia and normal classes almost equal to the COVID-19 number of images.
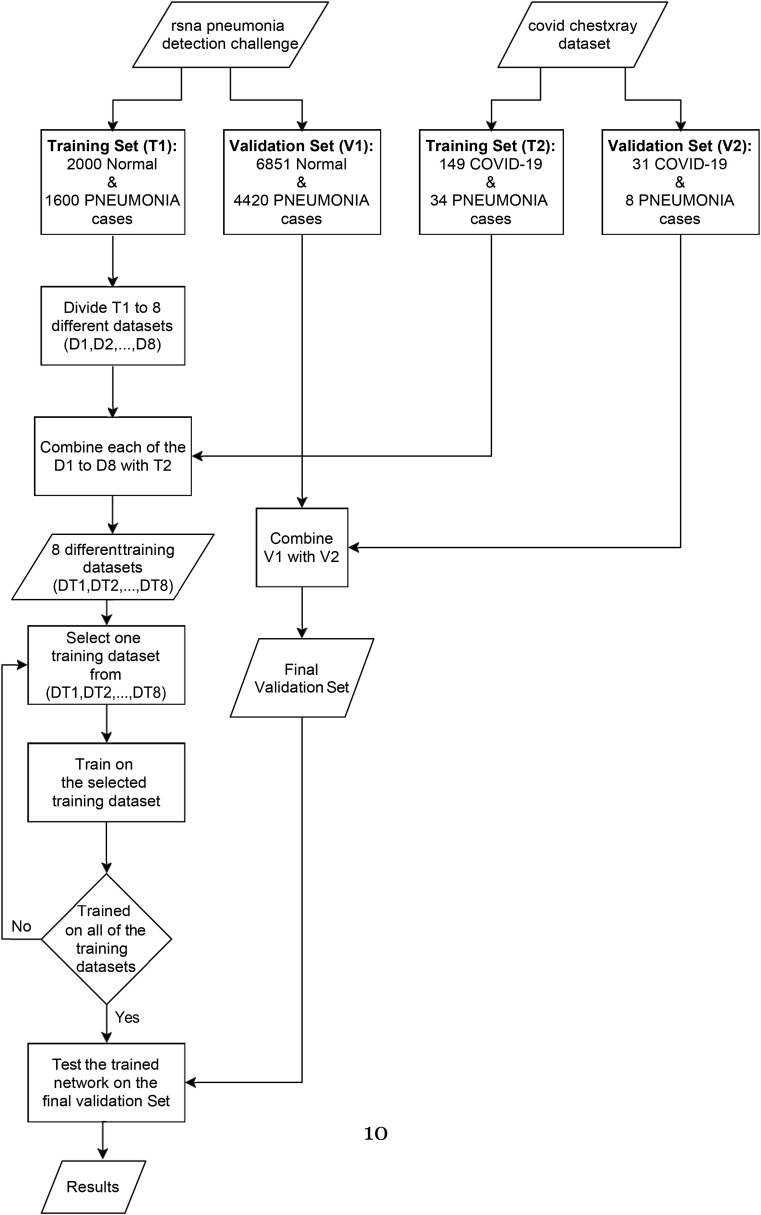
We decided to train the networks for 8 consecutive phases. In each phase, we selected 250 cases of normal class and 234 cases of pneumonia class along with the 149 COVID-19 cases. In total, we had 633 cases for each training phase. All of the COVID-19 images and 34 of the pneumonia images were common between each training phase and 250 normal cases, and 200 pneumonia cases were unique in each training phase. The common 149 COVID-19 and 34 pneumonia cases between all the training phases were from the covid chestxray dataset [37]; and the rest of the data were from the other dataset. Based on this categorizing, our training set includes 8 phases and 3783 images.
