Abstract
The integration of molecular and cell biology with materials science has led to strategies to improve the interface between dental implants with the surrounding soft and hard tissues in order to replace missing teeth and restore mastication. More than 3 million implants have been placed in the US alone and this number is rising by 500,000/year. Peri-implantitis, an inflammatory response to oral pathogens growing on the implant surface threatens to reduce service life leading to eventual implant failure, and such an outcome will have adverse impact on public health and create significant health care costs. Here we report a predictive approach to peptide design, which enabled us to engineer a bifunctional peptide to combat bacterial colonization and biofilm formation, reducing the adverse host inflammatory immune response that destroys the tissue surrounding implants and shortens their lifespans. This bifunctional peptide contains a titanium-binding domain that recognizes and binds with high affinity to titanium implant surfaces, fused through a rigid spacer domain with an antimicrobial domain. By varying the antimicrobial peptide domain, we were able to predict the properties of the resulting bifunctional peptides in their entirety by analyzing the sequence-structure-function relationship. These bifunctional peptides achieve: 1) nearly 100% surface coverage within minutes, a timeframe suitable for their clinical application to existing implants; 2) nearly 100% binding to a titanium surface even in the presence of contaminating serum protein; 3) durability to brushing with a commercially available electric toothbrush; and 4) retention of antimicrobial activity on the implant surface following bacterial challenge. A bifunctional peptide film can be applied to both new implants and/or repeatedly applied to previously placed implants to control bacterial colonization mitigating peri-implant disease that threatens dental implant longevity.
Keywords: Peri-implant disease, Bifunctional Peptides, Antimicrobial, Structure, Function, Titanium/Ti Alloy Implant, Dental Implant, Stability, Peptide Film
Graphical Abstract

INTRODUCTION
There is a continuing need to improve patient treatment to ensure the longevity of the therapeutic advantages offered by titanium dental implants 1–8. The creative fusion of molecular and cell biology with materials science and engineering has resulted in improved understanding of implant-tissue interfaces 9. These advances have been translated clinically to improve oral and systemic health through the replacement of diseased teeth by titanium dental implants, among numerous examples 10–12. Titanium and its alloys have the requisite toughness needed to resist the repeated loading that occurs with food mastication. Titanium is biocompatible, and more importantly, inherently promotes its own integration into host bone by activating the Wnt/integrin signaling pathway for osteogenesis 13.
In recent years, increased reliance on dental implants to restore missing teeth has resulted in a concomitant rise in the prevalence of peri-implant disease, a host-mediated immune response to bacteria which can shorten implant life and culminate in the loss of the implant 14–15. A growing consensus suggests that peri-implant disease is similar to periodontal disease: bacterial plaque accumulation and microbiome dysbiogenesis trigger a host immune inflammatory response that destroys soft- and hard-peri-implant tissues 16–18. Within weeks after implant placement, a biofilm develops consisting of the typical subgingival bacterial species, including keystone periodontal pathogens such as A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, and P. intermedia 19–21. Dysbiosis shifts the relative abundance of commensal species to pathogens. For many patients, this shift induces the host to mount an inflammatory response leading to peri-implant disease, starting with peri-implant mucositis and progressing to peri-implantitis, which is accompanied by dramatic bone loss that can necessitate implant removal 22. A treatment protocol that can slow and/or prevent bacterial infection may help to mitigate the host immune response and slow peri-implant disease progression.
The incidence of peri-implantitis is reported to be as high as 14.5% after 9 years of service although clinically significant, non-linear loss of bone support around the implants may be present as early as after 3 years after placement in more than 80% of patients 23. With over 3 million implants placed in the US alone and growing by 500,000 implants/year 7, a reduced service life ending in implant failure will adversely impact public health, trigger increased health care costs and precipitate a loss of public confidence in the dental profession. Furthermore, this outcome may dissuade many patients to avoid this therapy who would benefit from the health benefits of implants. Currently, the goal of implant disease treatment is to control bacterial infection through mechanical debridement and bacterial plaque control in order to mitigate the host immune response, in order to slow disease progression 24. However, debridement, which is often performed with titanium brushes 25 results in implant surface damage and loss of biocompatibility. This hastens inevitable apical movement of the implant-supporting tissues and worsens the loss of supporting hard and soft tissue.
An additional challenge associated with prevention of implant infection is the increased incidence of bacterial resistance, which represents a major public health concern 26–29. Novel antibacterial agents and strategies are needed to ensure future therapeutic efficacy 30. While systemic antibiotics can treat infections that result from non-resistant bacteria, the peri-implant environment poses many challenges. Implant surfaces are susceptible to biofilm development as bacteria attach to the surface and synthesize an extracellular biofilm matrix 31. An estimated 80% of human infections are associated with biofilms 32. Biofilms respond differently to antibiotics than planktonic bacteria and are difficult for antibiotics to penetrate 31, 33. Poor antibiotic penetration into biofilms results in subtherapeutic antibiotic concentrations and increases the likelihood of developing antibiotic resistance 34. Preventing the attachment and killing of planktonic bacteria to the implant surface, while killing them via antimicrobial agents that do not lead to the development of resistance represents a novel strategy for reducing biofilm formation and preventing persistent infection that leads to implant failure 35–36.
One approach to addressing bacterial resistance is the use of antimicrobial peptides (AMPs). AMPs are natural antimicrobials that form part of the innate immune defense peptides of both invertebrates and vertebrates. Most AMPs contain fewer than 50 amino acids and more than 2800 AMPs have been discovered from natural resources 29. Integration of computer-assisted peptide design methods has increased the number of in silico designed antimicrobial peptides 37–39. The mechanisms of AMP action on bacteria includes membrane perturbation, disruption and/or translocation affecting diverse physiological events such as cell wall biosynthesis, pore formation, and cell division, as well as non-membrane-based pathways 37, 40. The mechanism for GL13K action on bacterial membranes involves localized removal of lipid from the membrane through peptide induced micellization. The mechanism for action of AMPA is not yet fully explored, it is also considered to have its action through membrane permeation41–42. Furthermore, their antimicrobial affects can mitigate biofilm formation when used alone or in combination with other AMPs, or even with antibiotics, to achieve the desired antimicrobial effect and preserve the health of the host tissues without triggering resistance.
Systemic delivery of AMPs has been a major limiting factor in their wider use as therapeutics because a high AMP concentrations is required to achieve effective antimicrobial activity and such levels can potentially result in in vivo damage to host cells 43. Local delivery of AMPs may overcome this challenge by reducing and focusing the required therapeutic concentration and thereby decrease the potential for deleterious effects on eukaryotic cells 43. The advantages of delivering AMPs locally have been explored using a variety of methods for their retention on implant surfaces, including physical adsorption and chemical immobilization strategies. Chemical immobilization strategies include covalently attaching AMPs to the implant surfaces using silane-, catechol- and phosphate-groups 44–47. Structural constraints introduced to the AMP during covalent coupling are known to limit antimicrobial activity, more so, the covalent coupling procedure can only be performed prior to implantation, as they are generally performed under harsh conditions which prevents their intraoral application. To this point, investigators recently demonstrated that amphipathic GL13K antimicrobial peptide can be used to coat dentin to resist recurrent caries around resin bonded dental restorations 48–49. Combinatorial screening of peptide sequences with affinity for various materials has identified hundreds of peptides with the ability to self-assemble on metal-, metal oxide-, mineral- and polymer-surfaces 50–54. As the number of the biocombinatorially selected peptides increased, computational methods have provided an understanding for the peptide-solid materials interactions, as well as for their self-assembly and molecular recognition mechanism(s) on nanostructured materials including metals, metal oxides, ceramics and others 55–56. By merging biocombinatorial and computationally methods, we have developed high affinity inorganic binding peptides for titanium and titanium alloys. Furthermore, we have engineered these peptides into bifunctional peptide molecules that incorporate biologically instructive signaling functions in order to create novel, bioactive biomaterial interfaces 57–61. We demonstrated that titanium binding peptides (TiBP) can be used to design a biomimetic interface for enhancing bioactivity in osteoblast and fibroblast cells when coupled with RGD peptides 62. We have further demonstrated that TiBP is an effective anchor for AMPs on implant surfaces, serving to localize the molecule effectively for repeated intraoral applications 63. When a TiBP anchor was combined with a Wnt signaling peptide 64–65, the resulting bifunctional peptide produced a peptide film on implant surfaces that led to enhanced osteogenesis in human stem cells, consistent with directed bone regenerative capacity 11. We next combined the TiBP with antimicrobial peptides and demonstrated their effective use against E. coli, S. epidermis and S. mutant strains 66 67. While our earlier studies utilized a simple flexible spacer of amino acids, e.g., “GGG”, to combine two distinctive AMPs, we have now analyzed sequence-structure-function relationships for optimal design of the spacer. By computationally studying the Dictionary of Protein Secondary Structure (DSSP) features and observed patterns, we proposed secondary structure “rules” to enhance antimicrobial activity of bifunctional peptides. By designing a rigid and longer amino acid spacer domain, “GSGGG”, between the TiBP and the AMP domains, we significantly improved the antibacterial efficacy against S. epidermis bacteria 68. We demonstrated the TiBP as an effective anchor for the AMPs on implant surfaces serving to localize the molecule effectively for repeated intraoral applications 63.
