Abstract
Objectives
In previous influenza pandemics, bacterial co-infections have been a major cause of mortality. We aimed to evaluate the burden of co-infections in patients with COVID-19.
Methods
We systematically searched Embase, Medline, Cochrane Library, LILACS and CINAHL for eligible studies published from 1 January 2020 to 17 April 2020. We included patients of all ages, in all settings. The main outcome was the proportion of patients with a bacterial, fungal or viral co-infection. .
Results
Thirty studies including 3834 patients were included. Overall, 7% of hospitalised COVID-19 patients had a bacterial co-infection (95% CI 3-12%, n=2183, I2=92·2%). A higher proportion of ICU patients had bacterial co-infections than patients in mixed ward/ICU settings (14%, 95% CI 5-26, I2=74·7% versus 4%, 95% CI 1-9, I2= 91·7%). The commonest bacteria were Mycoplasma pneumonia, Pseudomonas aeruginosa and Haemophilus influenzae. The pooled proportion with a viral co-infection was 3% (95% CI 1-6, n=1014, I2=62·3%), with Respiratory Syncytial Virus and influenza A the commonest. Three studies reported fungal co-infections.
Conclusions
A low proportion of COVID-19 patients have a bacterial co-infection; less than in previous influenza pandemics. These findings do not support the routine use of antibiotics in the management of confirmed COVID-19 infection.
Keywords: Coronavirus, COVID-19, Coinfection, Meta-Analysis
Introduction
Since December 2019, the COVID-19 pandemic has spread from its epicentre in Wuhan, China to infect over 3 million people, with over 200,000 deaths associated with the disease worldwide.1 The pathogen responsible, severe acute respiratory coronavirus 2 (SARS-CoV-2), is an enveloped RNA beta coronavirus2 which is related phylogenetically to SARS-CoV-1.3 The most common symptoms are fever and cough;4 more severe outcomes (requiring mechanical ventilation and intensive care) are associated with older age, a higher percentage of comorbidities and higher mortality.5 , 6
Other respiratory viruses, such as seasonal/pandemic influenza, Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-1, show differing levels of bacterial/fungal co-infection. For example, it has been suggested that "influenza‐related bacterial infections overall may account for up to 30% of CAP cases",7 whereas there is evidence that co-infections do not occur in patients infected with MERS-CoV8 and occur rarely in patients infected with SARS-CoV-1.9 Furthermore, co-infection has been associated with more severe outcomes in pandemic and seasonal influenza.7 Therefore, there is a clinical need for robust investigation into co-infection in patients with COVID-19.
Many studies of hospitalised patients with COVID-19 note the empiric use of antibiotics in a majority of patients;5 , 10 , 11 however, there is evidence that the inflammatory serological markers that are usually associated with bacterial infection, such as raised procalcitonin and C-reactive protein, may appear in patients with COVID-19 without a corresponding bacterial co-infection occurring.12 , 13 In the context of rising levels of antimicrobial resistance,14 this review aims to inform sustainable and judicious antibiotic use.
Furthermore, this review aims to identify the risk-factors, prevalence, characteristics, consequences and aetiological agents associated with COVID-19 co-infection. This pandemic is placing a strain on the resources of healthcare systems worldwide; the evidence presented in this review can inform not only better treatment, but also more efficient use of equipment, medication and time.
Methods
Search strategy and selection criteria
For this systematic review and meta-analysis, we searched databases for studies of any design which reported numeric data on co-infections in patients with simultaneous SARS-CoV-2 infection. We excluded single case reports and studies with fewer than ten participants.
We searched MEDLINE, EMBASE, CINAHL, LILACS and the Cochrane Central Register of Controlled Trials (CENTRAL) from 1 January 2020 up to 17 April 2020, with no language restriction using the search terms ((“Coronavirus infection/” OR “SARS coronavirus/” or “severe acute respiratory syndrome/”) OR (coronavirus or COVID* or SARS*)) AND ((“mixed infection/” or “bacterial pneumonia/” or “bacteremia/” or “”secondary infection/” or “mycosis/” or “bacterial infection/” or “superinfection/” or “ventilator associated pneumonia/”) or (“coinfect* or co-infect* or (secondary infect*) or (concomitant infect*) or (mixed infect*))). We also searched reference lists of identified articles and handsearched relevant peer-reviewed journals up to 17 April 2020. Two independent reviewers (LL and BL/VB) screened the abstracts of identified studies and reviewed the full texts of those which were potentially eligible, with disagreements resolved by consensus.
We conducted this systematic review meta-analysis in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.15
Data extraction and analysis
Two reviewers (LL and BL/VB) independently extracted data from individual studies using a predefined template. We collected data on study methodology, location and setting, study population, the proportion of patients with co-infections and the pathogens implicated, method of detection of co-infections, prognosis in co-infected patients, and antibiotic use. For observational studies, we assessed the risk of bias in the domains of study group selection and ascertainment of co-infection using a modification of the Newcastle-Ottawa Scale.16 The comparability domain was not considered relevant due to the design of the included studies. The Cochrane Risk of Bias Tool (version 2) was used to assess risk of bias in randomised controlled trials (RCTs).17
We principally sought to analyse the proportion of patients with confirmed COVID-19 disease who were co-infected simultaneously with other pathogens, and to describe the co-infecting pathogens. For hospitalised patients, studies were stratified into those which only included patients receiving intensive care unit (ICU) care, and those which also included non-ICU patients. Studies of patients from out-patient settings and those which focussed on deceased patients were described narratively. Separate analyses were conducted for studies reporting laboratory-confirmed bacterial and viral co-infections. Laboratory-confirmed co-infections were those identified by bacterial or fungal culture of respiratory samples or blood, or through antigen detection methods or PCR detection of respiratory pathogens. We also analysed separately those studies which reported data on co-infections but without mention of the method of detection or the pathogens implicated. Where appropriate, sensitivity analyses were conducted to study the proportion of co-infection in different age groups (children, all adults, older adults).