By doing so, the network sees an almost equal number of images for each class, so it helps to improve the COVID-19 detection along with detecting pneumonia and normal cases. But as we had more pneumonia and normal cases, we showed the network different pneumonia and normal cases with COVID-19 cases in each phase. Implementing this method results in two advantages. One is that the network learns COVID-19 class features better along with the other classes; second, the normal and pneumonia classes’ detection improves greatly. Better detecting pneumonia and normal cases means not detecting wrong COViD-19 cases, which is one of our goals. Running this method also helps the network better identify COVID-19 and not detect faulty COVID-19 cases.
This method can be used for all circumstances in which there is a highly unbalanced dataset. We presented our way of allocating the images of the datasets into eight different phases as a flowchart in Fig. 3 . Some of the images of our dataset are shown in Fig. 2.
Fig. 3.
The flowchart of the proposed method for training set preparation.
Fig. 2.
Examples of the images in our dataset.
2.3. Training phase
We described in the dataset subsection 2.2 that we allocated 8 phases for training. For reporting more reliable results, we chose five folds for training, where in every fold the training set was made of 8 phases as it is mentioned. We have trained ResNet50V2 [36]; Xception [32]; and a concatenation of Xception and ResNet50V2 neural networks based on the explained method. This concatenated Neural Network has shown higher accuracy compared to others. As we have tested several networks in our project, the Xception [32] and ResNet50V2 [36] networks work as well or better than others in extracting deep features. By concatenating the output features of both networks, we helped the network learn to classify the input image from both feature vectors, which resulted in better accuracy. The training parameters are described in Table 2 .
Table 2.
In this table, we have listed the parameters and functions we used in the training procedure.
| Training Parameters | Xception | ResNet50V2 | Concatenated Network |
|---|---|---|---|
| Learning Rate | 1e-4 | 1e-4 | 1e-4 |
| Batch Size | 30 | 30 | 20 |
| Optimizer | Nadam | Nadam | Nadam |
|
Loss Function |
Categorical Crossentopy |
Categorical Crossentopy |
Categorical Crossentopy |
| Epochs per each Training Phase |
100 |
100 | 100 |
| Horizontal/Vertical flipping |
Yes |
Yes | Yes |
| Zoom Range | 5% | 5% | 5% |
| Rotation Range | 0–360° | 0–360° | 0–360° |
| Width/Height Shifting |
5% |
5% | 5% |
| Shift Range | 5% | 5% | 5% |
| Re-scaling | 1/255 | 1/255 | 1/255 |
Based on Table 2, we trained the networks using the Categorical cross-entropy loss function and Nadam optimizer. The learning rate was set to 1e-4. We trained the network for 100 epochs in each training phase and, because of having 8 training phases, the models were trained for 800 epochs. For the Xception and ResNet50V2, we selected the batch size equal to 30. But as the concatenated network had more parameters than Xception and ResNet50V2, we set the batch size equal to 20. Data augmentation methods were also implemented to increase training efficiency and prevent the model from overfitting.
We implemented the neural networks with Keras [38] library on a Tesla P100 GPU and 25 GB RAM that were provided by Google Colaboratory Notebooks.
3. Results
We validated our networks on 31 cases of COVID-19, 4420 cases of pneumonia, and 6851 normal cases. The reason our training data was less than the validation data is that we had a few cases of COVID-19 among many normal and pneumonia cases. Therefore, we could not use many images from the two other classes with COVID-19 fewer cases for training, because it would have made the network not learn COVID-19 features. To solve this issue, we selected 3783 images for training in 8 different phases. We evaluated our network on the remainder of the data so that our trained network's ultimate performance would be clear. It must be noticed that exceptionally, in fold3, we had 30 cases of COVID-19 for validation, and 150 other cases were allocated for training.
It is noteworthy that we used transfer learning in the training process. For all of the networks, we used the pre-trained ImageNet weights [33] at the beginning of the training and then resumed training based on the explained conditions on our dataset. We also used the accuracy metric for monitoring the network results on the validation set after each epoch to find the best and most converged version of the trained network.