Here, we report a novel antimicrobial medicinal approach to slowing or halting the progression of peri-implant disease by furthering the design of bifunctional peptides that deliver a local antimicrobial peptide. This film can be applied in two minutes and can be repeated at follow up appointments 63. The renewable effects of the bifunctional peptides upon successive reapplication were evaluated on bacteria-fouled and -cleaned dental implant surfaces, mimicking the re-treatment of implants affected by peri-implant disease in a dental office 63. We systematically studied the sequence-structure-function relationships of two bifunctional peptides that incorporated structurally distinctive antimicrobial peptides combined with the same anchoring domain using a newly developed longer, more rigid peptide spacer. Our secondary structure prediction suggested that greater helical content could improve antimicrobial activity while preserving the intrinsically disordered behavior of TiBP for effective surface binding. The resulting bifunctional peptides were evaluated for their suitability for clinical deployment using tests of peptide binding, stability, antimicrobial function and durability in vitro on titanium implant discs (see “Schematic”). Our computational predictions were merged with experimental structural analyses and showed enhanced design of bifunctional peptides with the best candidate molecules outperforming other peptides in promoting antimicrobial film activity. Overall, we demonstrate that our engineered small bifunctional peptide selectively binds to titanium/titanium alloy implant surfaces to deliver an antimicrobial peptide film in as little as two minutes. This non-surgical approach has the potential to improve oral health by controlling microbial dysbiogenesis and reducing peri-implant disease progression. This approach could be more widely beneficial for the design of a range of bioactive biomaterials interfaces that could for the basis for next generation therapeutics.
MATERIALS AND METHODS
Peptide Synthesis, Purification, and FITC-derivatization
Peptides were synthesized by standard solid phase peptide synthesis technique using Fmoc chemistries and Wang resins (AAPPTec Focus XC solid phase peptide synthesizer, Louisville, KY). Fmoc protecting groups were removed by 20% piperidine in dimethylformamide (DMF). Following deprotection, piperidine was removed by DMF and the samples were solubilized in DMF at a concentration of 0.2M and added in 7-fold excess. The amino acids were activated with 0.4M O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (HBTU) and 1M 4-methyl morpholine (NMM) in DMF. Reactions were performed with mechanical mixing under nitrogen gas. Resin-bound peptides were dried with ethanol and cleaved using a cleavage cocktail. Reagent K (TFA/thioanisole/phenol/ethanedithiol at a ratio of 87.5:5:5:2.5) was used to deprotect side-chain and peptides were precipitated with cold ether. The crude peptides were purified using reverse phase-HPLC to greater than 98% purity, lyophilized and stored at −20°C.
Fluorescein 5(6)-isothiocyanate (FITC) was used to fluorescently derivatize bifunctional peptides on their free C’-termini for experiments requiring visualization of bifunctional peptide molecules bound to the surface of titanium implant discs.
Peptide Property Calculations
Physicochemical data on the peptides including molecular weight, isoelectric point, charge and GRand AVerage of hydropathY (GRAVY) scores were obtained using the ExPasy ProtPram Server 69. Hydrophobicity was evaluated by the antimicrobial peptide database (APD3) 38.
Peptide CD Data Collection and Secondary Structure Prediction
Secondary structure estimation was accomplished using a Jasco J-810 circular dichroism (CD) spectrophotometer. Solutions of 40μM peptide in 100mM Tris-HCL buffer with varying volumes of 2,2,2-trifluoroethanol (TFE) were prepared for CD analysis. A minimum of 8 scans over wavenumber 190–260nm with a scan rate of 0.5 nm/min were collected on a calibrated spectrophotometer and averaged. The background was subtracted, and the spectra smoothed using the Savitzky-Golay algorithm. The resulting CD spectra were deconvoluted using the BeStSel web server for accurate prediction of protein secondary structure and folding 70.
Predicted secondary structure contents for helical (α, 310 and π-helix), beta (β-bridge, bonded turn), and irregular (bend and loop) features were determined using the Chou-Fasman algorithm. The Chou-Fasman algorithm was applied for each bifunctional peptide after uploading their CD spectra and amino acid sequences using the online server available through the CD Analysis and Plotting Tool (CAPITO) 71.
Peptide Structure Analysis
The de novo 3D structural modeling algorithm, PEP-FOLD 3.5, was implemented to generate Protein Data Base (PDB) models for a minimum of five of the best predictions for each bifunctional peptide sequences 72–73. PEP-FOLD 3.5 was used to generate 3D-structural conformations of linear peptides. PEP-FOLD 3.5 generates peptide structures by assigning one of 27 structural alphabets where fragments of four amino acid residues overlap with three residues. The structural alphabet generalizes the secondary structure by assigning geometric descriptors created by the Hidden Markov model as described by Maupetit, et. al., 74. 3D models were ultimately generated from the fragments using a coarse-grained representation and refined by 30,000 Monte-Carlo steps using the PEP-FOLD 3.5 online service on an average of 200 simulations executed assuming aqueous conditions and neutral pH. Once generated, the models were clustered and sorted using sOPEP (Optimized Potential for Efficient Structure Prediction) with non-biased modeling.
Similarity among the predicted secondary structure models for the bifunctional peptides was compared for each of the constituent domains: titanium binding domain (TiBP), spacer, and each of two unique antimicrobial domains using the MatchMaker tool. The individual constituent domain structures were superimposed on the corresponding segments of the bifunctional peptide structure. The Match-Align tool was used with a 5Å threshold and the percent identity or degree of relatedness was recorded.
Backbone rigidity of the bifunctional peptides was predicted using the DynaMine Webserver following their amino acid sequence in FASTA format 75. The server segmented the sequence and the fragments were used as the input for the DynaMine predictor for the given segment length. The predictions for each segment were reassembled to produce a dynamics profile from the amino acid sequence.
Protein Data Bank files containing the secondary structure models generated by PEP-FOLD 3.5 were visualized and further analyzed by the UCSF Chimera program 76. The theoretical “footprint” for each bifunctional peptide was calculated using the measure tool in Chimera. The footprint was determined by obtaining the distance from the α-carbon of amino acid residues to obtain length and width values. These measurements were converted to corresponding area and the number of peptide molecules required to saturate a 10 mm disc surface area serving as an implant mimic was determined. The number of peptides was converted to a molecular mass required to deliver the corresponding surface coverage to the titanium implant disc surface.
A web interface program DichroCalc 77 was used to predict the theoretical circular dichroism spectra from secondary structures models predicted with PEP-FOLD 3.5. Spectra were requested in ellipticity units [(deg. cm2)/dmol] over wavenumber 190nm to 260nm and compared to the corresponding experimentally collected spectra. The Hirst ab initio parameter set was used for backbone chromophores.
Titanium Implant Disc Preparation
Coin-shaped titanium implant discs were punched from grade 4 titanium by the USC Engineering Shop. The discs were 10mm in diameter and 0.5mm thick, lap-polished and air-blasted with 180–220 micron titanium dioxide particles. Following manufacturing, the discs were cleaned as stated in a published protocol used for producing surfaces optimal for osseous integration 4. The protocol included sonication in DI H2O for 5 minutes, ethanol for 30 seconds, DI H2O for 30 seconds, 40% sodium hydroxide for 10 minutes, washed in DI H2O for 5 minutes, 50% nitric acid for 10 minutes followed by rinsing with DI H2O for 5 minutes. The discs were autoclaved prior to use.
Peptide Binding to Implant Discs
Titanium implant disc functionalization with bifunctional peptides was accomplished by incubating 100μL of a specified fold concentration of the theoretical “footprint” concentration onto clean, sterile discs for different time periods at 37°C for 2 minutes. Following incubation, the discs were transferred to a sterile well in a 24-well plate containing 500μL of DI H2O and washed multiple times to remove unbound peptide. The discs were transferred to a clean glass microscope slide for imaging using a fluorescent microscope. All experiments were repeated a minimum of three times and images were recorded at 10X magnification.
Determination of Surface Coverage
A custom MatLab script was developed to determine the percent surface coverage of fluorescently labeled bifunctional peptides on the implant disc surface. The color fluorescent images were read into MatLab using the imread function. The images were converted to black and white and the total number of black and white pixels quantitated. The number of white pixels corresponding to the fluorescently labeled peptides, was divided by the total number of pixels to determine the percent surface coverage. The theoretical footprint was used solely to determine an ab initio concentration for achieving an optimal bifunctional peptide interface in a clinically relevant time interval and did not include lateral resolution.
Serum Competition Assay
Serum competition binding assay was completed with 1.0wt%, and 5.0wt% of bovine serum albumin (BSA) at selected theoretical “footprint” concentrations of FITC-labeled bifunctional peptide. Prior to functionalization of sterile titanium implant discs, a solution of BSA and the bifunctional peptide was made in a sterile centrifuge tube. In a sterile 24 well plate, 100μL of the BSA/bifunctional peptide solution was pipetted onto a titanium implant disc and incubated at 37°C for 2 minutes. The discs with BSA/bifunctional peptide were transferred to a new well containing 500μL sterile deionized water for 1 minute and washed to remove unbound peptide. The discs were transferred onto a clean glass microscope slide and imaged as previously described 63.
Mechanical Durability Assay
Sterile titanium implant discs were functionalized with FITC-labeled bifunctional peptides and brushed using a commercially available electric toothbrush, as previously described 63, 78–79. Deionized water was applied to the functionalized discs and an electric toothbrush with a round head the same size as the implant disc was applied to the disc for 1 minute. A 100g weight was secured to the toothbrush 10cm from the brush head to ensure consistent force during brushing. Following brushing the implant disc was imaged.
Bacteria Culture
Streptococcus mutans bacteria (ATCC 700610) were cultured according to an ATCC protocol. Frozen stocks were streak plated on agar and incubated for 24 hours at 37°C in atmosphere with 5% CO2. A single bacterial colony was used to inoculate 5mL of Brain Heart Infusion (BHI) broth in a sterile 50mL conical tube and incubated overnight in the same conditions. Following incubation, 1mL of culture was added to 9mL of fresh media and grown to mid-log phase with a final concentration of 105 CFU/mL.