We estimated the pooled proportion of co-infected patients using a random effects model (DerSimonian Laird weights method), stabilising the variances using the Freeman-Tukey double arcsine transformation so that studies with proportions close to 0% or 100% were approximately estimated, with computation of exact binomial 95% confidence intervals. We assessed heterogeneity using the I2 statistic. As we anticipated a high level of heterogeneity, an a priori decision was made not to abandon meta-analyses due to high heterogeneity.
The relative contribution of each named pathogen to the total number of either bacterial or viral co-infections was described. Where data were reported, we also estimated the effect of co-infection on the risk of death by random effects model meta-analysis of crude odds ratios of co-infected versus those not coinfected, using a generic inverse effects model.
All analyses were conducted in Stata 16·0 software (StataCorp. 2019. Stata Statistical Software: Release 16. College Station, TX:StatCorp LLC.). The study protocol is registered with the National Institute for Health Research international prospective register of systematic reviews (PROSPERO).18
Role of the funding source
Nottingham NIHR Biomedical Research Centre had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
After deduplication, our search retrieved 795 titles, of which 654 were excluded in the initial screen. The abstracts of the remaining 141 studies were scrutinised and 74 were retained for review of the full-text. A further 44 studies were excluded at this stage as they failed to meet the inclusion criteria, leaving 30 studies which were included in the analyses (Figure 1 ). Twenty-three (77%) of the included studies were from China, three from the USA (10%), two from Spain (6·7%) and one each (3·3%) from Thailand and Singapore. Data on 3834 patients from 29 observational studies and one RCT were included (see Table 1 and Table 2 for characteristics of included studies). The RCT was an open-label comparison of lopinavir-ritonavir plus standard care versus standard care alone in patients with COVID-19 pneumonia, in which the primary endpoint was time to clinical improvment19. Of the 27 studies reporting data for hospitalised patients,5 , 6 , 10 , 12 , 13 , 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40six had separate data for patients receiving ICU care.6 , 20, 21, 22 , 25 , 35 Two studies reported data on deceased COVID-19 patients11 , 41 and one study included non-hospitalised patients.42 Most data were for adults (range of median ages 42 to 63 years), with only three small studies reporting data exclusively from children (n=86).13 , 31 , 39 One study included data from older adults (≥60 years) only, with a median age of 69 years (IQR 65 to 76).32 Antibiotic use was reported in 17 studies, with >90% of patients receiving empirical antibiotics in 10 studies.5 , 10 , 11 , 19 , 24 , 25 , 34 , 35 , 40 , 41
Figure 1.
PRISMA flow diagram for study selection
Table 1.
Characteristics of 22 included studies with data on either specific detected pathogens or negative microbiology testing
| Author | Country | Study type | N participants | Age group | ICU | MV | Deaths | % receiving antibiotic | % receiving antiviral | % Patients with bacterial co-infection | % Patients with fungal co-infection | % Patients with respiratory viral co-infection |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zheng F39 | China | Retrospective case series, Hospital | 25 | Children | 8% | 8% | 0% | 56% | 48% (IFN/arbidol/oseltamivir/ lopinavir/litonovir) | 16% | ·· | 8% |
| Zhao38 | China | Prospective cohort, Hospital | 19 | Adult | 0 | 0 | 0 | 0 | 100 (lopinavir/ritonavir) | 5% | ·· | 5% |
| Zhang J37 | China | Case series, Hospital | 140 | Adult | ·· | ·· | ·· | ·· | ·· | 3·6% | ·· | 1·4% |
| Young36 | Singapore | Case series, Hospital/ICU | 18 | Adult | 11% | 5·6% | 0 | Empirical in patients with suspected CAP | 28% (lopinavir/ritonivir) | ·· | ·· | 0 |
| Yang X35 | China | Case series, ICU | 52 | Adult | 100% | 71% | 61·5% | 94% | 44% | 7·7% | 5·8% | ·· |
| Xia13 | China | Case series, Hospital | 20 | Children | 0 | 0 | 0 | ·· | ·· | 20% | 0 | 25% |
| Wu J34 | China | Case series, Hospital | 80 | Adult/children | ·· | 0 | 0 | 91.2% (Mostly moxifloxacin) | 100% ribavirin |
0 | 0 | 0 |
| Wu C5 | China | Retrospective cohort, Hospital/ICU | 201 | Adult | 26·4% | 33·3% | 21·9% | 97·5% (empirical) | 84·6% (oseltamivir/ganciclovir/lopinavir/ ritinovir/IFN alpha) |
0 | 0 | 0·5% |
| Wang Z10 | China | Case series, Hospital | 69 | Adult | ·· | ·· | 7·2% | 98·5% (empirical) |
98·5% |
13·8% | 6·9% | 7·1% |
| Wang Y 33 | China | Case series, Hospital | 55 | Adult/children | 0 | 0 | 0 | ·· | 100% (lopinavir/ritinovir) |
5·4% | ·· | 1·8% |
| Mo29 | China | Retrospective cohort Hospital/ICU |
155 | Adult | 23·9% | 23·2% | 14·2% | ·· | 29% (arbidol/lopinavir & ritonavir/IFN) | 1·3% | ·· | 3·9% |
| Lin D27 | China | Case series, Hospital | 92 | Adult | ·· | ·· | ·· | ·· | ·· | ·· | ·· | 3·2% |
| Lian26 | China | Retrospective cohort, Hospital/ICU | 788 | Adult | 2·4% | 2·3% | 0 | ·· | 84·8% | 0 | ·· | ·· |
| Huang25 | China | Case series, Hospital/ICU | 41 | Adult | 32% | 10% | 15% | 100% (empirical) | 93% (oseltamivir) | 10% | ·· | ·· |
| Chen 23 | China | Case series, Hospital/ICU | 99 | Adult | 23% | 4% | 11% | 71% (single antibiotic 25%; multiple 45%. Cephalosporins, quinolones, carbapenems, tigecycline, linezolid, antifungals) |
76% | 1% | 4% | 0 |
| Bhatiraju22 | USA | Case series, ITU | 24 | Adult | 100% | 75% | 50% | ·· | 33% | 0 | ·· | 0 |
| Arentz20 | USA | Case series, ITU | 21 | Adult | 100% | 71% | 67% | ·· | ·· | 4·8% | ·· | 14·3% |
| Pongipurul30 |
Thailand | Case series, Hospital | 11 | Adult | 0 | 0 | 0 | 54·4% (ceftriaxone or amoxicillin) |
27·3% |
45·4% | ·· | 18·2% |
| Barassa21 | Spain | Case series, ITU | 48 | Adult | 100% | 93·5% | 12·5% | 87·5% (beta-lactams +linezolid, levofloxacin, ceftriaxone, azithromycin, linezolid) |
94 (lopinavir, ritonavir, IFN) |
12·5% | ·· | ·· |
| Tagarro31 | Spain | Case series Hosp/non-hosp/PIC | 41 | Adult | 9·7% | 2% | 0 | ·· | ·· | ·· | ·· | 10·8% |
| Kim42 | USA | Path-lab database, Non-hospitalised |
115 | Adult/children |
0 | 0 | 0 | ·· | ·· | ·· | ·· | 20% |
| Feng6 | China | Case series, Hospital/ICU | 476 | Adult | 14·7% | 8·2% | 8% | 67% | 60·1% | 8·5% | ·· | ·· |
Key: ICU: Intensive care unit; MV: Mechanical ventilation; IFN: interferon
Table 2.