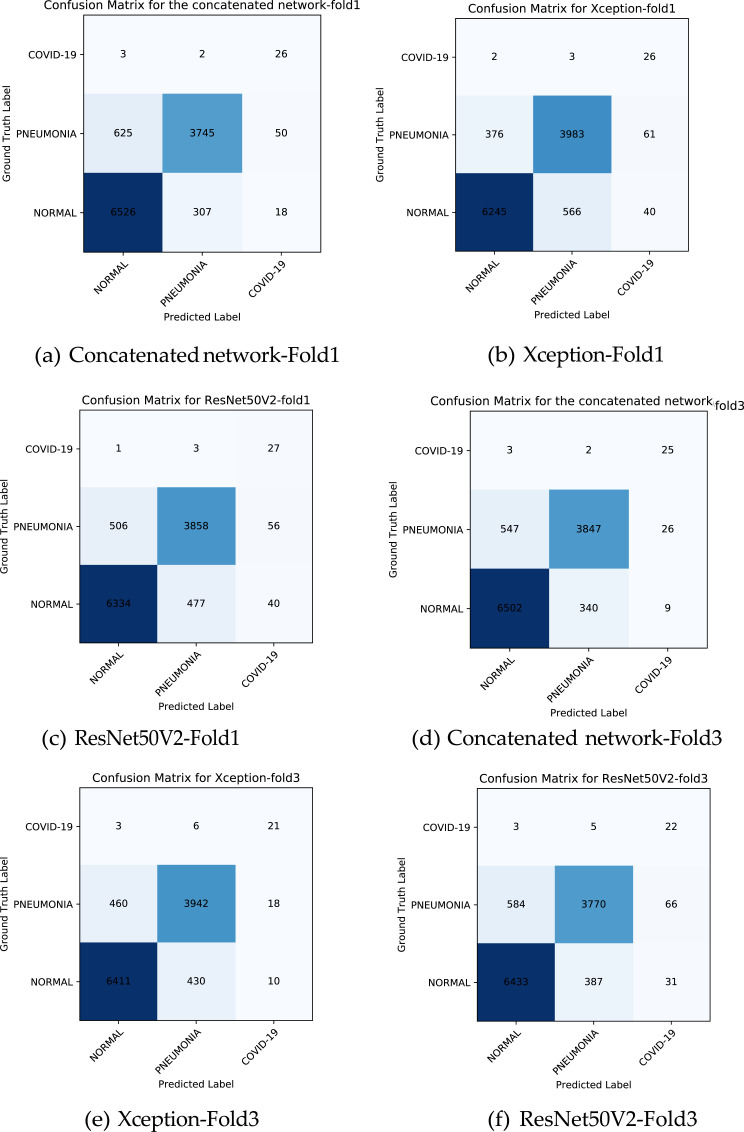
The evaluation results of the neural networks are presented in Fig. 4 which shows the confusion matrices of each network for fold one and three. Table 3 and Table 4 show the details of our results. We reported the four different metrics for evaluating our network for each of the three class as follows:
Fig. 4.
This figure shows the confusion matrix of the network for fold 1 and 3.
Table 3.
This table reports the number of true and false positives and false negatives for each class.
