Abstract
We report a non-cytotoxic resin compatible with and designed for use in custom high-resolution 3D printers that follow the design approach described in Gong et al., Lab Chip 17, 2899 (2017). The non-cytotoxic resin is based on a poly(ethylene glycol) diacrylate (PEGDA) monomer with avobenzone as the UV absorber instead of 2-nitrophenyl phenyl sulfide (NPS). Both NPS-PEGDA and avobenzone-PEGDA (A-PEGDA) resins were evaluated for cytotoxicity and cell adhesion. We show that NPS-PEGDA can be made effectively non-cytotoxic with a post-print 12-hour ethanol wash, and that A-PEGDA, as-printed, is effectively non-cytotoxic. 3D prints made with either resin do not support strong cell adhesion in their as-printed state; however, cell adhesion increases dramatically with a short plasma treatment. Using A-PEGDA, we demonstrate spheroid formation in ultra-low adhesion 3D printed wells, and cell migration from spheroids on plasma-treated adherent surfaces. Given that A-PEGDA can be 3D printed with high resolution, it has significant promise for a wide variety of cell-based applications using 3D printed microfluidic structures.
Keywords: 3D printing, microfluidics, cytotoxicity, biocompatibility, resin, spheroid
Graphical Abstract
Introduction
3D printing is an attractive fabrication method for microfluidic devices.1–10 Key advantages include full 3D layout of device components, avoidance of lengthy and tedious cleanroom processes, and rapid fabrication capability, which in turn enables a “fail fast and often” approach to device development.11 Over the last few years the Nordin group has focused on solving one of the major roadblocks to successful 3D printing of microfluidic devices, namely, reducing the achievable feature size so that fabricated devices operate in the truly microfluidic (as opposed to millifluidic) regime.12,13 This has required the development of custom 3D printers and materials,14 and has resulted in channels as small as 18 μm × 20 μm,14 high density (88 per mm2) chip-to-chip interconnects with integrated microgaskets,15 and highly integrated valves and pumps in an extremely small volume (2.6 mm3) that takes full advantage of the 3D layout possibilities enabled by 3D printing.16
However, we have not yet addressed the issue of biocompatible materials17 that are also suitable for use with and take advantage of our custom 3D printers. The purpose of this paper is to evaluate two candidate resins in terms of cytotoxicity and cell adhesion, followed by a demonstration using 3D cell culture (spheroids). We show that the main poly(ethylene glycol) diacrylate (PEGDA) resin that we reported in 2017,14 and have used in a number of our subsequent papers,13,15,16,18,19 can be made effectively non-cytotoxic by leaching out the cytotoxic component of the printed material in a post-3D printing process based on a 12-hour ethanol wash. More importantly, in this paper we demonstrate that a newly formulated resin (A-PEGDA) is essentially non-cytotoxic in its as-printed state, i.e., it facilitates cell culture with no required post-print processing. We also show that the surfaces of as-printed PEGDA polymer have very low cell adhesion, but that cell adhesion dramatically improves with a simple, short plasma treatment. We then show that the material properties of 3D printed surfaces for the A-PEGDA resin can facilitate spheroid nucleation and growth (non-adherent case) and cell migration from spheroids (adherent case). These data suggest that 3D printed devices based on A-PEGDA may be suitable for 3D tissue culture, which opens the possibility of creating complete organ-on-a-chip systems using 3D printing technology. Further applications include stem cell analysis, cell-drug interactions, and 3D cellular scaffolds as demonstrated by numerous researchers.20–25
Materials and Methods
3D Printer
The 3D printer used in this paper is a second-generation version of the custom 3D printer described in References 14–16. It has a 385 nm LED light source and a pixel pitch of 7.6 μm in the projected image plane. The custom python software that operates the 3D printer gives us the ability to control all aspects of 3D printing such that a single print job can comprise an arbitrary combination of layer thicknesses, exposure times, and multiple images for each layer, which enables complete dose control throughout the volume of a 3D printed device.