Characteristics of 8 included studies describing co-infections but without specific information on individual pathogens
| Author | Country | Study type | N participants | Age group | ICU (%) | MV (%) | Deaths (%) | Antibiotics (%) | Antivirals (%) | % Patients with co-infection |
|---|---|---|---|---|---|---|---|---|---|---|
| Zhou40 | China | Retrospective cohort, Hospital | 191 | Adult | 26% | 17% | 28·3% | 95% | 21% (lopinivir/ritonivir) | 15% |
| Wan S12 | China | Case series, Hospital/ICU | 135 |
Adult | 29·6% | 0·7% | 0·7% | 43·7% (not specified) |
100% Kaletra |
5·1% |
| Ruan41 | China | Retrospective database, Died vs discharged | 150 (68 deceased, 82 survivors) |
Not specified | n/a | n/aa | 16·2% | 93% | 49% | 16% |
| Du R11 | China | Case series, Deceased | 109 | Adult | 46·8% | 30·3% | 100% | 100% | 94·5% | 38·5% |
| Ding24 | China | Case Series. Hospital | 115 | Adult | 0 | 0 | 0 | 100% | 100% (including oseltamivir) |
4·4% |
| Wang L32 | China | Case series, Hospital/ICU | 339 | Older adults | 23·6% based on ‘critical’ definition |
23·6% | 19·2% | ·· | ·· | 42·8% |
| Liu Y28 | China | Case series, Hospital/ICU | 12 | Adult/children | ·· | 50% | 0 | ·· | 100% (Ribavirin +IFN), 33·3% oseltamivir |
16·6% |
| Cao19 | China | Randomised Control Trial | 199 | Adult | ·· | 16·1% | 22·1% (day 28) | 95% |
49·7% |
3·6% |
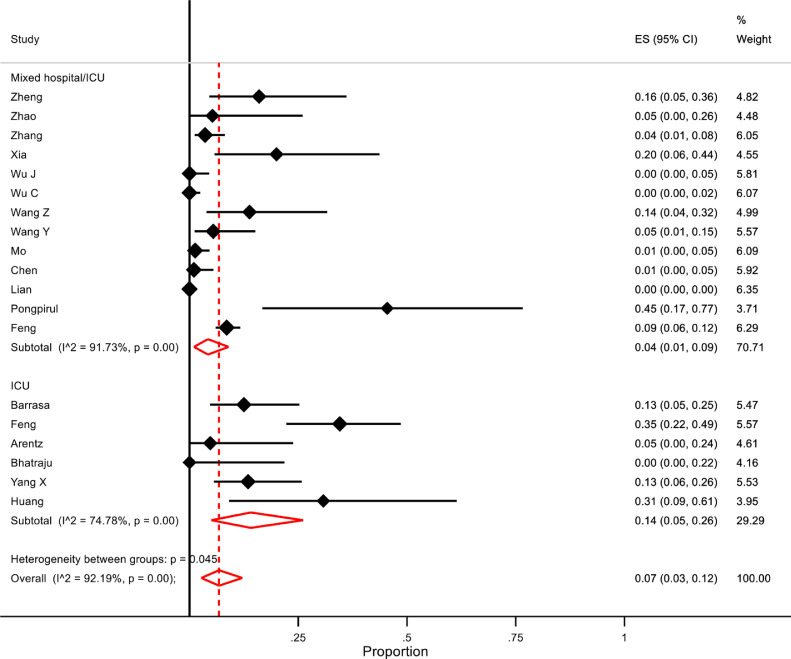
For hospitalised COVID-19 patients, the overall pooled proportion of patients who had laboratory-confirmed bacterial co-infections was 7% (95% confidence intervals (CI) 3 to 12, n=2183, 18 studies, 19 datasets, I2 92·2%). Subgroup analysis of studies with separate data for ICU patients only showed that a greater proportion of them had bacterial co-infections than patients from mixed hospitalised/ICU studies (14% (95% CI 5-26), n=204, I2=74.7% versus 4% (95% CI 1-9), n=1979, I2 91·7% respectively, p=0·05)(Figure 2 ). Sensitivity analysis excluding one study which only included children did not significantly affect the overall proportion of patients with bacterial co-infection, nor did it decrease the heterogeneity (6% (95% CI 2-12), I2=92·4%), although the difference between ICU patients and a mixed hospital population failed to reach statistical significance (p=0·08). One study25 defined secondary infection as developing ≥48 hours after admission with a positive culture of a new pathogen from a lower respiratory tract specimen or blood taken ≥48 hours after admission. There were no data in the remaining studies relating to the time from admission to detection of co-infection.