Visualizing Bacteria on Implant Discs
Following the functionalization of titanium discs, they were transferred to a sterile well in a 24-well plate and 400 μL of S. mutans bacteria at a concentration of 103 CFU/mL was added to the wells containing the discs and incubated for 24 hours at 37°C in atmosphere with 5% CO2. Following incubation, discs were imaged with a fluorescent microscope to visualize FTIC-labeled bifunctional peptides on the disc surface after bacterial challenge. The dead bacteria were stained with propidium iodide and imaged using a fluorescent microscope. Experiments were repeated in triplicate and images were recorded at 10X magnification.
Statistical Analysis
For all experimental groups, values are reported as mean ± standard deviations. One-way analysis of variance (ANOVA) was performed to assess statistical significance. Statistical significance was set at p<0.05.
RESULTS AND DISCUSSION
We explored the structure-function relationship of bifunctional peptides designed with two distinct antimicrobial peptide domains. Each bifunctional peptide molecule incorporated three peptide domains: an implant anchoring domain provided by the TiBP, an antimicrobial domain provided by the AMP, and a spacer domain to ensure the functionality of each of the two other domains when constrained within a single peptide chain. Computationally derived rules for predicting performance of antimicrobial bifunctional peptide films and experimentally evaluated for antimicrobial activity, extent of film coverage and binding, binding under competition from an interloper contaminant, and mechanical durability within clinically relevant parameters needed by dentists to treat peri-implant disease.
Design by Structure Prediction from Amino Acid Sequence
In our earlier work, we identified secondary structure rules that associate greater antimicrobial property with α-helix features adopted over 4- and 5-amino acid residues 67. Based on this, we screened the CAPITO Webserver provided in the Antimicrobial Peptide Database (APD) that contains the cationic AMPs identified with low minimum inhibitory concentrations (MIC) based on their percentage of α-helix secondary structure 71. The method implemented in CAPITO uses the Chou-Fasman algorithm to analyze the relative frequencies of amino acids adopting a specific secondary structure conformation based on protein structures previously solved by X-ray crystallography. The secondary structure prediction mainly relies on the probability parameters obtained for the occurrence of α-helix, β-sheet and turns. The Chou-Fasman method is roughly 60% accurate in predicting secondary structures compared to 80% accuracy achieved by some of the recent machine learning approaches 71; however computationally Chou-Fasman remains a simple and efficient method for approximating secondary structure content starting from an amino acid sequence. Chou-Fasman was used as an initial estimation tool in developing rules for the antimicrobial peptide film property. We identified two AMPs with low MIC: AMPA80, comprised of 60% predicted α-helix forming amino acids and GL13K81 containing no predicted α-helix forming amino acids (Table 1). The MIC for each AMP is given is Supplemental Information Table S1. Titanium binding peptides were selected using phage display and characterized for their binding affinity using Quartz Crystal Microbalance Spectroscopy (QCM) 62 67. Based upon this earlier work, we selected one of the strong titanium binding peptides as a promising candidate for the bifunctional peptide film 62. When AMPA was combined with a spacer to the titanium binding peptide (TiBP) domain, the α-helix content of the resulting bifunctional TiBP-AMPA increased to 69%. Whereas combining TiBP with GL13K resulted in a drastic change to the α-helix content for the bifunctional TiBP-GL13K molecule, with α-helix content as low as 50%.
Table 1: Chou-Fasman secondary structure predictions from amino acid sequences for the bifunctional peptide and its constitutive domains.
Secondary structure features including helix (α, 310 and π-helix), beta (β-bridge, bonded turn), and irregular (bend and loop) features.
| α-helix | β-strand | irregular | |
|---|---|---|---|
| TiBP | 0% | 0% | 100% |
| AMPA | 60% | 0% | 40% |
| GL13K | 0% | 0% | 100% |
| TiBP-AMPA | 69% | 0% | 31% |
| TiBP-GL13K | 50% | 0% | 50% |
Native chimeric proteins containing multiple functional domains often are separated by inter-domain sequences called “spacers” that enable multiple domains to coexist on a single polypeptide chain. Inspired by this, we studied different spacer sequences when designing the bifunctional peptides described here. The goal of the spacer design was to preserve and enhance the function of each of the functional domains within the molecule. We previously tested the effects of spacers on the overall bifunctional peptide by testing them with a single antimicrobial peptide, AMP1, linked with a strong titanium binding peptide sequence (TiBP) 62, 66–68. Here, a five-amino-acid spacer, i.e. GSGGG, resulted in drastic improvement of the antimicrobial efficacy against S. epidermis, compared to a three amino acid, GGG spacer. We therefore elected to combine the TiBP domain to each of the two selected AMPs using the GSGGG spacer.
Table 2 provides the physicochemical properties for AMPs and the related bifunctional peptide. Both of the AMPs selected are cationic with net positive charge of 5 and 4, respectively for AMPA and GL13K. In contrast, the net charges of the corresponding bifunctional peptides, TiBP-AMPA and TiBP-GL13K increased to 8 and 7, respectively.
Table 2:
Physicochemical properties of peptides.
| # AA | MW | pI | Charge | GRAVY | ||
|---|---|---|---|---|---|---|
| TiBP | RPRENRGRERGL | 12 | 1496 | 12 | +3 | −2.6 |
| AMPA | KWKLWKKIEKWGQGIGAVLKWLTTW | 25 | 3085 | 10 | +5 | −0.4 |
| GL13K | GKIIKLKASLKLL | 13 | 1429 | 11 | +4 | 0.7 |
| TiBP-AMPA | RPRENRGRERGL GSGGG KWKLWKKIEKWGQGIGAVLKWLTTW | 43 | 4991 | 12 | +8 | −1 |
| TiBP-GL13K | RPRENRGRERGL GSGGGG KIIKLKASLKLL | 30 | 3218 | 12 | +7 | −0.8 |
The Chou-Fasman secondary structure algorithm predicted greater helical content in the bifunctional peptides compared to either of the AMP domains in isolation. Further, the distribution of secondary structure between the AMPs in isolation compared to the bifunctional peptides suggested that the AMPA domain retained a greater percentage of secondary structure than GL13K. Thus, we predicted that a “design with AMPA would have greater antimicrobial potential than one containing GL13K. We recognize the limitations of Chau-Fasman in predicting secondary structure; however, the GL13K α-helix prediction differed only by 10% from a recently reported estimate on the secondary structure of GL13K using a complementary approach 48–49.
Design by Hydrophobicity and Amphipathicity
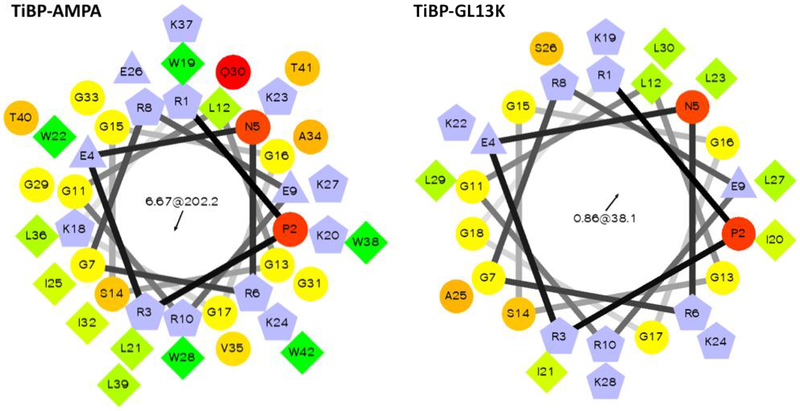
Hydrophobicity and amphipathicity are believed to allow the AMP to penetrate a bacterial lipid bilayer and disrupt the cell membrane 82–83. Using the Calculate and Predict tool from the Antimicrobial Peptide Database (APD), it was determined that TiBP-AMPA has a hydrophobic ratio of 30% with 10 hydrophobic residues aligned along the same surface of the α-helix. TiBP-GL13K revealed a hydrophobic ratio of 26%, slightly less than that of TiBP-AMPA, with only 5 hydrophobic residues aligned on the same surface (Supplemental Information Figure 1). This sequence analysis revealed that the majority of the hydrophobic amino acids in both bifunctional peptides were located in the AMP region. The TiBP region contains only one hydrophobic residue on the C’-terminus of the binding peptide, immediately before the spacer. The hydrophobic nature of the bifunctional peptide is attributed to the AMP portion, which may increase the likelihood of the AMP interacting with the bacterial membrane while the binding domain remains anchored on the implant surface. To visually demonstrate this distribution of residues, we generated helical wheels using an online tool (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). The helical wheel diagrams represented in Figure 1 show that 10 hydrophobic residues reside on the same surface of the α-helix for TiBP-AMPA compared to only 5 for TiBP-GL13K.
Figure 1: Helical wheel predictions of bifunctional peptides.
Hydrophilic amino acid residues are represented as circles, hydrophobic amino acid residues as diamonds, potentially negatively charged residues as triangles, and potentially positively charged residues as pentagons. The most hydrophobic amino acid residue is shown in green with the chroma intensity decreasing proportionally to hydrophobicity, with zero hydrophobicity coded as yellow. Hydrophilic residues are coded red, with intense red chroma being the most hydrophilic (uncharged) residue, and the chroma decreasing proportionally to the hydrophilicity. Potentially charged residues are shown as blue.