3D Printer Resin Formulations
We create custom photopolymerizable resins that consist of poly(ethylene glycol) diacrylate (PEGDA, MW258) as the monomer, 1% (w/w) phenylbis(2,4,6-trimethylbenzoyl) phosphine oxide (Irgacure 819) as the photoinitiator, and a UV absorber to control light penetration during 3D printing. Two resin formulations are considered in this paper, each with a different UV absorber. The first uses 2% (w/w) 2-nitrophenyl phenyl sulfide (NPS),14–16 and the second uses 0.38% (w/w) avobenzone. We refer to these resin formulations as NPS-PEGDA and A-PEGDA, respectively. The UV absorber concentrations are such that NPS-PEGDA and A-PEGDA have comparable optical penetration depths. This was necessary because the two UV absorbers have different molar absorptivities over the emission spectrum of the 3D printer’s 385 nm LED light source as shown in Ref. 14. The PEGDA and Irgacure 819 were purchased from Sigma-Aldrich, the NPS from TCI America, and the avobenzone from MakingCosmetics Inc. All materials were used as-received.
Sample Preparation for Cytotoxicity Testing
We used 1 mm thick, 25 mm square glass slides as 3D printing substrates. Before use, slides were rinsed with acetone and isopropyl alcohol (IPA). Slides intended for NPS-PEGDA were silanized by immersion in toluene mixed with 10% 3-(trimethoxysilyl)propyl methacrylate for 2 hours to facilitate better attachment of the 3D printed device. After silanization, glass slides were stored in fresh toluene inside a closed container until use, which ranged from under an hour to several weeks.
All 3D prints reported in this paper were fabricated with a layer thickness of 10 μm. Except where otherwise noted, each layer had an exposure time of 550 ms for both resin formulations. The image plane irradiance was 21.2 mW/cm2 with an LED source spectrum as reported in Ref. 14.
As shown in Fig. 1, substrates for cellular cytotoxicity and adherence testing were designed as 14 mm diameter, 150 μm thick samples. The circular design is cut along a chord on one side because the maximum print area is 19.35 × 12.16 mm2. After printing, samples were rinsed with IPA before optical curing, which consisted of a 30-minute exposure in a custom curing station using a 430 nm LED (Thorlabs, Newton, New Jersey) having a measured irradiance of 11.3 mW/cm2 in the curing plane. After curing, samples were soaked in DI water, which enabled them to be more easily separated from their glass substrates using a razor blade. Samples were then allowed to air dry before use.
Figure 1:
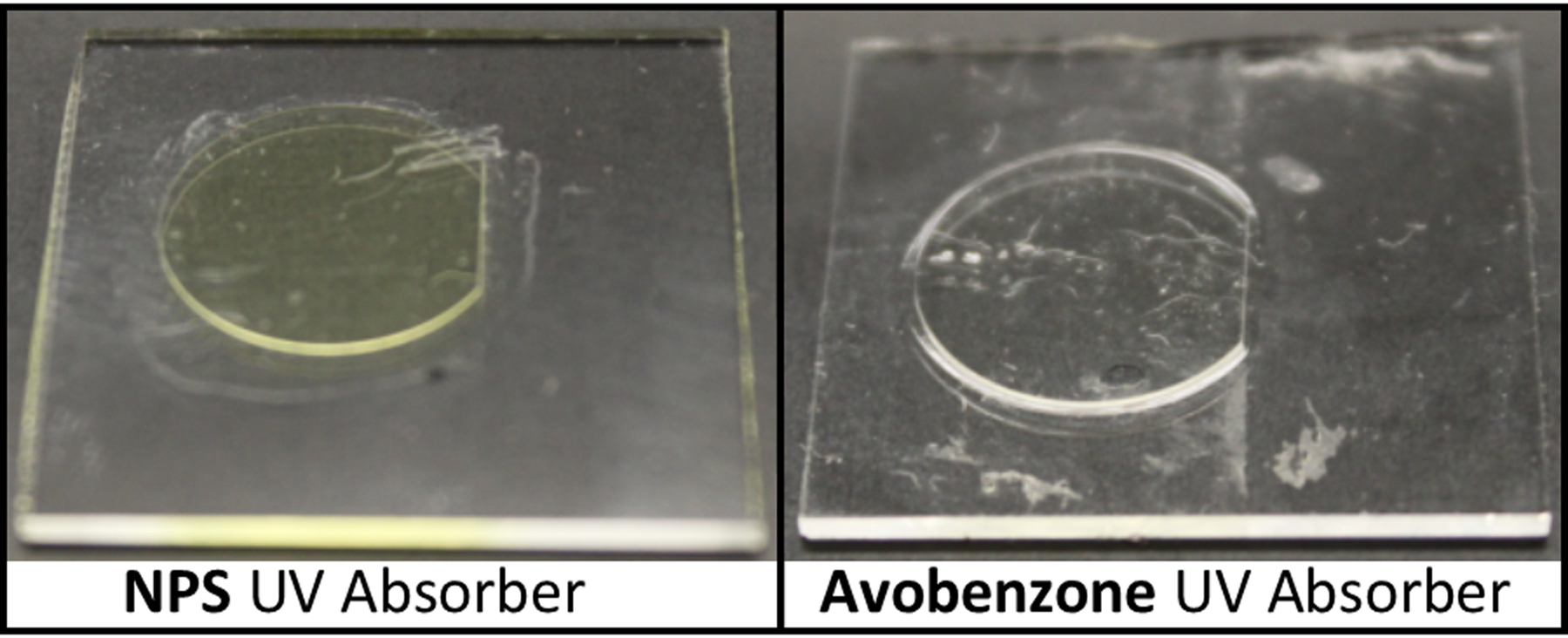
3D printed NPS-PEGDA (left) and A-PEGDA (right) substrates for cellular cytotoxicity and adherence testing. Samples are still attached to their glass substrates from 3D printing.
Cytotoxicity Testing
The PEGDA material was tested using a direct contact technique as outlined in ISO 10993–5, which consisted of growing a cell layer on tissue-culture polystyrene (TCPS) and placing the material being tested on top of the cell layer. After 24 hours, the cells were stained with a Calcein AM/Ethidium homodimer live/dead assay (Biotium Inc.) and imaged to detect changes indicative of cytotoxicity. Stained cells were imaged using a FLoid Cell Imaging Station (Life Technologies) and images were analyzed and viability assessed using CellProfiler (CellProfiler.org). According to ISO 10993–5, a cell response is considered cytotoxic if there is a 30% decrease in surface area coverage versus a control. Replicates were performed with two cell culture wells for each wash time; three images were taken and analyzed per well.
Cytotoxicity tests were performed with an endothelial cell line, EA.hy926 (ATCC CRL-2922), which were grown and passaged according to protocols established by the manufacturer. These cells were chosen over a primary or stem cell line because of their ease of use and low cost of maintenance. It is worth noting, however, that this cell line is likely less sensitive to cytotoxicity than other cell lines, but should show general cell behavior in the presence of strong cytotoxic agents.
Cell Adherence Testing
After testing the cytotoxicity of both the NPS-PEGDA and A-PEGDA resins, adherence testing was performed on PEGDA substrates with TCPS as a control to see how well anchoring proteins secreted from the cells adhered to the PEGDA material. After printing, the substrates were sterilized and placed in a sterile 12-well plate. Sterilization involved soaking the substrates in ethanol for ~10 min and then drying in an oven at 50C for 3–4 hours. Endothelial cells (EA.hy926) were then plated into the wells so that the cells could attach and proliferate on the PEGDA material. The cells were viewed after a 24-hour exposure time and stained using Calcein AM/ethidium homodimer-1 (EthD) live/dead assay. Adherence was measured by image analysis using CellProfiler and calculating the fractional cell surface coverage of the adherent cells in comparison to the TCPS control, which was passaged at the same cell density. Replicates were performed with three cell culture wells and five pictures per well for analysis.