| Fold | Network | COVID-19 Correct detected |
COVID-19 Not detected |
COVID-19 Wrong detected |
PNEUMONIA Correct detected |
PNEUMONIA Not detected |
PNEUMONIA Wrong detected |
NORMAL Correct detected |
NORMAL Not detected |
NORMAL Wrong detected |
|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
Xception | 26 | 5 | 101 | 3983 | 437 | 569 | 6245 | 606 | 378 |
| ResNet50V2 | 27 | 4 | 96 | 3858 | 562 | 480 | 6334 | 517 | 507 | |
| Concatenated | 26 | 5 | 68 | 3745 | 675 | 309 | 6526 | 325 | 628 | |
|
2 |
Xception | 23 | 8 | 42 | 3874 | 546 | 409 | 6426 | 425 | 528 |
| ResNet50V2 | 22 | 9 | 67 | 3659 | 761 | 501 | 6340 | 511 | 713 | |
| Concatenated | 23 | 8 | 27 | 3913 | 507 | 434 | 6413 | 438 | 492 | |
|
3 |
Xception | 21 | 9 | 28 | 3942 | 478 | 436 | 6411 | 440 | 463 |
| ResNet50V2 | 22 | 8 | 97 | 3770 | 650 | 392 | 6433 | 418 | 587 | |
| Concatenated | 25 | 5 | 35 | 3847 | 573 | 342 | 6502 | 349 | 550 | |
|
4 |
Xception | 22 | 9 | 42 | 3818 | 602 | 433 | 6411 | 440 | 576 |
| ResNet50V2 | 22 | 9 | 78 | 4015 | 405 | 758 | 6065 | 786 | 364 | |
| Concatenated | 26 | 5 | 77 | 3860 | 560 | 480 | 6340 | 511 | 519 | |
|
5 |
Xception | 21 | 10 | 41 | 4041 | 379 | 502 | 6335 | 516 | 362 |
| ResNet50V2 | 21 | 10 | 42 | 3604 | 816 | 284 | 6549 | 302 | 802 | |
| Concatenated | 24 | 7 | 43 | 3941 | 479 | 390 | 6442 | 409 | 462 |
Table 4.
Some of the evaluation metrics have been reported in this table.
| Fold | Network | Accuracy | COVID-19 Sensitivity |
PNEUMONIA Sensitivity |
NORMAL Sensitivity |
COVID-19 Specificity |
PNEUMONIA Specificity |
NORMAL Specificity |
COVID-19 Accuracy |
PNEUMONIA Accuracy |
NORMAL Accuracy |
COVID-19 Precision |
PNEUMONIA Precision |
NORMAL Precision |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
Xception | 90.72 | 83.87 | 90.11 | 91.15 | 99.1 | 91.73 | 91.51 | 99.06 | 91.10 | 91.29 | 20.47 | 87.50 | 94.29 |
| ResNet50V2 | 90.41 | 87.09 | 87.28 | 92.45 | 99.15 | 93.03 | 88.61 | 99.12 | 90.78 | 90.94 | 21.95 | 88.93 | 92.58 | |
| Concatenated | 91.10 | 83.87 | 84.72 | 95.25 | 99.4 | 95.51 | 85.89 | 99.35 | 91.29 | 91.57 | 27.65 | 92.37 | 91.22 | |
|
2 |
Xception | 91.33 | 74.19 | 87.64 | 93.79 | 99.63 | 94.06 | 88.14 | 99.56 | 91.55 | 91.57 | 35.38 | 90.45 | 92.40 |
| ResNet50V2 | 88.66 | 70.96 | 82.78 | 92.54 | 99.41 | 92.72 | 83.98 | 99.33 | 88.83 | 89.17 | 24.71 | 87.95 | 89.89 | |
| Concatenated | 91.56 | 74.19 | 88.52 | 93.60 | 99.76 | 93.69 | 88.95 | 99.69 | 91.67 | 91.77 | 46 | 90.01 | 92.87 | |
|
3 |
Xception | 91.79 | 70 | 89.18 | 93.57 | 99.75 | 93.66 | 89.6 | 99.67 | 91.91 | 92.01 | 42.85 | 90.04 | 93.26 |
| ResNet50V2 | 90.47 | 73.33 | 85.29 | 93.89 | 99.14 | 94.30 | 86.81 | 99.07 | 90.78 | 91.11 | 18.48 | 90.58 | 91.63 | |
| Concatenated | 91.79 | 83.33 | 87.03 | 94.90 | 99.69 | 95.03 | 87.64 | 99.65 | 91.90 | 92.04 | 41.66 | 91.83 | 92.20 | |
|
4 |
Xception | 90.70 | 70.96 | 86.38 | 93.57 | 99.63 | 93.71 | 87.06 | 99.55 | 90.84 | 91.01 | 34.37 | 89.81 | 91.75 |
| ResNet50V2 | 89.38 | 70.96 | 90.83 | 88.52 | 99.31 | 88.99 | 91.82 | 99.23 | 89.71 | 89.82 | 22 | 84.11 | 94.33 | |
| Concatenated | 90.47 | 83.87 | 87.33 | 92.54 | 99.32 | 93.03 | 88.34 | 99.27 | 90.8 | 90.89 | 25.24 | 88.94 | 92.43 | |