Dynamics Prediction in Bifunctional Peptide Design
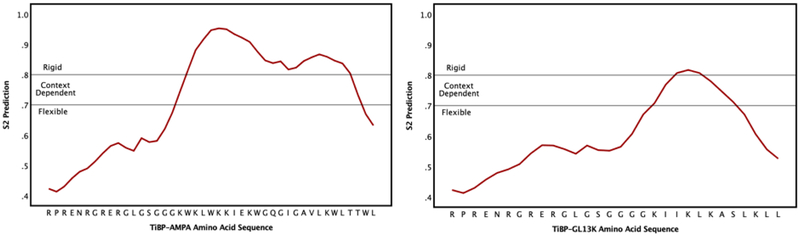
Next, the dynamics of the peptide backbone were studied to determine the disorder of the regions within the whole bifunctional peptide relative to their constitutive binding, spacer, and AMP domains. DynaMine, a tool that leverages chemical shift data to make predictions about backbone dynamics at the amino acid residue level, was used for this purpose. The dynamics of the residues are essential for peptide function, so evaluating the backbone dynamics in relation to the bifunctional peptide function is important in considering the design of these peptides 75. Given a protein sequence, DynaMine predicts backbone flexibility at the level of amino acid residue in the form of backbone N-H S2 order parameter values. These S2 values represent how restricted the movement of the atomic bond vector is with respect to the molecular reference frame. The results from the DynaMine analysis are depicted in Figure 2.
Figure 2: DynaMine classification for backbone dynamics of amino acids comprising each bifunctional peptide.
The AMP domains located on the C’-terminus represent a more ordered region relative to the TiBP binding domain located on the N’-terminus. The TiBP domain is an intrinsically disordered peptide. AMPA has more order than GL13K, which could contribute to its greater predicted antimicrobial function.
The more hydrophilic amino acids located in the AMP domain of the bifunctional peptide correspond to the more ordered region of the bifunctional peptide. The Gly and Ser residues comprising the spacer region are known to be more disordered serving to promote the propensity for a more dynamic backbone that improves overall function of the tethered molecule, namely binding to the implant surface while presenting an active antimicrobial domain. This is important for dental implants as the accumulation of a biofilm on the implant is believed to lead to an adverse host immune response to the bacterial antigens, resulting in host directed inflammatory destruction of soft and hard tissues surrounding the implant 18, 22, 84–85. Consistent with this interpretation, the binding domain for the bifunctional peptides showed the least variation in order. Intrinsically disordered proteins (IDPs) function in a wide spectrum of biological situations due to their ability to adapt their structure by adopting a conformation over a small number of amino acid residues 75, 86–88. Thus, it would be expected that the dynamics of the binding domain may resemble the range of conformational structure observed within IDPs.
Secondary Structure Modeling and Analysis
A more in depth secondary structure prediction was accomplished by developing structural models from the amino acid sequences using PEP-FOLD 3.5 89. Secondary structure models generated for the individual domains and the bifunctional peptide appear in Figure 3. Secondary structural analysis of the antimicrobial peptide domains, AMPA and GL13K, revealed that the AMPA domain is composed of two short α-helixes joined by a turn while GL13K is composed of one short α -helix. The Chou-Fasman analysis did not identify helicity exclusive to the GL13K domain, although helicity was predicted for the bifunctional peptide, TiBP-GL13K.
Figure 3: Secondary structure models and structural similarity analysis.
Each TiBP domain is colored purple, the AMPA domain is colored orange, the GL13K domain is colored red, and the spacer domain linking the antimicrobial and binding domain is colored black. The chart depicts the structural similarity determined by superimposing the domain model over the bifunctional model and calculating the percent identity.
The mechanism by which amphipathic α-helical AMPs kill bacteria may involve their creation of trans-bilayer pores which serve to disrupt the bacterial membrane by separating the polar from the non-polar parts90. Thus, we predicted TiBP-AMPA would have greater antimicrobial activity than TiBP-GL13K due to the greater number of membrane-disrupting helical features present in AMPA (two features) compared to GL13K (one feature).
Using Chimera, the PDB file could be compared using the MatchMaker tool and the structure models superimposed 91. Similarity among each of the individual functional domains was evaluated by superimposing the structural model for the AMP and TiBP domain alone on the corresponding portion of the bifunctional peptide. The superimposed structures were further studied to determine the percent identity or the degree of relatedness. This was useful in determining the preservation of the TiBP domain and each antimicrobial domain when linked by the spacer in the whole bifunctional peptide. This analysis revealed that 91.7% of the TiBP domain identity was preserved when combined with either AMPA or GL13K through the GSGGG spacer. The GL13K domain retained 92.3% identity, while the AMPA domain retained only 60% identity. This suggests that most of the secondary structure of the TiBP and GL13K domain are preserved by the GSGGG spacer, while further engineering of the spacer could improve the antimicrobial activity of TiBP-AMPA. Preservation of the TiBP domain is postulated to be critical for binding to the implant surface in the presence of competing proteins while contributing to the durability of the bifunctional peptide in the oral environment both of which are an essential property for effective clinical deployment..
Experimental Determination of Secondary Structure
Next, secondary structure of the bifunctional peptide was experimentally determined using CD spectroscopy in an aqueous environment with increasing concentrations of TFE to mimic peptide film behavior. Theoretical spectra were determined using DichroCalc 77 from the PDB files generated from the secondary structure models. Secondary structures of the bifunctional peptides were evaluated experimentally using CD spectroscopy in an aqueous environment. The presence of ordered or disordered conformational state was assayed for each bifunctional peptide. We next evaluated the folding propensity of the peptide sequences in the presence of increasing TFE concentration. TFE is used as a structure stabilizing solvent to mimic the restricted mobility of the peptides due to inherited function of the bifunctional peptide. As the bifunctional peptide interacts with the titanium surface through its anchoring domain it also interacts with the bacterial membrane via the antimicrobial domain. Theoretical spectra were determined using DichroCalc from the PDB files generated from the secondary structure models 90. Analyses performed by the Dynamine program for dynamic behavior of the TiBP domain suggest the TiBP behaves as an intrinsically disordered peptide. While these structural states are representatives of the peptide in aqueous environments, circular dichroism data supported this behavior. In the absence of the TFE, both of the peptides exhibited a strong negative ellipticity band around 198nm representing the pi-pi* transition. This is a characteristic band for random coil conformation which is in equilibrium with other secondary structures. Based upon our prior TiBP related work, we conclude that the titanium binding features of the bifunctional peptide was preserved 66. Overall, addition of TFE to each peptide resulted in reduced intensity of the pi-pi* transition ellipticity band and an observed ~10 nm red shift in absorption wavelength. These results suggest a shift in secondary structure population and both peptides undergoing some degree of conformational reordering in the presence of TFE. Relating such transitions to the bifunctionality of the peptide may provide insights for rational design for bioactive interactions at the interfaces by the peptide.
The CD spectra were deconvoluted using Beta Sheet Selection (BeStSel), a method for secondary structure determination from CD spectra 70. BeStSel links the CD spectra structural findings to the computational Dictionary of Protein Secondary Structure (DSSP) patterns 92. Our previously described “Rule Induction” method also relied on DSSP structure patterns and identified a pattern of 4- and 5- amino acid α-helix structures as being linked to antimicrobial activity in bifunctional peptides 67–68. The BeStSel tool allows for deconvolution of experimental CD spectra into the structural feature patterns that are used to inform the “Rule Induction” method. Both bifunctional peptides were found to switch their conformation from an unordered state in aqueous buffers to their functionally relevant α-helical conformation in the presence of TFE. The theoretical CD spectra determined from the PDB model files more closely represented the 90% TFE environment. This suggests that the computational structural predictions are more accurate for environments similar to the conditions in which the peptides act as a film. The results from the deconvolution using BeStSel for TiBP-AMPA and TiBP-GL13K are depicted in Figure 4.
Figure 4: Theoretical and experimental CD spectra with deconvolution using Beta Sheet Selection (BeStSel).
Experimental CD spectra were collected in aqueous environment and with increasing concentrations of 2,2,2-trifluoroethanol. The pie charts represent BeStSel’s deconvolution of the CD spectra.
Theoretical Surface Coverage Determination
The concentration of bifunctional peptides theoretical needed to provide 100% surface coverage when applied onto an implant-mimicking titanium disc surface was estimated by measuring the dimensions of the binding peptide domain to obtain a theoretical “footprint” area. The resulting area was used to determine the concentration of peptide molecules required to cover a 10 mm diameter titanium implant disc (Table 3). One limitation of the theoretical surface coverage concentration calculation is that the surface is assumed to be smooth; however, the surface roughness produced by blasting an implant with titanium dioxide to promote osseous integration would result in a greater surface area than what would be calculated. This limitation was overcome by using a multiple of the theoretical binding concentration, up to 6-times (6X), to achieve near 100% surface coverage after a two-minutes binding period. We focused on minimizing the time required to achieve complete surface functionalization by the bifunctional peptide because this will be important in translating this technology to a clinical application. The two-minute binding time frame represents a reasonable working time for application of the bifunctional peptide film in a clinical environment.
Table 3:
Theoretical “footprint” calculation and concentrations.
| Theoretical Footprint | Conc. | |||
|---|---|---|---|---|
| Length (Å) | Width (Å) | Area (Å2) | μM | |
| TiBP-AMPA | 19.8 | 16.6 | 329 | 111 |
| T1BP-GL13K | 14.7 | 11.8 | 173 | 211 |
Evaluation of Binding, Stability and Durability
The theoretical footprint concentration of each bifunctional peptide with a multiple of the binding concentration up to 6X was determined to result in near 100% surface coverage after incubation with a titanium implant disc for only two minutes at 37°C. Following incubation, the discs were washed to remove unbound or non-specifically bound peptide from the surface prior to imaging with a fluorescent microscope. The fluorescent images were then analyzed using a MATLAB script to determine the percentage of the implant disc covered by the bifunctional peptides. The initial binding for TiBP-AMPA resulted in 99% surface coverage after two minutes compared to 96% for TiBP-GL13K. This indicates that in a clinically achievable application, the bifunctional peptide are able to form an antibacterial film with near complete coverage of the implant surface. Representative fluorescent images for each bifunctional peptide are depicted in Figure 5, while the chart indicates the mean with standard deviation error bars for three replicate experiments.