Adherence was tested on both treated and untreated 3D printed PEGDA surfaces. Treated surfaces were subjected to a 1–2 minute plasma in a Harrick Plasma Cleaner (Model PDC-32G) using standard lab atmosphere. After plasma treatment, the substrates were sterilized and tested for cell adherence according to the procedure outlined above.
3D-Printed Wellplate Preparation for Spheroid Testing
A-PEGDA resin was used to 3D print wells for spheroid testing. Each device contained 6 identical cylindrical wells. The wells were 3 mm deep with slightly sloping sidewalls (top and bottom well diameters of 3.8 mm and 3.6 mm, respectively). The resultant 32 μL well volume is similar to a single well in a 384-well plate. Before use for cell culture, the 3D printed plates were sterilized using 70% ethanol for 10 minutes and were dried overnight in an oven.
3D Spheroid Growth
Lung epithelial carcinoma cell line A549 (ATCC CCL-185)), endothelial fusion cells EA.hy926, and lung fibroblasts (HFL1, ATCC CCL-153) were maintained in 2D culture as specified by the manufacturers but using DMEM Ham’s F-12 media (Corning, 10–092-CV). Epithelial cell 3D growth was initiated through forced aggregation on 1% agarose coated 96-well plates made in-house and through supplementation of 0.024% methylcellulose (Millipore Sigma, M7027), and 10μM Y-27632 ROCK inhibitor (Cayman Chemical Company, 10005583). The seeded cells were maintained in a 37°C, 5% CO2 incubator. Media was changed every 2–3 days but without the addition of Y-27632. Endothelial cells and fibroblasts were cocultured through 1:1 seeding of 104 cells per well for each cell line cultured in media supplemented with methylcellulose and 50μg/mL Collagen, Type I rat tail (Corning, 354236). The cocultures were seeded on the 3D printed devices or on a flat-bottom TCPS plate (Sarstedt, NC9624222) to show the capability of the devices to induce 3D growth.
Spheroid Migration
At least three epithelial cell line (A549) spheroids were transferred to either a 96-well flat-bottom TC plate (Sarstedt, NC9624222) or 3D printed A-PEGDA with or without plasma treatment. The media was maintained to be the same as the growth media. After 24 hours the spheroids were imaged using Thermo Evos XL Core with a 10X or 40X objective as stated in each figure.
Image analysis and statistics
Cell migration was quantified using ImageJ-FIJI.26 The spheroid area, As, and total cell area, At, are defined in Fig. 2. The dell migration area is At − As and the normalized cell migration area is (At − As)/As. Pairwise mean comparisons were made using the Kruskal-Wallis non-parametric test with no false discovery rate (FDR) corrections for multiple t-tests using Graphpad Prism 8.