|
5 |
Xception | 91.99 | 67.74 | 91.42 | 92.46 | 99.64 | 92.71 | 91.87 | 99.55 | 92.20 | 92.23 | 33.87 | 88.95 | 94.59 |
| ResNet50V2 | 90.01 | 67.74 | 81.53 | 95.59 | 99.63 | 95.87 | 81.98 | 99.54 | 90.27 | 90.23 | 33.33 | 92.69 | 89.08 | |
| Concatenated | 92.08 | 77.41 | 89.16 | 94.03 | 99.62 | 94.33 | 89.62 | 99.56 | 92.31 | 92.29 | 35.82 | 90.99 | 93.30 | |
|
Average |
Xception | 91.31 | 73.35 | 88.95 | 92.91 | 99.55 | 93.17 | 89.63 | 99.48 | 91.52 | 91.62 | 33.39 | 89.35 | 93.26 |
| ResNet50V2 | 89.79 | 74.02 | 85.54 | 92.60 | 99.33 | 92.98 | 86.64 | 99.26 | 90.07 | 90.25 | 24.09 | 88.85 | 91.50 | |
| Concatenated | 91.40 | 80.53 | 87.35 | 94.06 | 99.56 | 94.32 | 88.09 | 99.50 | 91.60 | 91.71 | 35.27 | 90.83 | 92.40 |
We also reported the overall accuracy metric, defined as:
In these equations, TP (True Positive) is the number of correctly classified images of a class, FP (False Positive) is the number of the wrong classified images of a class, FN (False Negative) is the number of images of a class that have been detected as another class, and TN (True Negative) is the number of images that do not belong to a class and were not classified as belonging to that class.
4. Discussion
It can be understood from the confusion matrices and the tables that the concatenated network performs better in detecting COVID-19 and not detecting false cases of COVID-19 and outputs better overall accuracy. Although we had an unbalanced dataset and a few cases of COVID-19, by using the proposed technique, we could have improved COVID-19 detection along with the other classes detection. The reason the precision of the COVID-19 class is low is that in our work, despite some other researches that worked on detecting COVID-19 from X-ray images, we tested our neural networks on a massive number of images. Our test images were much more than our train images. As is explained above, because we had only 31 cases of COVID-19 and 11271 cases from the other two classes, the false positives of the COVID-19 class would become more than true positives. For example, in the first fold, the concatenated network detected 26 cases correctly out of 31 COVID-19 cases, and from 11271 other cases, only mistakenly identify 68 cases as COVID-19. If we had equal samples from the COVID-19 class as from the other classes, the precision would become high value. Still, because of having few COVID-19 cases and many other cases for validation, the precision would become low in value.
In another study, the results were presented in two forms, 2 and 3 classes, that due to the imbalance in the dataset, there are several meaningless results [39]. We have presented the results for each class and for all the classes with meaningful results that are more practical. We could have tested our network on a few cases like some of the other researches have done recently, but we wanted to show the real performance of our network with few COVID-19 cases. As mentioned, mistakenly detecting 68 cases from 11271 cases to be infected with COVID-19 is not very much but not very well also, and we hope that by using much-provided data from patients infected, COVID-19, the detection accuracy will rise much more.