Figure 5: Fluorescent microscopy images of bifunctional peptide binding to titanium implant discs, binding with competition from BSA, and durability following 1 minute of brushing with an electric toothbrush.
The chart depicts the means and standard deviations of three replicate experiments for each bifunctional peptide in each condition. TiBP-AMPA binding was statistically significant compared to binding in competition with BSA and durability after 1-minute of brushing (p < 0.05). Statistical significance was determined for all conditions of TiBP-GL13K bifunctional peptide (p < 0.05). Statistical analysis was conducted using a one-way ANOVA.
The bifunctional peptide film could be applied to a new dental implant prior to implantation and subsequently during recall appointments for treatment to previously placed implants. Recently we demonstrated in vitro, a bifunctional peptide that retained ability to bind to the implant surface after overnight incubation in vitro after bacterial fouling and cleaning using a commercially available electric toothbrush 68. The re-binding of the bifunctional peptide to a fouled and cleaned surface represents the feasibility of applying this technology to existing implants at recall appointments where the bifunctional peptide can be reapplied. However, when rebinding the bifunctional peptide in the oral environment, the peptide will compete for the implant surface with serum and saliva proteins, even after the implant is cleaned using standard dental practices. Thus, the ability of the peptide film to functionalize the implant surface in the presence of serum proteins was determined by pre-mixing the peptide with varying concentrations of bovine serum albumin (BSA) followed by incubating the mixture on the titanium disc for 2 minutes at 37°C. The results of the competitive binding of the bifunctional peptide in the presence of 0.01% BSA are depicted in Figure 5. TiBP-AMPA achieved 80% surface coverage while TiBP-GL13K achieved 73% surface coverage. There was no statistical difference between the coverage achieved by the two bifunctional peptides coverage in competition with BSA.
The durability of the bifunctional peptide films was evaluated by brushing the functionalized implant discs with a commercially available electric toothbrush with a round head slightly larger than the implant disc for one minute. The presence of bifunctional peptides was determined by fluorescently imaging the discs with FITC-labeled peptides before and after brushing. The durability of the TiBP-AMPA peptide film was significantly (p < 0.05) greater than that of the TiBP-GL13K peptide film. For TiBP-AMPA, 75% of the bifunctional peptide coating was retained compared to 27% for TiBP-GL13K (Figure 5).
Bifunctional Activities of the Designed Peptides
The binding and antimicrobial domain activities were evaluated by challenging the bifunctional peptide film applied to titanium implant discs with S. mutans bacteria for 24 hours. The FITC-labeled bifunctional peptides were visualized on the disc surface following 24 hours of bacterial challenge using a fluorescent microscope and the surface coverage was determined using MATLAB. The surface coverage was 84% for TiBP-AMPA and 60% for TiBP-GL13K. Representative fluorescent images of FITC-labeled bifunctional peptide on the implant disc and quantification of the percent surface coverage of three replicate experiments are contained in Figure 6.
Figure 6:
Visualization of FITC labeled bifunctional peptides using fluorescence microscopy after challenge by S. mutans for 24 hours. The percentage of peptide coverage was determined by evaluating images with a MATLAB script. The chart represents results obtained during three replicate experiments, of which, the fluorescence images are selected as representative of the whole. A statistically significant difference (p < 0.05) was found between the means for TiBP-AMPA and TiBP-GL13K coverage using a one-way ANOVA.
The percentage of α-helical secondary structure computationally predicted by the Chou-Fasman method, the secondary structure modeling and the experimental determination of secondary structure using CD supported our design prediction that TiBP-AMPA would outperform TiBP-GL13K in promotion of an antibacterial implant interface. We relied upon the previously established “rule” method 39 for the design of the bifunctional peptides in this manuscript took into the structural composition of the entire bifunctional peptide, not just the binding- or antimicrobial-domains. The rule method was trained on antimicrobial function with experimentally determined antimicrobial functions as the to identify secondary structural features in bifunctional peptides that promote formation of an effective interface for the prevention of implant associated infection.
The antimicrobial functional efficacy of TiBP-AMPA compared to TiBP-GL13K supported our design prediction. The use of propidium iodide (PI) staining to identify dead bacteria on the titanium disc surface showed 46% dead bacteria coverage for TiBP-AMPA, compared to 10% dead bacteria coverage for TiBP-GL13K. Sterilized bare discs were used as controls showed no dead bacteria. Representative fluorescent images and quantification of three replicate experiments are depicted in Figure 7.
Figure 7: Fluorescence microscopy images and quantification of propidium iodide (PI) staining of dead S. mutans bacteria on implant discs after challenge for 24 hours.
Dead bacteria appear with red fluorescence. The means and standard deviations are depicted in the chart for bare, sterilized titanium discs and discs functionalized by 2 minutes of bifunctional peptide binding at 37°C prior to bacterial challenge. Three replicate experiments were performed and a statistically significant difference (p < 0.05) was observed between means using ANOVA.
While one might postulate that this increase in antimicrobial function could due to the amount of bifunctional peptide remaining on the surface, e.g. 84% for TiBP-AMPA compared to 60% for TiBP-GL13K, the conservation of structure in the TiBP domain by the superimposed predicted secondary structures shown in Figure 3 suggests equal percentages of identity or relatedness for the binding domains. However, the analysis of backbone dynamics revealed that the antimicrobial domain of TiBP-AMPA was more highly ordered than that of TiBP-GL13K. The expected disordered region represented by the TiBP in both bifunctional peptides extends through the spacer, and much of the antimicrobial domain is disordered for TiBP-GL13K. This finding could affect the anchoring of the bifunctional peptide on the disc, as the disordered region of GL13K may be more available to interact with the implant surface nonspecifically. Additionally, the proposed mechanism of antimicrobial function is more dependent on the hydrophobicity and amphipathicity of the peptide without the additional effects attributed to the increased stability and greater number of α-helical secondary structure features in the AMPA compared to Gl13K. Secondary structure modeling revealed that the AMPA domain comprised two α-helical features joined by a turn compared to only one α-helical feature for GL13K.
The hydrophobic ratio of TiBP-AMPA was slightly greater (30%) than that of TiBP-GL13K (26%). More hydrophobic residues were aligned on the same face of TiBP-AMPA, with 10 residues being aligned for TiBP-AMPA compared to only 5 residues for TiBP-GL13K. With regard to hydrophobicity and amphipathicity, the design prediction based on the sequence and structure supported the experimental finding that TiBP-AMPA has greater antimicrobial activity than TiBP-Gl13K.
Overall, the antimicrobial peptide film property is observed to be dependent on the extent of the α-helical secondary structural features. Peptide stability under competitive binding environment was observed to be related to the ordered structures observed from our analysis of backbone dynamics. Molecular recognition based peptide self-assembly domain prevented removal of the peptide even under harsh washing conditions. Ordering seems to provide the TiBP-AMPA peptide a competitive advantage. It may be plausible to design TiBP-GL13K with a different spacer combination which would induce an ordering in the structure and enhance its competitive binding. Conformational design parameters are postulated to play a critical role in the peptide stability considering the anchoring domain of the bifunctional peptides are the same. Secondary structure may undergo major changes on the metal surfaces, however functional assembly behavior under the biologically challenged material interfaces may still have a folding preferential.
Taken together, our experimental results demonstrate that using computationally efficient, less resource intensive methods can be used to successfully predict the properties of bifunctional peptide prior to more costly and time-consuming experimental evaluation. The computational design approach was validated experimentally by assessing the binding and antimicrobial function of bifunctional peptides on a simulated implant surface. This technology and approach to design represents a novel strategy to improving and developing bifunctional peptide films to combat bacterial infection and prevent/treat peri-implant disease. Prior to clinical trials, the cytocompatibility and biocompatibility of the bifunctional peptides to human cells will need to be examined. Additionally, the incorporation of AMP sequences not previously explored as antimicrobial agents in bifunctional peptides represents an opportunity to develop more broad-spectrum antibacterial efficacy, as well as options for treatment of antibiotic resistant bacterial strains. The predictive design approach developed here offers a method for evaluating the functional efficacy of AMP, spacer, and binding domain combinations based on analyses of the relationship between sequence, structure and function. Combining this approach with the recently developed soft epitaxial fit for the binding domain of the peptide to match the epitaxial sites on the implant surfaces would be another area to explore in order to tune the functionality at the solid-interface. This approach may also provide predictable-, rational-, peptide design approaches for a wide range of hybrid interfaces by combining conformational and interface design aspects 93.
CONCLUSION
We developed a predictive computational approach for designing bifunctional peptides that sought to correlate structure and function, e.g. antimicrobial activity and demonstrated that the resulting peptides show promise as a medicinal approach to addressing bacterial dysbiogenesis94–95 that leads to peri-implant disease with a shortening of the useful life of dental implants. With over 3 million implants placed in the US alone and growing by 500,000 implants/year 7, a reduced service life ending in implant failure will adversely impact public health. The rapid (two minute) delivery of antimicrobial bifunctional peptide films was tested on dental implant surfaces mimicking the application sequence necessary for re-treatment of peri-implantitis in a dental office 63. The designed functional peptides consisted of three domains, from the N’-terminus: titanium binding, spacer and antimicrobial.