Figure 2:
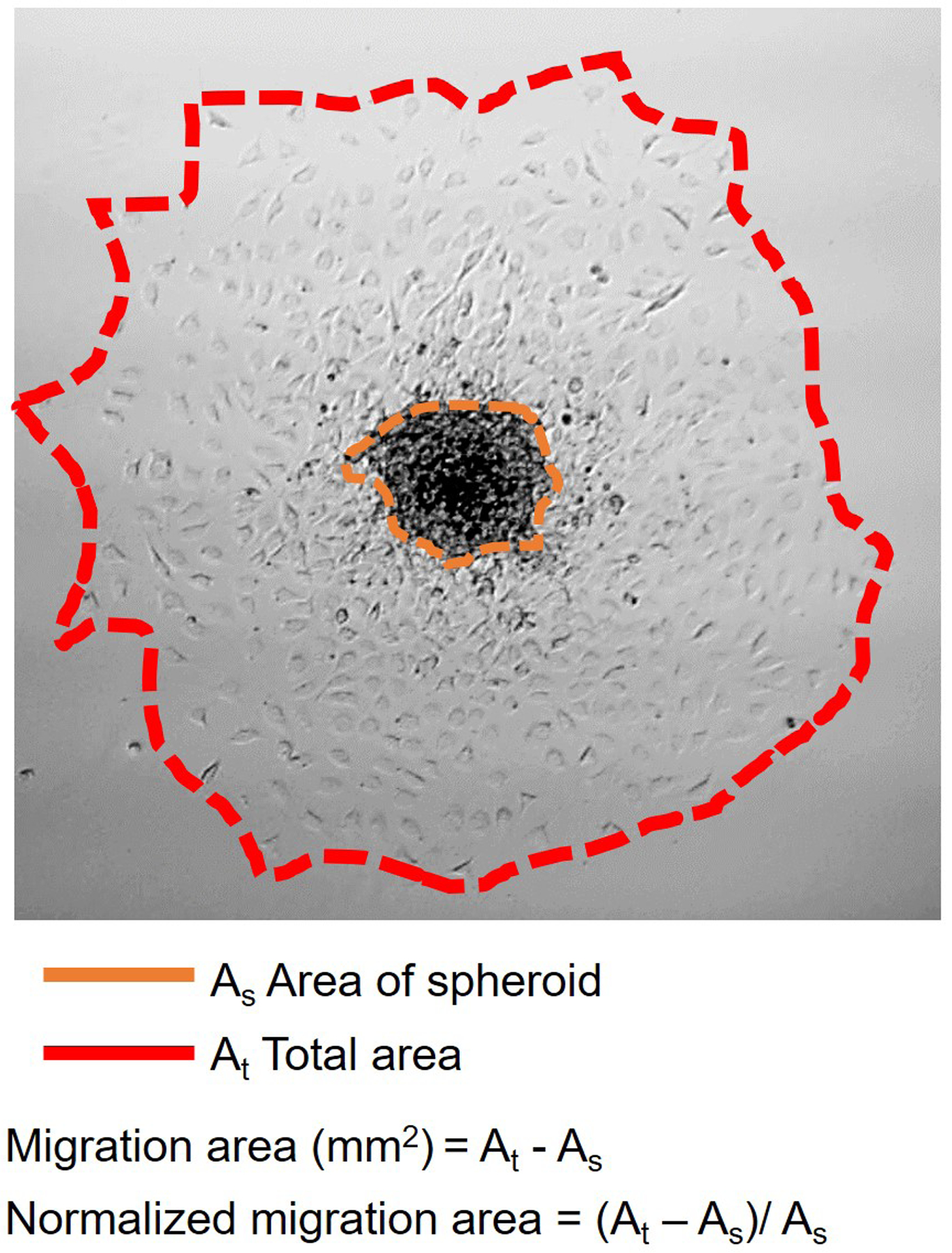
Spheroid migration characterization.
Results and Discussion
Cytotoxicity
For the initial cytotoxicity investigation, a resin comprised of only PEGDA and Irgacure 819 (i.e., no UV absorber) was used, and was 3D printed the same as described for NPS-PEGDA and A-PEGDA. When tested with cells, no cytotoxic response was observed, thereby confirming the cell culture compatibility of the monomer and photoinitiator components of the resin.
Next, the NPS-PEGDA 3D printed polymer was tested. As shown in Fig. 3, a severe cytotoxic response from the cell layer was induced by the as-printed NPS-PEGDA polymer, indicating that NPS is cytotoxic. To overcome this effect, we evaluated the possibility of leaching NPS out of the polymer matrix as a post-print process.27 Both water and ethanol were tested as possible eluents, the former because higher molecular weight PEGDA is commonly used as a hydrogel, and the latter because of the high solubility of NPS in ethanol and ethanol’s compatibility with 3D printed PEGDA. PEGDA substrates were washed for up to 96 hours. Given the low solubility of NPS in water, the water was changed out every 24 hours to avoid saturating the solution. Ethanol was not changed out during washings.