5. Conclusion
In this paper, we presented a concatenated neural network based on Xception and ResNet50V2 networks for classifying the chest X-ray images into three categories of normal, pneumonia, and COVID-19. We used two open-source datasets that contained 180 and 6054 images from patients infected with COVID-19 and pneumonia, respectively, and 8851 images from normal people. As we had a few images of the COVID-19 class, we proposed a method for training the neural network when the dataset is unbalanced. We separated the training set into 8 successive phases, in which there were 633 images (149 COVID-19, 234 pneumonia, 250 normal) in each phase. We selected the number of each class almost equal to each other in each phase so that our network also learns COVID-19 class characteristics, not only the features of the other two classes. In each phase, the images from normal and pneumonia classes were different, so that the network can distinguish COVID-19 from other classes better. Our training set included 3783 images, and the rest of the images were allocated for evaluating the network. We tried to test our model on a large number of images so that our real achieved accuracy would be clear. We achieved an average accuracy of 99.50%, and 80.53% sensitivity for the COVID-19 class, and an overall accuracy equal to 91.4% between five folds. We hope that our trained network that is publicly available will be helpful for medical diagnosis. We also hope that in the future, larger datasets from COVID-19 patients become available, and by using them, the accuracy of our proposed network increases further.
Code availability
In this GitHub profile (https://github.com/mr7495/covid19), we have shared the trained networks and all the used code in this paper. We hope our work be useful to help in future researches.
Author agreement statement
We declare that this manuscript is original, has not been published before and is not currently being considered for publication elsewhere. We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us. We understand that the Corresponding Author is the sole contact for the Editorial process. He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
Declaration of competing interest
The authors declare no competing interest.
Acknowledgment
We would like to appreciate Joseph Paul Cohen and the others who provided these X-ray images from patients infected to COVID-19. We thank Linda Wang and Alexander Wong for making their code available, in which we have used a part of it on our research for preparing our dataset. We also thank Google Colab server for providing free GPU.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100360.
Contributor Information
Mohammad Rahimzadeh, Email: mh_rahimzadeh@elec.iust.ac.ir.
Abolfazl Attar, Email: attar.abolfazl@ee.sharif.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pirouz B., Shaffiee Haghshenas S., Shaffiee Haghshenas S., Piro P. Investigating a serious challenge in the sustainable development process: analysis of confirmed cases of covid-19 (new type of coronavirus) through a binary classification using artificial intelligence and regression analysis. Sustainability. 2020;12(6):2427. [Google Scholar]
- 2.Pirouz B., Shaffiee Haghshenas S., Pirouz B., Shaffiee Haghshenas S., Piro P. Development of an assessment method for investigating the impact of climate and urban parameters in confirmed cases of covid-19: a new challenge in sustainable development. Int J Environ Res Publ Health. 2020;17(8):2801. doi: 10.3390/ijerph17082801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.univers1 Everything about the Corona virus. 2020-04-12. https://medicine-and-mental-health.xyz/archives/4510
- 4.McIntosh K. 2020-04-10. Coronavirus disease 2019 (COVID-19): epidemi- ology, virology, clinical features, diagnosis, and prevention. [Google Scholar]
- 5.Jiang F., Deng L., Zhang L., Cai Y., Cheung C.W., Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (covid-19) J Gen Intern Med. 2020:1–5. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO 2020-04-10. https://www.who.int
- 7.Sun D., Li H., Lu X.-X., Xiao H., Ren J., Zhang F.-R., Liu Z.-S. Clinical features of severe pediatric patients with coronavirus disease 2019 in wuhan: a single center's observational study. World J. Pediatr. 2020:1–9. doi: 10.1007/s12519-020-00354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An P., Chen H., Jiang X., Su J., Xiao Y., Ding Y., Ren H., Ji M., Chen Y., Chen W. 2020. Clinical features of 2019 novel coronavirus pneumonia presented gastrointestinal symptoms but without fever onset. [arxiv] [Google Scholar]