While the antimicrobial peptides were varied, the binding and spacer domains were kept constant with the goal of demonstrating that a computational approach can predict the antimicrobial properties of the resulting bifunctional peptide films. The percentage of α-helicity of the bifunctional peptides and their individual constituting domains were computationally predicted by the Chou-Fasman algorithm. The peptides were further analyzed using chimera secondary structure models and these secondary structure predictions were compared experimentally using CD spectroscopy. This predictive design approach considered the structure and function of the entire bifunctional peptide molecule. Three design methods were used here, namely sequence-based, de novo modeling, and experimental evaluation; all indicated that peptide incorporating the AMPA anti-microbial domain had greater helical content than the one containing the GL13K antimicrobial domain. This prediction was confirmed by the backbone dynamic data. This ensemble of structural analysis techniques formed the basis for the prediction that TiBP-AMPA would functionally outperform TiBP-GL13K as an antimicrobial peptide film. In particular, amphipathie and α-helicity were shown to be more prominent in ordered regions, which contribute to greater antimicrobial peptide film activity. The bifunctional peptides were evaluated experimentally for their potential to prevent and treat peri-implant disease. The bifunctional peptides were delivered in the clinically relevant manner (2 minute binding period), under competition with serum proteins. Their mechanical durability was tested, and they were empirically challenged with bacteria to confirm our computational predictions. The peptide films have been shown capable of rebinding ability through up to five cycles of bacterial fouling, cleaning and reapplication. These results demonstrate the success of our computational design approach and suggest that the TiBP-AMPA peptide has strong potential as a treatment for peri-implant disease due to its ability mitigate bacterial biofilm formation.
Supplementary Material
Figure S1: Bifunctional peptide hydrophobicity analysis using the Antimicrobial Peptide Database Calculator and Predictor tool.
Table S1. The minimum inhibitory concentration of the AMP domain in isolation. Peptides requiring amidation at the C’-terminus were synthesized using Rink-amide resin.
Table S2. Fluorescence image quantification for percentage surface coverage of the images using ImageJ software.
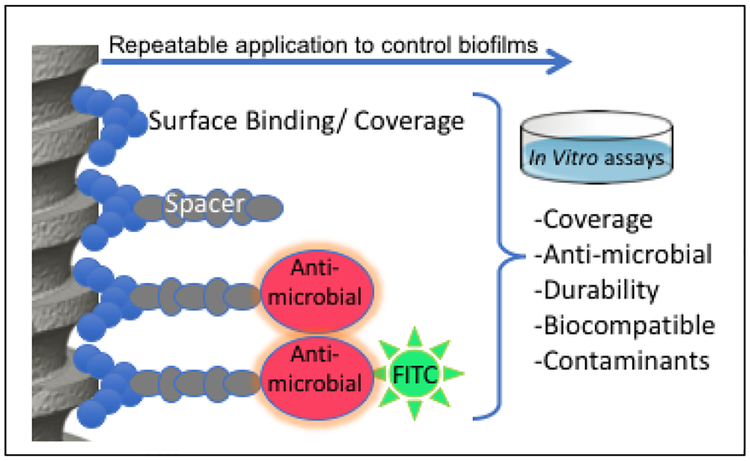
Scheme 1.
Our approach includes an antimicrobial peptide film based upon an engineered bifunctional peptide composed of peptide domains for implant binding and antimicrobial activity separated by a spacer. The peptide was tested using a variety of in vitro assays to demonstrate its suitability.
REFERENCES
- 1.Aparicio C; Gil FJ; Fonseca C; Barbosa M; Planell JA, Corrosion behaviour of commercially pure titanium shot blasted with different materials and sizes of shot particles for dental implant applications. Biomaterials 2003, 24 (2), 263–73. [DOI] [PubMed] [Google Scholar]
- 2.Ronold HJ; Ellingsen JE; Lyngstadaas SP, Tensile force testing of optimized coin-shaped titanium implant attachment kinetics in the rabbit tibiae. J Mater Sci Mater Med 2003, 14 (10), 843–9. [DOI] [PubMed] [Google Scholar]
- 3.Ronold HJ; Lyngstadaas SP; Ellingsen JE, A study on the effect of dual blasting with TiO2 on titanium implant surfaces on functional attachment in bone. J Biomed Mater Res A 2003, 67 (2), 524–30. DOI: 10.1002/jbm.a.10580. [DOI] [PubMed] [Google Scholar]
- 4.Ronold HJ; Lyngstadaas SP; Ellingsen JE, Analysing the optimal value for titanium implant roughness in bone attachment using a tensile test. Biomaterials 2003, 24 (25), 4559–64. [DOI] [PubMed] [Google Scholar]
- 5.Norowski PA Jr.; Bumgardner JD, Biomaterial and antibiotic strategies for peri-implantitis: a review. J Biomed Mater Res B Appl Biomater 2009, 88 (2), 530–43. DOI: 10.1002/jbm.b.31152. [DOI] [PubMed] [Google Scholar]
- 6.Aparicio C; Padros A; Gil FJ, In vivo evaluation of micro-rough and bioactive titanium dental implants using histometry and pull-out tests. Journal of the mechanical behavior of biomedical materials 2011, 4 (8), 1672–82. DOI: 10.1016/j.jmbbm.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Achermann G, How will dentistry look in 2020? In Vision 2020: Simply doing more for dental professionals, The Straumann Group: Amsterdam, 2012; p http://www.straumann.com. [Google Scholar]
- 8.Monjo M; Ramis JM; Ronold HJ; Taxt-Lamolle SF; Ellingsen JE; Lyngstadaas SP, Correlation between molecular signals and bone bonding to titanium implants. Clinical oral implants research 2013, 24 (9), 1035–43. DOI: 10.1111/j.1600-0501.2012.02496.x. [DOI] [PubMed] [Google Scholar]
- 9.Orsini G; Pagella P; Mitsiadis TA, Modern Trends in Dental Medicine: An Update for Internists. Am J Med 2018, 131 (12), 1425–1430. DOI: 10.1016/j.amjmed.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 10.Gungormus M; Oren EE; Horst JA; Fong H; Hnilova M; Somerman MJ; Snead ML; Samudrala R; Tamerler C; Sarikaya M, Cementomimetics-constructing a cementum-like biomineralized microlayer via amelogenin-derived peptides. International journal of oral science 2012, 4 (2), 69–77. DOI: 10.1038/ijos.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y; Snead ML; Tamerler C, Bio-inspired hard-to-soft interface for implant integration to bone. Nanomedicine 2015, 11 (2), 431–4. DOI: 10.1016/j.nano.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Piriz R; Cabal B; Goyos-Ball L; Fernandez A; Bartolome JF; Moya JS; Torrecillas R, Current state-of-the-art and future perspectives of the three main modern implant-dentistry concerns: Aesthetic requirements, mechanical properties, and peri-implantitis prevention. J Biomed Mater Res A 2019, 107 (7), 1466–1475. DOI: 10.1002/jbm.a.36661. [DOI] [PubMed] [Google Scholar]
- 13.Olivares-Navarrete R; Hyzy SL; Park JH; Dunn GR; Haithcock DA; Wasilewski CE; Boyan BD; Schwartz Z, Mediation of osteogenic differentiation of human mesenchymal stem cells on titanium surfaces by a Wnt-integrin feedback loop. Biomaterials 2011, 32 (27), 6399–411. DOI: 10.1016/j.biomaterials.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarnow DP, Increasing Prevalence of Peri-implantitis: How Will We Manage? J Dent Res 2016, 95 (1), 7–8. DOI: 10.1177/0022034515616557. [DOI] [PubMed] [Google Scholar]
- 15.Valente NA; Andreana S, Peri-implant disease: what we know and what we need to know. J Periodontal Implant Sci 2016, 46 (3), 136–51. DOI: 10.5051/jpis.2016.46.3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salvi GE; Cosgarea R; Sculean A, Prevalence and Mechanisms of Peri-implant Diseases. J Dent Res 2017, 96 (1), 31–37. DOI: 10.1177/0022034516667484. [DOI] [PubMed] [Google Scholar]
- 17.Berglundh T; Armitage G; Araujo MG; Avila-Ortiz G; Blanco J; Camargo PM; Chen S; Cochran D; Derks J; Figuero E; Hammerle CHF; Heitz-Mayfield LJA; Huynh-Ba G; Iacono V; Koo KT; Lambert F; McCauley L; Quirynen M; Renvert S; Salvi GE; Schwarz F; Tarnow D; Tomasi C; Wang HL; Zitzmann N, Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Periodontol 2018, 89 Suppl 1, S313–S318. DOI: 10.1002/JPER.17-0739. [DOI] [PubMed] [Google Scholar]
- 18.Caton JG; Armitage G; Berglundh T; Chapple ILC; Jepsen S; Kornman KS; Mealey BL; Papapanou PN; Sanz M; Tonetti MS, A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. Journal of Periodontology 2018, 89, S1–S8. DOI: 10.1002/Jper.18-0157. [DOI] [PubMed] [Google Scholar]
- 19.Listgarten MA; Lai CH, Comparative microbiological characteristics of failing implants and periodontally diseased teeth. Journal of Periodontology 1999, 70 (4), 431–437. DOI: DOI 10.1902/jop.1999.70.4.431. [DOI] [PubMed] [Google Scholar]
- 20.Hultin M; Gustafsson A; Hallstrom H; Johansson LA; Ekfeldt A; Klinge B, Microbiological findings and host response in patients with peri-implantitis. Clinical oral implants research 2002, 13 (4), 349–358. DOI: DOI 10.1034/j.1600-0501.2002.130402.x. [DOI] [PubMed] [Google Scholar]
- 21.Quirynen M; Vogels R; Peeters W; Steenberghe D; Naert I; Haffajee A, Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clinical oral implants research 2006, 17 (1), 25–37. DOI: 10.1111/j.1600-0501.2005.01194.x. [DOI] [PubMed] [Google Scholar]
- 22.Dutzan N; Kajikawa T; Abusleme L; Greenwell-Wild T; Zuazo CE; Ikeuchi T; Brenchley L; Abe T; Hurabielle C; Martin D; Morell RJ; Freeman AF; Lazarevic V; Trinchieri G; Diaz PI; Holland SM; Belkaid Y; Hajishengallis G; Moutsopoulos NM, A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Science translational medicine 2018, 10 (463). DOI: 10.1126/scitranslmed.aat0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derks J; Schaller D; Hakansson J; Wennstrom JL; Tomasi C; Berglundh T, Peri-implantitis - onset and pattern of progression. J Clin Periodontol 2016, 43 (4), 383–8. DOI: 10.1111/jcpe.12535. [DOI] [PubMed] [Google Scholar]