Figure 3:
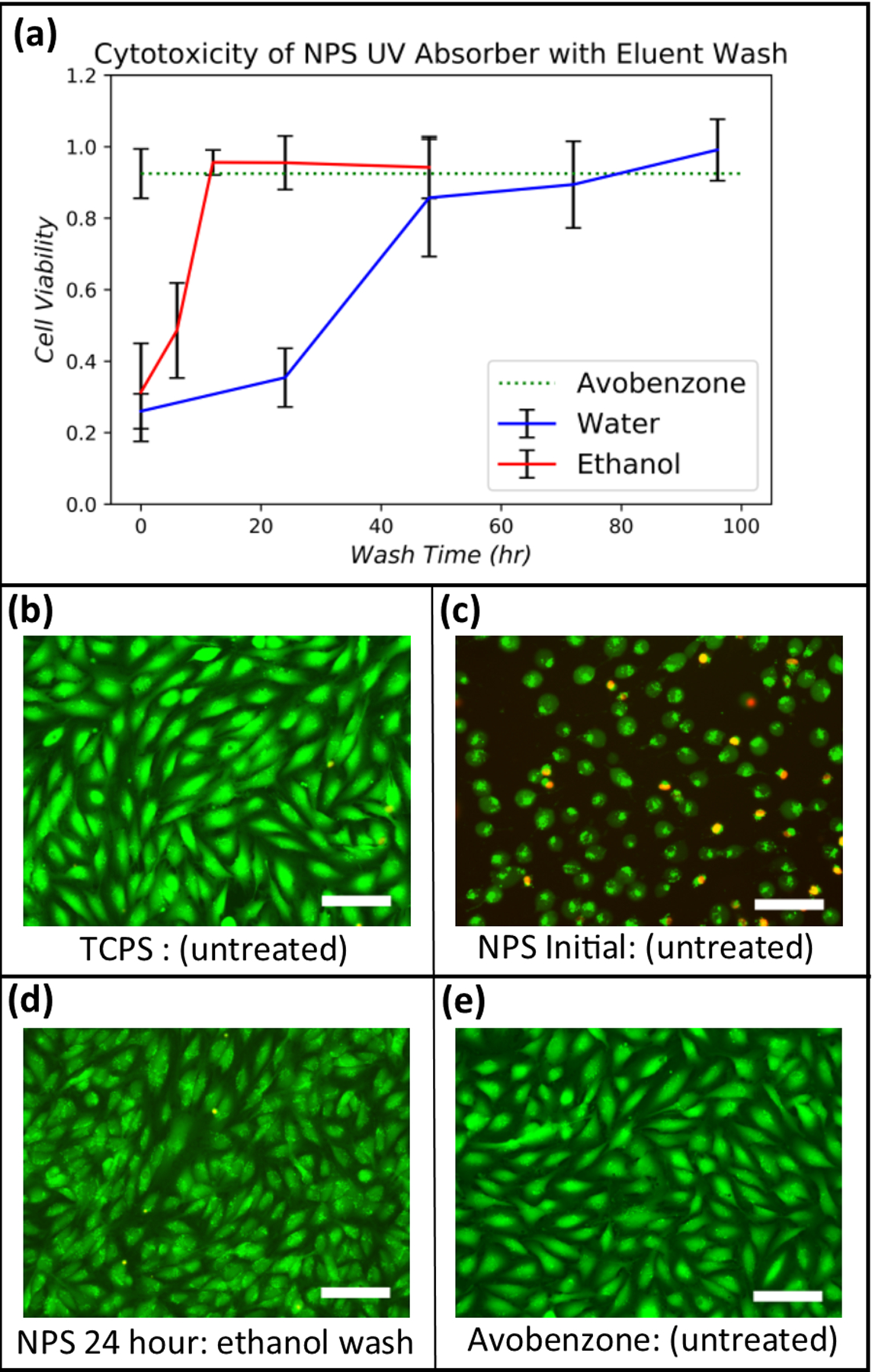
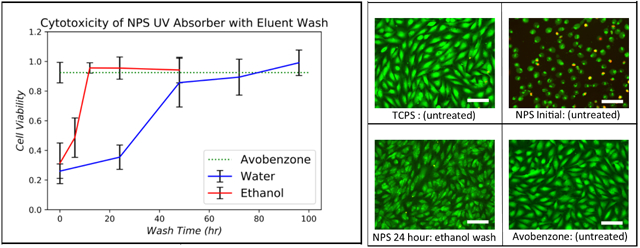
(a) Cell viability for NPS-PEGDA as a function of wash time for water and ethanol. The result for unwashed A-PEGDA is also shown (corresponding error bar is at the left of the dashed green line). Data were normalized against a TCPS control seeded at the same density. Error bars indicate standard deviation of replicates (n≥3). (b)-(e) Stained EA.hy926 cells for (b) TCPS (control), (c) as-printed NPS-PEGDA (i.e., no washing) (d) NPS-PEGDA washed in ethanol for 24 hours, and (e) as-printed A-PEGDA. Scale bars are 100 μm.
The results are shown in Fig. 3(a), where the fractional surface coverage of cells normalized to TCPS seeded at the same cell density is plotted as a function of wash time. Using water as eluent, a non-cytotoxic response (>70% cell coverage), was achieved after a 48 hour wash time, with results continuing to improve up to the maximum tested wash time of 96 hours. Note that a much shorter wash time, 12 hours, was required with ethanol to achieve a non-cytotoxic response comparable to a water 96 hour wash time. Ethanol is, therefore, a better eluent choice.
Since NPS-PEGDA requires post-print processing to combat cytotoxicity, which is ultimately undesirable, we evaluated avobenzone, which is used in commercially available consumer products such as sunscreen, as an alternate UV absorber. As observed in Fig. 3(e), A-PEGDA exhibits excellent cytocompatibility in its as-printed state. The normalized fractional surface coverage of cells for A-PEGDA