Figure 2.
Forest plot of proportion of COVID-19 patients with bacterial co-infections. Subgroup analysis for ICU versus mixed ward/ICU settings.
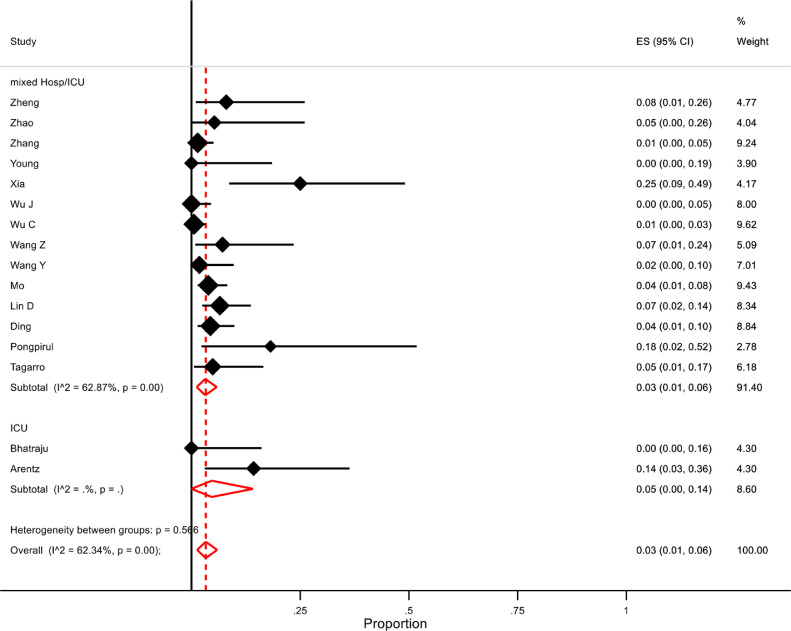
From pooled analysis, an estimated 3% of patients had a confirmed viral co-infection (95% CI 1-6, n=1014, 16 studies, I2=62·3%). Subgroup analysis did not show a significant difference in proportions between patients on ICU and the mixed hospitalised population (5% (95% CI 1-14, n=42, 2 studies) and 3% (95% CI 1-5%, n=972, 14 studies, I2=62·9%) respectively)(Figure 3 ). Excluding three studies which only included paediatric patients resulted in a slight decrease in overall proportion of viral co-infected patients to 2% (95% CI 1-5%, n=886, 13 studies, I2=55·5%). One study42, which included SARS-CoV-2 positive patients seen in outpatients and the emergency department reported viral co-infection in 23/115 patients (20%).
Figure 3.
Forest plot of proportion of hospitalised COVID-19 patients with viral co-infections. Subgroup analysis for ICU versus mixed ward/ICU settings
Eight studies described the occurrence of co-infections in participants but did not describe the pathogens involved. For hospitalised patients, the overall proportion of co-infected patients was 12% (95% CI 2-29, n=991, 6 studies, I2=97·3%). One study, which only included older adults ≥60 years, indicated that secondary infections were diagnosed on the basis of inflammatory biomarkers.32 In a sensitivity analysis excluding this study, the proportion of co-infected patients fell to 7% (95% CI 2-12, n=652, I2=79·5%), similar to the proportion of patients with specific bacterial co-infections. Two studies which examined the characteristics of deceased patients enumerated secondary infections, with one specifying that lung bacterial and fungal disease occurred at a late stage of the disease.11 , 41
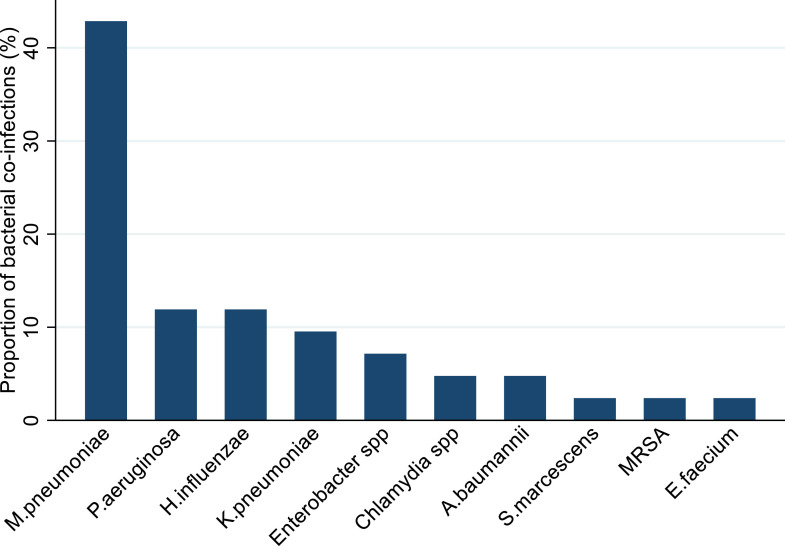
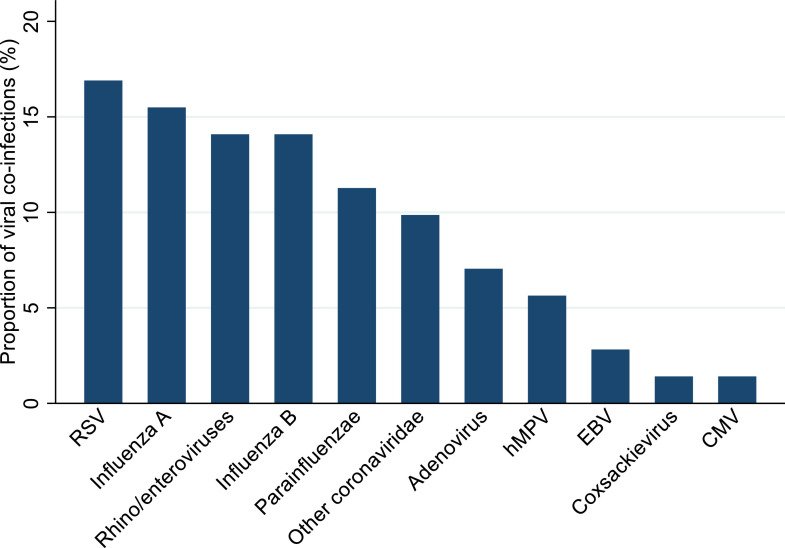
Specific co-infecting pathogens were identified in 17 studies.5 , 10 , 13 , 20 , 21 , 23 , 24 , 27 , 29, 30, 31 , 33 , 35 , 37, 38, 39 , 42 The most frequently detected bacterial pathogen was Mycoplasma pneumoniae (42% of 27 confirmed bacterial pathogen detections), followed by Pseudomonas aeruginosa (12%, including one patient with bacteraemia) and Haemophilus influenzae (12%). Klebsiella pneumoniae was isolated from four patients, including a carbapenem resistant isolate from one patient with hospital-acquired pneumonia (HAP) and bacteraemia, and an extended spectrum beta lactamase positive isolate from another patient with HAP. Other bacteria detected were Enterobacter species (three patients), Acinetobacter baumannii and Chlamydia species (two patients each), and Enterococcus faecium, methicillin resistant Staphylococcus aureus (MRSA), and Serratia marscecens (one patient each)(Figure 4 ). Four fungal pathogens were identified from three studies.10 , 23 , 35 Candida