- 24.Lang NP; Wilson TG; Corbet EF, Biological complications with dental implants: their prevention, diagnosis and treatment. Clinical oral implants research 2000, 11 Suppl 1, 146–55. [DOI] [PubMed] [Google Scholar]
- 25.Carral C; Munoz F; Permuy M; Linares A; Dard M; Blanco J, Mechanical and chemical implant decontamination in surgical peri-implantitis treatment: preclinical “in vivo” study. J Clin Periodontol 2016, 43(8), 694–701. DOI: 10.1111/jcpe.12566. [DOI] [PubMed] [Google Scholar]
- 26.Wimley WC; Hristova K, Antimicrobial peptides: successes, challenges and unanswered questions. J Membr Biol 2011, 239 (1–2), 27–34. DOI: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hvistendahl M, Public health. China takes aim at rampant antibiotic resistance. Science 2012, 336 (6083), 795 DOI: 10.1126/science.336.6083.795. [DOI] [PubMed] [Google Scholar]
- 28.Pruden A, Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ Sci Technol 2014, 48 (1), 5–14. DOI: 10.1021/es403883p. [DOI] [PubMed] [Google Scholar]
- 29.Torres MDT; Sothiselvam S; Lu TK; de la Fuente-Nunez C, Peptide Design Principles for Antimicrobial Applications. Journal of molecular biology 2019. DOI: 10.1016/j.jmb.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Mao J; Kuranaga T; Hamamoto H; Sekimizu K; Inoue M, Rational design, synthesis, and biological evaluation of lactam-bridged gramicidin A analogues: discovery of a low-hemolytic antibacterial peptide. ChemMedChem 2015, 10 (3), 540–5. DOI: 10.1002/cmdc.201402473. [DOI] [PubMed] [Google Scholar]
- 31.Costerton JW; Stewart PS; Greenberg EP, Bacterial biofilms: a common cause of persistent infections. Science 1999, 284 (5418), 1318–22. [DOI] [PubMed] [Google Scholar]
- 32.Salwiczek M; Qu Y; Gardiner J; Strugnell RA; Lithgow T; McLean KM; Thissen H, Emerging rules for effective antimicrobial coatings. Trends in Biotechnology 2014, 32 (2), 82–90. DOI: 10.1016/j.tibtech.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Raphel J; Holodniy M; Goodman SB; Heilshorn SC, Multifunctional coatings to simultaneously promote osseointegration and prevent infection of orthopaedic implants. Biomaterials 2016, 84, 301–314. DOI: 10.1016/j.biomaterials.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tobin EJ, Recent coating developments for combination devices in orthopedic and dental applications: A literature review. Adv Drug Deliver Rev 2017, 112, 88–100. DOI: 10.1016/j.addr.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Zilberman M; Elsner JJ, Antibiotic-eluting medical devices for various applications. Journal of controlled release : official journal of the Controlled Release Society 2008, 130 (3), 202–15. DOI: 10.1016/j.jconrel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Pritchard EM; Valentin T; Panilaitis B; Omenetto F; Kaplan DL, Antibiotic-Releasing Silk Biomaterials for Infection Prevention and Treatment. Adv Funct Mater 2013, 23 (7), 854–861. DOI: 10.1002/adfm.201201636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fjell CD; Hiss JA; Hancock REW; Schneider G, Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 2012, 11 (1), 37–51. DOI: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 38.Wang G; Li X; Wang Z, APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 2016, 44 (D1), D1087–93. DOI: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boone K; Camarda K; Spencer P; Tamerler C, Antimicrobial peptide similarity and classification through rough set theory using physicochemical boundaries. BMC Bioinformatics 2018, 19, 469 DOI: 10.1186/s12859-018-2514-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayramov DF; Neff JA, Beyond conventional antibiotics - New directions for combination products to combat biofilm. Adv Drug Deliv Rev 2017, 112, 48–60. DOI: 10.1016/j.addr.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji S; Li W; Baloch AR; Wang M; Li H; Cao B; Zhang H, Efficient biosynthesis of a Cecropin A-melittin mutant in Bacillus subtilis WB700. Sci Rep 2017, 7, 40587 DOI: 10.1038/srep40587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balhara V; Schmidt R; Gorr SU; Dewolf C, Membrane selectivity and biophysical studies of the antimicrobial peptide GL13K. Biochim Biophys Acta 2013, 1828 (9), 2193–203. DOI: 10.1016/j.bbamem.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 43.Eckert R, Road to clinical efficacy: challenges and novel strategies for antimicrobial peptide development. Future Microbiol 2011, 6 (6), 635–51. DOI: 10.2217/fmb.11.27. [DOI] [PubMed] [Google Scholar]
- 44.Holmberg KV; Abdolhosseini M; Li Y; Chen X; Gorr SU; Aparicio C, Bio-inspired stable antimicrobial peptide coatings for dental applications. Acta Biomater 2013, 9 (9), 8224–31. DOI: 10.1016/j.actbio.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godoy-Gallardo M; Mas-Moruno C; Fernandez-Calderon MC; Perez-Giraldo C; Manero JM; Albericio F; Gil FJ; Rodriguez D, Covalent immobilization of hLf1–11 peptide on a titanium surface reduces bacterial adhesion and biofilm formation. Acta Biomater 2014, 10 (8), 3522–34. DOI: 10.1016/j.actbio.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Chen R; Willcox MD; Ho KK; Smyth D; Kumar N, Antimicrobial peptide melimine coating for titanium and its in vivo antibacterial activity in rodent subcutaneous infection models. Biomaterials 2016, 85, 142–51. DOI: 10.1016/j.biomaterials.2016.01.063. [DOI] [PubMed] [Google Scholar]
- 47.Chouirfa H; Bouloussa H; Migonney V; Falentin-Daudre C, Review of titanium surface modification techniques and coatings for antibacterial applications. Acta Biomater 2019, 83, 37–54. DOI: 10.1016/j.actbio.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 48.Moussa DG; Fok A; Aparicio C, Hydrophobic and antimicrobial dentin: A peptide-based 2-tier protective system for dental resin composite restorations. Acta Biomater 2019, 88, 251–265. DOI: 10.1016/j.actbio.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moussa DG; Kirihara JA; Ye Z; Fischer NG; Khot J; Witthuhn BA; Aparicio C, Dentin Priming with Amphipathic Antimicrobial Peptides. J Dent Res 2019, 10.1177/0022034519863772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarikaya M; Tamerler C; Jen AK; Schulten K; Baneyx F, Molecular biomimetics: nanotechnology through biology. Nat Mater 2003, 2 (9), 577–85. DOI: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 51.Peelle BR; Krauland EM; Wittrup KD; Belcher AM, Design criteria for engineering inorganic material-specific peptides. Langmuir 2005, 21 (15), 6929–33. DOI: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- 52.Heinz H; Farmer BL; Pandey RB; Slocik JM; Patnaik SS; Pachter R; Naik RR, Nature of molecular interactions of peptides with gold, palladium, and Pd-Au bimetal surfaces in aqueous solution. J Am Chem Soc 2009, 131 (28), 9704–14. DOI: 10.1021/ja900531f. [DOI] [PubMed] [Google Scholar]
- 53.Puddu V; Slocik JM; Naik RR; Perry CC, Titania binding peptides as templates in the biomimetic synthesis of stable titania nanosols: insight into the role of buffers in peptide-mediated mineralization. Langmuir 2013, 29 (30), 9464–72. DOI: 10.1021/la401777x. [DOI] [PubMed] [Google Scholar]
- 54.Walsh TR; Knecht MR, Biointerface Structural Effects on the Properties and Applications of Bioinspired Peptide-Based Nanomaterials. Chemical reviews 2017, 117 (20), 12641–12704. DOI: 10.1021/acs.chemrev.7b00139. [DOI] [PubMed] [Google Scholar]
- 55.Heinz H; Ramezani-Dakhel H, Simulations of inorganic-bioorganie interfaces to discover new materials: insights, comparisons to experiment, challenges, and opportunities. Chemical Society reviews 2016, 45 (2), 412–448. DOI: 10.1039/c5cs00890e. [DOI] [PubMed] [Google Scholar]
- 56.Feng J; Slocik JM; Sarikaya M; Naik RR; Farmer BL; Heinz H, Influence of the Shape of Nanostructured Metal Surfaces on Adsorption of Single Peptide Molecules in Aqueous Solution. Small 2012, 8 (7), 1049–1059. DOI: 10.1002/smll.201102066. [DOI] [PubMed] [Google Scholar]
- 57.Tamerler C; Sarikaya M, Genetically designed Peptide-based molecular materials. ACS nano 2009, 3 (7), 1606–15. DOI: 10.1021/nn900720g. [DOI] [PubMed] [Google Scholar]