- 9.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel coronavirus (2019-ncov) pneumonia. Radiology. 2020:200274. doi: 10.1148/radiol.2020209021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., Van Der Laak J.A., Van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Cheng J.-Z., Ni D., Chou Y.-H., Qin J., Tiu C.-M., Chang Y.-C., Huang C.-S., Shen D., Chen C.-M. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in ct scans. Sci Rep. 2016;6(1):1–13. doi: 10.1038/srep24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lakshmanaprabu S., Mohanty S.N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on ct images. Future Generat Comput Syst. 2019;92:374–382. [Google Scholar]
- 13.Zreik M., Lessmann N., van Hamersvelt R.W., Wolterink J.M., Voskuil M., Viergever M.A., Leiner T., Išgum I. Deep learning analysis of the myocardium in coronary ct angiography for identification of patients with functionally significant coronary artery stenosis. Med Image Anal. 2018;44:72–85. doi: 10.1016/j.media.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Rahimzadeh M., Attar A. 2020. Sperm detection and tracking in phase-contrast microscopy image sequences using deep learning and modified csr-dcf.2002.04034 [Google Scholar]
- 15.Yang R., Li X., Liu H., Zhen Y., Zhang X., Xiong Q., Luo Y., Gao C., Zeng W. Chest ct severity score: an imaging tool for assessing severe covid-19. Radiology: Cardiothor. Imag. 2020;2(2) doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis.2003.05037 [Google Scholar]
- 17.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X. 2020. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) [medRxiv] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Wong A. 2020. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images.2003.09871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coron- avirus disease (covid-19) using x-ray images and deep convolutional neural networks.2003.10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronneberger O., Fischer P., Brox T. International Conference on Medical image computing and computer-assisted intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 22.Shan+ F., Gao+ Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. 2020. Lung infection quantification of covid-19 in ct images with deep learning.2003.04655 [Google Scholar]
- 23.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Chen Y., Su J., Lang G. 2020. Deep learning system to screen coronavirus disease 2019 pneumonia.2002.09334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahimzadeh M., Attar A. 2020. Introduction of a new dataset and method for detecting and counting the pistachios based on deep learning.2005.03990 [Google Scholar]
- 25.Lih O.S., Jahmunah V., San T.R., Ciaccio E.J., Yamakawa T., Tanabe M., Kobayashi M., Faust O., Acharya U.R. Comprehensive electrocardiographic diagnosis based on deep learning. Artif Intell Med. 2020;103:101789. doi: 10.1016/j.artmed.2019.101789. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Qian H., Ciaccio E.J., Lewis S.K., Bhagat G., Green P.H., Xu S., Huang L., Gao R., Liu Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput Methods Progr Biomed. 2020;187:105236. doi: 10.1016/j.cmpb.2019.105236. [DOI] [PubMed] [Google Scholar]
- 27.Dekhtiar J., Durupt A., Bricogne M., Eynard B., Rowson H., Kiritsis D. Deep learning for big data applications in cad and plm–research review, opportunities and case study. Comput Ind. 2018;100:227–243. [Google Scholar]
- 28.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- 29.He K., Zhang X., Ren S., Sun J. 2015. Deep residual learning for image recognition. [Google Scholar]
- 30.Huang G., Liu Z., van der Maaten L., Weinberger K.Q. 2016. Densely connected convolutional networks. [Google Scholar]
- 31.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D. Going deeper with convolutions. IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Boston, MA, 2015,: 2015. pp. 1–9. [Google Scholar]
- 32.Chollet F. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Xception: deep learning with depthwise separable con- volutions; pp. 1251–1258. [Google Scholar]
- 33.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009. Ima- genet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 34.Szegedy C., Ioffe S., Vanhoucke V., Alemi A. 2016. Inception-v4, inception-resnet and the impact of residual connections on learning. [Google Scholar]
- 35.Zoph B., Vasudevan V., Shlens J., Le Q.V. 2017. Learning transfer- able architectures for scalable image recognition. [Google Scholar]
- 36.He K., Zhang X., Ren S., Sun J. European conference on computer vision. Springer; 2016. Identity mappings in deep residual networks; pp. 630–645. [Google Scholar]
- 37.Cohen J.P., Morrison P., Dao L. 2020. Covid-19 image data collection.2003.11597 [Google Scholar]
- 38.Chollet, F. and Others (2015). keras.
- 39.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.