albicans was isolated from the respiratory tract in five patients and urinary tract of a sixth. Other fungal co-infections from respiratory samples were Aspergillus flavus (2 patients), Aspergillus fumigatus and Candida glabrata (one patient each). Viral co-infecting respiratory pathogens were identified in 14 studies5 , 10 , 13 , 20 , 24 , 27 , 29, 30, 31 , 33 , 37, 38, 39 , 42; respiratory syncytial virus (RSV) was the most prevalent respiratory virus (16·9% of detections), followed by influenza A (15·5%).
Figure 4.
Bacterial pathogens detected in COVID-19 patients, as a proportion (%) of the total number of detections (n=27) Key: M pneumoniae - Mycoplasma pneumoniae; P aeruginosa – Pseudomonas aeruginosa; H influenzae – Haemophilus influenzae; K pneumoniae – Klebsiella pneumoniae, A baumannii – Acinetobacter baumannii, S marcescens - Serratia marcescens, MRSA – Methicillin-resistant Staphylococcus aureus; E faecium – Enterococcus faecium.
Figure 5.
Viral pathogens as a proportion (%) of the total number of viral detections (n=71). Key: RSV – Respiratory Syncytial Virus, hMPV – human Metapneumovirus, EBV – Epstein-Barr Virus, CMV - Cytomegalovirus.
Pooled analysis of crude odds ratios for death indicated that COVID-19 patients with a co-infection were more likely to die than patients who did not have a co-infection (pooled OR 5·82, 95% CI 3·4 – 9·9, n=733, 4 studies, I2=85·4%).10 , 35 , 40 , 41 One study reported that bacterial infection was a predictor of death in older patients based on univariate regression analysis (hazard ratio (HR) 7·01 (95% CI 3·65-13·5, p<001), but it was not a significant predictor on multivariate analysis (adjusted HR 1·52 (95% CI 0·71-3·24, p=0·28).32 Antimicrobial resistant gram-negative bacteria were isolated from patients in one of the studies35; the co-infecting pathogens were not specified in the other studies which reported mortality data.
In the risk of bias assessment 11 of the 29 (38%) observational studies were judged to have an element of selection bias as the patients they reported may not have been truly representative of patients with COVID-19. In the ascertainment of outcome domain, 21 of 29 studies (72%) were at some risk of bias, which was generally attributable to incomplete follow-up of patients, with most studies reporting that many patients were still hospitalised at the censor date. The one RCT was judged to be at low risk of bias in the domains relating to the randomisation process, deviations from the intended interventions and outcome measurement. In the missing outcome and selection of reported result domains there were some concerns due to lack of information on how secondary infections were ascertained and continued hospitalisation of some participants at the censor date.
Discussion
To our knowledge, our study is the first systematic review to evaluate the burden of co-infections in patients with confirmed SARS-CoV-2 infection.