- 58.Kacar T; Zin MT; So C; Wilson B; Ma H; Gul-Karaguler N; Jen AK; Sarikaya M; Tamerler C, Directed self-immobilization of alkaline phosphatase on micro-patterned substrates via genetically fused metal-binding peptide. Biotechnol Bioeng 2009, 103 (4), 696–705. DOI: 10.1002/bit.22282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamerler C; Khatayevich D; Gungormus M; Kacar T; Oren EE; Hnilova M; Sarikaya M, Molecular biomimetics: GEPI-based biological routes to technology. Biopolymers 2010, 94 (1), 78–94. DOI: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- 60.Gungormus M; Branco M; Fong H; Schneider JP; Tamerler C; Sarikaya M, Self assembled bi-functional peptide hydrogels with biomineralization-directing peptides. Biomaterials 2010, 31 (28), 7266–74. DOI: 10.1016/j.biomaterials.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.VanOosten SK; Yuca E; Karaca BT; Boone K; Snead ML; Spencer P; Tamerler C, Biosilver nanoparticle interface offers improved cell viability. Surf Innov 2016, 4 (3), 121–132. DOI: 10.1680/jsuin.16.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yazici H; Fong H; Wilson B; Oren EE; Amos FA; Zhang H; Evans JS; Snead ML; Sarikaya M; Tamerler C, Biological response on a titanium implant-grade surface functionalized with modular peptides. Acta Biomater 2013, 9 (2), 5341–5352. DOI: 10.1016/j.actbio.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wisdom C; Chen C; Yuca E; Zhou Y; Tamerler C; Snead ML, Repeatedly Applied Peptide Film Kills Bacteria on Dental Implants. JOM (1989) 2019, 71 (4), 1271–1280. DOI: 10.1007/s11837-019-03334-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warotayanont R; Zhu D; Snead ML; Zhou Y, Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun 2008, 367 (1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warotayanont R; Frenkel B; Snead ML; Zhou Y, Leucine-rich amelogenin peptide induces osteogenesis by activation of the Wnt pathway. Biochem Biophys Res Commun 2009, (25), 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yazici H; O’Neill MB; Kacar T; Wilson BR; Oren EE; Sarikaya M; Tamerler C, Engineered Chimeric Peptides as Antimicrobial Surface Coating Agents toward Infection-Free Implants. ACS Appl Mater Interfaces 2016, 8 (8), 5070–81. DOI: 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yucesoy DT; Hnilova M; Boone K; Arnold PM; Snead ML; Tamerler C, Chimeric peptides as implant functionalization agents for titanium alloy implants with antimicrobial properties. JOM 2015, 67 (4), 754–766. DOI: 10.1007/s11837-015-1350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wisdom C; VanOosten SK; Boone KW; Khvostenko D; Arnold PM; Snead ML; Tamerler C, Controlling the Biomimetic Implant Interface: Modulating Antimicrobial Activity by Spacer Design. J Mol Eng Mater 2016, 4 (1). DOI: 10.1142/S2251237316400050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilkins MR; Gasteiger E; Bairoch A; Sanchez JC; Williams KL; Appel RD; Hochstrasser DF, Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 1999, 112, 531–52. DOI: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 70.Micsonai A; Wien F; Bulyaki E; Kun J; Moussong E; Lee YH; Goto Y; Refregiers M; Kardos J, BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res 2018, 46 (W1), W315–W322. DOI: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiedemann C; Bellstedt P; Gorlach M, CAPITO-a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics 2013, 29 (14), 1750–1757. DOI: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
- 72.Thevenet P; Shen YM; Maupetit J; Guyon F; Derreumaux P; Tuffery P, PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Research 2012, 40 (W1), W288–W293. DOI: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen YM; Maupetit J; Derreumaux P; Tuffery P, Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J Chem Theory Comput 2014, 10 (10), 4745–4758. DOI: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 74.Maupetit J; Derreumaux P; Tuffery P, PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Research 2009, 37, W498–W503. DOI: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cilia E; Pancsa R; Tompa P; Lenaerts T; Vranken WF, The DynaMine Webserver: predicting protein dynamics from sequence. Nucleic Acids Res 2014, 42 (Web Server issue), W264–70. DOI: 10.1093/nar/gku270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE, UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 2004, 25 (13), 1605–1612. DOI: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Bulheller BM; Hirst JD, DichroCalc--circular and linear dichroism online. Bioinformatics 2009, 25 (4), 539–40. DOI: 10.1093/bioinformatics/btp016. [DOI] [PubMed] [Google Scholar]
- 78.Bizhang M; Riemer K; Arnold WH; Domin J; Zimmer S, Influence of Bristle Stiffness of Manual Toothbrushes on Eroded and Sound Human Dentin--An In Vitro Study. PLoS One 2016, 11 (4), e0153250 DOI: 10.1371/journal.pone.0153250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mazumdar P; Chowdhury D; Chatterjee S; Jajoo N, Effect of four different dentifrices applied by customized automated brushing device on enamel surface abrasion: An in vitro profilometric study. J Conserv Dent 2019, 22 (2), 191–95. DOI: 10.4103/JCD.JCD_392_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji S; Li W; Zhang L; Zhang Y; Cao B, Cecropin A-melittin mutant with improved proteolytic stability and enhanced antimicrobial activity against bacteria and fungi associated with gastroenteritis in vitro. Biochem Biophys Res Commun 2014, 451 (4), 650–5. DOI: 10.1016/j.bbrc.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 81.Chen X; Hirt H; Li Y; Gorr SU; Aparicio C, Antimicrobial GL13K peptide coatings killed and ruptured the wall of Streptococcus gordonii and prevented formation and growth of biofilms. PLoS One 2014, 9 (11), e111579 DOI: 10.1371/journal.pone.0111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu X; Zhang L; Wang J; Ma Z; Xu W; Li J; Shan A, Characterization of antimicrobial activity and mechanisms of low amphipathic peptides with different alpha-helical propensity. Acta Biomater 2015, 18, 155–67. DOI: 10.1016/j.actbio.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 83.Wang J; Dou X; Song J; Lyu Y; Zhu X; Xu L; Li W; Shan A, Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med Res Rev 2019, 39 (3), 831–859. DOI: 10.1002/med.21542. [DOI] [PubMed] [Google Scholar]
- 84.Schwarz F; Derks J; Monje A; Wang HL, Peri-implantitis. J Periodontol 2018, 89 Suppl 1, S267–S290. DOI: 10.1002/JPER.16-0350. [DOI] [PubMed] [Google Scholar]
- 85.Chapple ILC; Mealey BL; Van Dyke TE; Bartold PM; Dommisch H; Eickholz P; Geisinger ML; Genco RJ; Glogauer M; Goldstein M; Griffin TJ; Holmstrup P; Johnson GK; Kapila Y; Lang NP; Meyle J; Murakami S; Plemons J; Romito GA; Shapira L; Tatakis DN; Teughels W; Trombelli L; Walter C; Wimmer G; Xenoudi P; Yoshie H, Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Periodontol 2018, 89 Suppl 1, S74–S84. DOI: 10.1002/JPER.17-0719. [DOI] [PubMed] [Google Scholar]
- 86.Delak K; Harcup C; Lakshminarayanan R; Sun Z; Fan Y; Moradian-Oldak J; Evans JS, The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry 2009, 48 (10), 2272–81. DOI: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lacruz RS; Lakshminarayanan R; Bromley KM; Hacia JG; Bromage TG; Snead ML; Moradian-Oldak J; Paine ML, Structural analysis of a repetitive protein sequence motif in strepsirrhine primate amelogenin. PLoS One 2011, 6 (3), e18028 DOI: 10.1371/journal.pone.0018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wald T; Osickova A; Sulc M; Benada O; Semeradtova A; Rezabkova L; Veverka V; Bednarova L; Maly J; Macek P; Sebo P; Slaby I; Vondrasek J; Osicka R, Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J Biol Chem 2013, 288 (31), 22333–45. DOI: 10.1074/jbc.M113.456012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lamiable A; Thevenet P; Rey J; Vavrusa M; Derreumaux P; Tuffery P, PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016, 44 (W1), W449–54. DOI: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wimley WC, Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 2010, 5 (10), 905–17. DOI: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25 (13), 1605–12. DOI: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 92.Kabsch W; Sander C, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22 (12), 2577–637. DOI: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 93.Zhu EB; Wang SY; Yan XC; Sobani M; Ruan LY; Wang C; Liu Y; Duan XF; Heinz H; Huang Y, Long-Range Hierarchical Nanocrystal Assembly Driven by Molecular Structural Transformation. J. Am. Chem. Soc 2019, 141 (4), 1498–1505. DOI: 10.1021/jacs.8b08023. [DOI] [PubMed] [Google Scholar]
- 94.Socransky SS; Haffajee AD, The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol 1992, 63 (4 Suppl), 322–31. [DOI] [PubMed] [Google Scholar]
- 95.Haffajee AD; Socransky SS, Microbial etiological agents of destructive periodontal diseases. Periodontol 2000 1994, 5, 78–111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Bifunctional peptide hydrophobicity analysis using the Antimicrobial Peptide Database Calculator and Predictor tool.
Table S1. The minimum inhibitory concentration of the AMP domain in isolation. Peptides requiring amidation at the C’-terminus were synthesized using Rink-amide resin.
Table S2. Fluorescence image quantification for percentage surface coverage of the images using ImageJ software.