Our meta-analysis indicated that overall 7% of hospitalised COVID-19 patients had a bacterial co-infection, increasing to 14% in studies that only included ICU patients. These analyses, arising from the earliest cases of the SARS-CoV-2 pandemic, suggest that bacterial co-infections are less prevalent in COVID-19 patients than in patients with influenza. In the 2009 influenza pandemic, 1 in 4 severe or fatal cases of influenza A(H1N1)pdm09 had a bacterial infection, with an apparent association with morbidity and mortality.43 The bacteria most commonly associated with influenza infection have been reported to be those which commonly colonise the nasopharynx, such as Streptococcus pneumoniae, Staphylococcus aureus, and Streptococcus pyogenes, with secondary infection usually occurring in the first 6 days of influenza.43, 44, 45 This is in contrast to the bacterial pathogens which we identified in this review, in which the most commonly detected bacterial pathogen was M.pneumoniae, followed by P.aeruginosa, H.influenzae and K.pneumoniae. We found only one report of MRSA and no cases of co-infection with S.pneumoniae nor S.pyogenes. Our findings are similar to studies reporting the bacteria implicated in secondary infections in patients with MERS-CoV and SARS-CoV.8 , 46 , 47
It is noteworthy that in the studies where the diagnostic method was described, all the M.pneumoniae co-infections were diagnosed serologically through detection of IgM. This, on its own, is not a highly specific test and may result in overestimation of infections.48 In a study of patients with SARS, those who tested positive for M.pneumoniae and C.pneumoniae on serology were PCR- negative at the time of respiratory specimen collection.9 The identification of co-infections with gram-negative organisms is consistent with the types of pathogens frequently associated with hospital–acquired pneumonia (HAP) or ICU-HAP as a complication of ICU care49 and does not necessarily suggest a specific predilection for Gram-negative co-infections in COVID-19. Although there were no specific data on antimicrobial resistance patterns in the bacteria identified in the studies included in this review, one study reported the detection of extended spectrum β-lactamase (ESBL)-positive K.pneumoniae, ESBL-positive P.aeruginosa, and carbapenem-resistant K.pneumoniae in patients on ICU.35 A highly resistant Acinetobacter baumannii was isolated in a further study.23
Only three of the included studies in our review reported co-infections with fungal pathogens. However, the prevalence of fungal infections in patients with COVID-19 warrants further investigation and at the time of writing there are an increasing number of reports from Europe of patients with probable or possible COVID-19 associated pulmonary aspergillosis.50, 51, 52, 53, 54 Patients with acute respiratory distress syndrome caused by influenza have been reported to be at increased risk of invasive aspergillosis (IA), even in the absence of predisposing immunocompromising conditions.55 , 56 Early diagnosis of IA is crucial for successful treatment yet conventional microscopy and culture of respiratory tract sample has only low sensitivity and specificity of around 50%.57 Detection of galactomannan (a polysaccharide antigen found primarily in the cells walls of Aspergillus species) from bronchoalveolar lavage fluid (BALF), has been shown to be a useful and rapid tool for identification of IA in both immunocompromised and immunocompetent patients.58, 59, 60, 61 However, as bronchoscopy is an aerosol-generating procedure that poses a substantial risk to staff and patients, it has been recommended that bronchoscopy should have an extremely limited role in COVID-19 patients and only be considered when upper respiratory samples are negative and another diagnosis would significantly change clinical management.62 Serum galactomannan detection for diagnosis of aspergillosis in COVID-19 patients is less sensitive than in influenza patients for currently unknown reasons, and galactomannan testing is not validated for upper respiratory tract specimens.63 In light of the current diagnostic difficulties and the uncertainties relating to the risks associated with IA in COVID-19 pneumonia, clinicians should maintain a high level of suspicion for this infection in critically-ill patients.
We estimated that 3% of patients hospitalised with COVID-19 were also co-infected with another respiratory virus; respiratory syncytial virus (RSV) and influenza A being the most common viral pathogens identified in this review. Influenza has been shown to have dual seasonality in China, with the incidence in northern China following a winter pattern typically seen in northern hemisphere countries, whereas in southern China the virus is prevalent throughout the year.64 Surveillance of children over eight seasons in Beijing has indicated that typically the RSV season lasts from mid-October to mid-May. With the emergence of the COVID-19 pandemic during the winter months, it is unsurprising that studies in our review were also detecting viral co-infection in patients with COVID-19. From available data, we are unable to draw conclusions as to whether patients who have a concurrent viral infection have a worse prognosis than those in whom SARS-CoV-2 is the only detected pathogen.
The strengths of our study include our use of a comprehensive search strategy to identify potentially eligible studies from multiple databases as well as handsearching relevant journals for recently published articles up to our search cut-off date. However, we did encounter some methodological limitations. More than three-quarters of the studies we included were from China describing patients at the start of the pandemic. The majority of studies were case series reporting the clinical characteristics of patients infected with a previously unencountered virus, and details on identified co-infections were generally limited. Testing for co-infecting pathogens during the course of a pandemic is important but most of the studies screened did not report on this. It is possible that patients with a suspected secondary infection may not have had thorough microbiological investigations given the unprecedented circumstances and enormous strain on the hospital systems. Furthermore, there were very few data on the timing of co-pathogen detections which is important for understanding their aetiology. Administration of broad-spectrum antibiotics to a large proportion of the patients included in this review may also have decreased the sensitivity of bacterial culture methods, which could have resulted in underestimation of the true numbers of co-infections. Additionally, some of the bacteria reported may have been merely colonising a normally non-sterile site rather than being causative agents of secondary infections, but it was not possible to differentiate between these possibilities from the data provided. Differences between the healthcare systems in China and other parts of the world mean the overall estimate of co-infected patients may not be representative globally. Although there were more than 3,800 patients included in this review, this is only a small representation of the total number of SARS-CoV-2 infections, which currently stands at around 3 million cases worldwide.65 Additionally, the majority of patients with COVID-19 patients do not require hospitalisation but patients in the studies included in this review were predominantly hospitalised.
There was significant heterogeneity among studies, particularly in the meta-analyses of bacterial co-infections which was not accounted for by age group or setting. This unexplained heterogeneity may be due to differences between studies in disease severity, patient comorbidities, treatment differences (such as corticosteroid administration), use of antibiotics prior to and during hospitalisation, or other unidentified covariates. We also found that a considerable number of included participants remained hospitalised at the censor date of most studies; underestimation of secondary bacterial or fungal infections developing later in the course of the disease is likely.
We did not assess publication bias in this review as the usefulness of standard publication bias tests for proportional meta-analyses has been questioned, with funnel plots and statistical tests potentially yielding misleading results.66 Although publication bias may cause inflated estimates in meta-analyses of studies of treatment effect, this is an unlikely scenario in the context of studies reporting the proportion of patients with co-infections in COVID-19.
In conclusion, we found that the overall proportion of COVID-19 patients who have a bacterial coinfection is lower than in previous influenza pandemics, with little evidence of S.aureus, S.pneumoniae or S.pyogenes having a major role. Overall, these finding support stopping empirical antibiotics in the vast majority of patients when COVID-19 infection is diagnosed. As the pandemic evolves around the world, and as more publications emerge from countries outside China,67 these findings will need to be constantly reviewed. We intend to update our findings from this review every two months to identify any emergent changes.
Funding
LL and VB are funded by the National Institute for Health Research (NIHR) Nottingham Biomedical Research C entre, UK.
Contributors
LL and WSL designed the study. LL, BL and VB reviewed the literature, extracted the data and checked the extracted data. LL planned and conducted the analysis. LL and BL wrote the first draft and all authors reviewed and approved the final manuscript.
Declaration of Competing Interest
LL, BL and VB declare no competing interests. WSL's institution has received unrestricted investigated-initiated research funding from Pfizer for an unrelated pneumonia cohort study in which he is the Chief Investigator.
References
- 1.World Health Organization. Coronavirus Disease (COVID-19) pandemicAvailable from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 13 May 2020
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020 Mar 13 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J. COVID-19 with Different Severity: A Multi-center Study of Clinical Features. Am J Respir Crit Care Med. 2020 doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joseph C, Togawa Y, Shindo N. Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses. 2013;7(Suppl 2):105–113. doi: 10.1111/irv.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahariadis G, Gooley TA, Ryall P, Hutchinson C, Latchford MI, Fearon MA. Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: variable incidence of atypical bacteria coinfection based on diagnostic assays. Can Resp J. 2006;13(1):17–22. doi: 10.1155/2006/862797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical Features of 69 Cases with Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis. 2020. Mar 16 doi: 10.1093/cid/ciaa272. pii: ciaa272[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du RH, Liu LM, Yin W, Wang W, Guan LL, Yuan ML. Hospitalization and Critical Care of 109 Decedents with COVID-19 Pneumonia in Wuhan. China. 2020 Apr 7 doi: 10.1513/AnnalsATS.202003-225OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y. Clinical Features and Treatment of COVID-19 Patients in Northeast Chongqing. J Med Virol. 2020 Mar 21 doi: 10.1002/jmv.25783. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: Different points from adults. Pediatr Pulmonol. May 2020;55(5):1169–1174. doi: 10.1002/ppul.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Antimicrobial resistance factsheet 2018Available from: https://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 13 May 2020
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells G, Shea B, O'Connell D, Peterson J, V Welch, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analysesAvailable from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 13 May 2020
- 17.Sterne JAC, Savovic J, Page MJ, Elbers RG, Blencowe NS, Boutron I. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. doi: 10.1136/bmj.l4898. [DOI] [PubMed] [Google Scholar]
- 18.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. PROSPERO. 2020 doi: 10.1016/j.jinf.2020.05.046. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020178398 CRD42020178398 2020 Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M. Characteristics and Outcomes of 21 Critically Ill Patients With COVID-19 in Washington State. JAMA. 2020 Mar 19 doi: 10.1001/jama.2020.4326. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrasa H, Rello J, Tejada S, Martin A, Balziskueta G, Vinuesa C. SARS-Cov-2 in Spanish Intensive Care: Early Experience with 15-day Survival In Vitoria. Anaesth Crit Care Pain Med. 2020 Apr 9 doi: 10.1016/j.accpm.2020.04.001. pii: S2352-5568(20)30064-3[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N Engl J Med. 2020 Mar 30 doi: 10.1056/NEJMoa2004500. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol. 2020 Mar 20 doi: 10.1002/jmv.25781. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lian J, Jin X, Hao S, Cai H, Zhang S, Zheng L. Analysis of Epidemiological and Clinical features in older patients with Corona Virus Disease 2019 (COVID-19) out of Wuhan. Clin Infect Dis. 2020 Mar 25 doi: 10.1093/cid/ciaa242. pii: ciaa242[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J. Co-infections of SARS-CoV-2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. Apr 2020;63(4):606–609. doi: 10.1007/s11427-020-1668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan. China. Clin Infect Dis. Mar 16 2020 doi: 10.1093/cid/ciaa270. pii: ciaa270[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pongpirul WA, Mott JA, Woodring JV, Uyeki TM, MacArthur JR, Vachiraphan A. Clinical Characteristics of Patients Hospitalized with Coronavirus Disease, Thailand. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200598. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagarro A, Epalza C, Santos M, Sanz-Santaeufemia FJ, Otheo E, Moraleda C. Screening and Severity of Coronavirus Disease 2019 (COVID-19) in Children in Madrid, Spain. JAMA Pediatr. Apr 8 2020 doi: 10.1001/jamapediatrics.2020.1346. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, He W, Yu X, Hu D, Bao M, Liu H. Coronavirus Disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect. Mar 30 2020 doi: 10.1016/j.jinf.2020.03.019. pii: S0163-4453(20)30146-8[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Liu Y, Liu L, Wang X, Luo N, Li L. Clinical Outcomes in 55 Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Who Were Asymptomatic at Hospital Admission in Shenzhen, China. J Infect Dis. May 11 2020;221(11):1770–1774. doi: 10.1093/infdis/jiaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D. Clinical Characteristics of Imported Cases of COVID-19 in Jiangsu Province: A Multicenter Descriptive Study. Clin Infect Dis. Feb 29 2020 doi: 10.1093/cid/ciaa199. pii: ciaa199[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. May 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J. Epidemiologic Features and Clinical Course of Patients Infected with SARS-CoV-2 in Singapore. JAMA. Mar 3 2020 doi: 10.1001/jama.2020.3204. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. Feb 19 2020 doi: 10.1111/all.14238. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. Mar 12 2020 doi: 10.1093/cid/ciaa247. pii: ciaa247[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng F, Liao C, Fan QH, Chen HB, Zhao XG, Xie ZG. Clinical Characteristics of Children with Coronavirus Disease 2019 in Hubei, China. Curr Med Sci. 2020;40(2):275–280. doi: 10.1007/s11596-020-2172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. JAMA. Apr 15 2020 doi: 10.1001/jama.2020.6266. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacIntyre CR, Chughtai AA, Barnes M, Ridda I, Seale H, Toms R. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect Dis. 2018;18(1):637. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chertow DS, Memoli MJ. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309(3):275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 45.Klein EY, Monteforte B, Gupta A, Jiang W, May L, Hsieh YH. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10(5):394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arabi YM, Balkhy HH, Hayden FG, Bouchama A, Luke T, Baillie JK. Middle East Respiratory Syndrome. N Engl J Med. 2017;376(6):584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang DM, Chamberlain DW, Poutanen SM, Low DE, Asa SL, Butany J. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol. 2005;18(1):1–10. doi: 10.1038/modpathol.3800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobieszczanska BM, Kasprzykowska U, Duda-Madej A, Secewicz A, Marciniak J, Gosciniak G. Relevance of serology for Mycoplasma pneumoniae infection among children with persistent cough. Adv Clin Exp Med. 2014;23(2):185–190. doi: 10.17219/acem/37046. [DOI] [PubMed] [Google Scholar]
- 49.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010;51(Suppl 1):S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 50.Alanio A, Dellière S, Fodil S, Bretagne S, B.M. High Prevalence of Putative Invasive Pulmonary Aspergillosis in Critically Ill COVID-19 Patients (April 14, 2020) Available from: SRN:https://ssrn.com/abstract=3575581 or 10.2139/ssrn.3575581. Accessed 13 May 2020 [DOI] [PMC free article] [PubMed]
- 51.Blaize M, Mayaux J, Nabet C, Lampros A, Marcelin AG, Thellier M. Fatal Invasive Aspergillosis and Coronavirus Disease in an Immunocompetent Patient. Emerg Infect Dis. Apr 28 2020;26(7) doi: 10.3201/eid2607.201603. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koehler P, Cornely OA, Bottiger BW, Dusse F, Eichenauer DA, Fuchs F. COVID-19 Associated Pulmonary Aspergillosis. Mycoses. Apr 27 2020 doi: 10.1111/myc.13096. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. Mar 27 2020 doi: 10.1016/S1473-3099(20)30200-0. pii: S1473-3099(20)30200-0[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Arkel ALE, Rijpstra TA, Belderbos HNA, van Wijngaarden P, Verweij PE, Bentvelsen RG. COVID-19 Associated Pulmonary Aspergillosis. Am J Respir Crit Care Med. 2020 May 12 doi: 10.1164/rccm.202004-1038LE. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koehler P, Bassetti M, Kochanek M, Shimabukuro-Vornhagen A, Cornely OA. Intensive care management of influenza-associated pulmonary aspergillosis. Clin Microbiol Infect. 2019;25(12):1501–1509. doi: 10.1016/j.cmi.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 56.Schauwvlieghe A, Rijnders BJA, Philips N, Verwijs R, Vanderbeke L, Van Tienen C. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med. 2018;6(10):782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 57.Hope WW, Walsh TJ, Denning DW. Laboratory diagnosis of invasive aspergillosis. Lancet Infect Dis. 2005;5(10):609–622. doi: 10.1016/S1473-3099(05)70238-3. [DOI] [PubMed] [Google Scholar]
- 58.D'Haese J, Theunissen K, Vermeulen E, Schoemans H, De Vlieger G, Lammertijn L. Detection of galactomannan in bronchoalveolar lavage fluid samples of patients at risk for invasive pulmonary aspergillosis: analytical and clinical validity. J Clin Microbiol. 2012;50(4):1258–1263. doi: 10.1128/JCM.06423-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meersseman W, Lagrou K, Maertens J, Wilmer A, Hermans G, Vanderschueren S. Galactomannan in bronchoalveolar lavage fluid: a tool for diagnosing aspergillosis in intensive care unit patients. Am J Respir Crit Care Med. 2008;177(1):27–34. doi: 10.1164/rccm.200704-606OC. [DOI] [PubMed] [Google Scholar]
- 60.Patterson TF, Thompson GR, 3rd Denning DW, Fishman JA, Hadley S, Herbrecht R. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;63(4):e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou W, Li H, Zhang Y, Huang M, He Q, Li P. Diagnostic Value of Galactomannan Antigen Test in Serum and Bronchoalveolar Lavage Fluid Samples from Patients with Nonneutropenic Invasive Pulmonary Aspergillosis. J Clin Microbiol. 2017;55(7):2153–2161. doi: 10.1128/JCM.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahidi MM, Lamb C, Murgu S, Musani A, Shojaee S, Sachdeva A. American Association for Bronchology and Interventional Pulmonology (AABIP) Statement on the Use of Bronchoscopy and Respiratory Specimen Collection in Patients with Suspected or Confirmed COVID-19 Infection. J Bronchology Interv Pulmonol. Mar 18 2020 doi: 10.1097/LBR.0000000000000681. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verweij PE, Gangneux J-P, Bassetti M, Brüggemann RJM, Cornely OA, Koehler P. Diagnosing COVID-19-associated pulmonary aspergillosis. Lancet Microbe. May 08 2020 doi: 10.1016/S2666-5247(20)30027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shu YL, Fang LQ, de Vlas SJ, Gao Y, Richardus JH, Cao WC. Dual seasonal patterns for influenza, China. Emerg Infect Dis. 2010;16(4):725–726. doi: 10.3201/eid1604.091578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report – 99 Available from:https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200428-sitrep-99-covid-19.pdf?sfvrsn=119fc381_2. Accessed 13 May 2020
- 66.Hunter JP, Saratzis A, Sutton AJ, Boucher RH, Sayers RD, Bown MJ. In meta-analyses of proportion studies, funnel plots were found to be an inaccurate method of assessing publication bias. J Clin Epidemiol. 2014;67(8):897–903. doi: 10.1016/j.jclinepi.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. Apr 22 2020 doi: 10.1001/jama.2020.6775. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]