is shown in Fig. 3(a) as the dashed green line to facilitate direct comparison with NPS-PEGDA. Note that A-PEGDA results are comparable to the best NPS-PEGDA results for both water and ethanol washes. Given that A-PEGDA is essentially non-cytotoxic as-printed, it is a more attractive material than NPS-PEGDA for cell-based studies.
Cell Adherence
We performed adherence testing on PEGDA substrates with TCPS as a control to determine how well anchoring proteins secreted from the cells adhered to the PEGDA material. After printing with either NPS-PEGDA (followed by ethanol wash) or A-PEGDA (no wash), the substrates were sterilized and placed in a sterile 12-well plate. Endothelial cells (EA.hy926) were then passaged into the wells of the 12-well plate such that the cells could attach and proliferate on the PEGDA material. The cells were viewed after a 24-hour exposure time and stained using a Calcein AM/EthD live/dead assay. Adherence was measured by image analysis using CellProfiler and calculating the fractional cell surface coverage of the adherent cells in comparison to the TCPS control which was passaged at the same cell density.
Initial testing showed that the cells did not adhere well to the printed PEGDA material in its native state (Fig. 4(d)), even after the previously discussed eluent treatments were performed to reduce the cytotoxicity of NPS-PEGDA. Low cell adhesion on PEGDA printed materials is consistent with our earlier results28,29 and those of other groups on the known overall protein-repellant nature of untreated PEG.30
Figure 4:
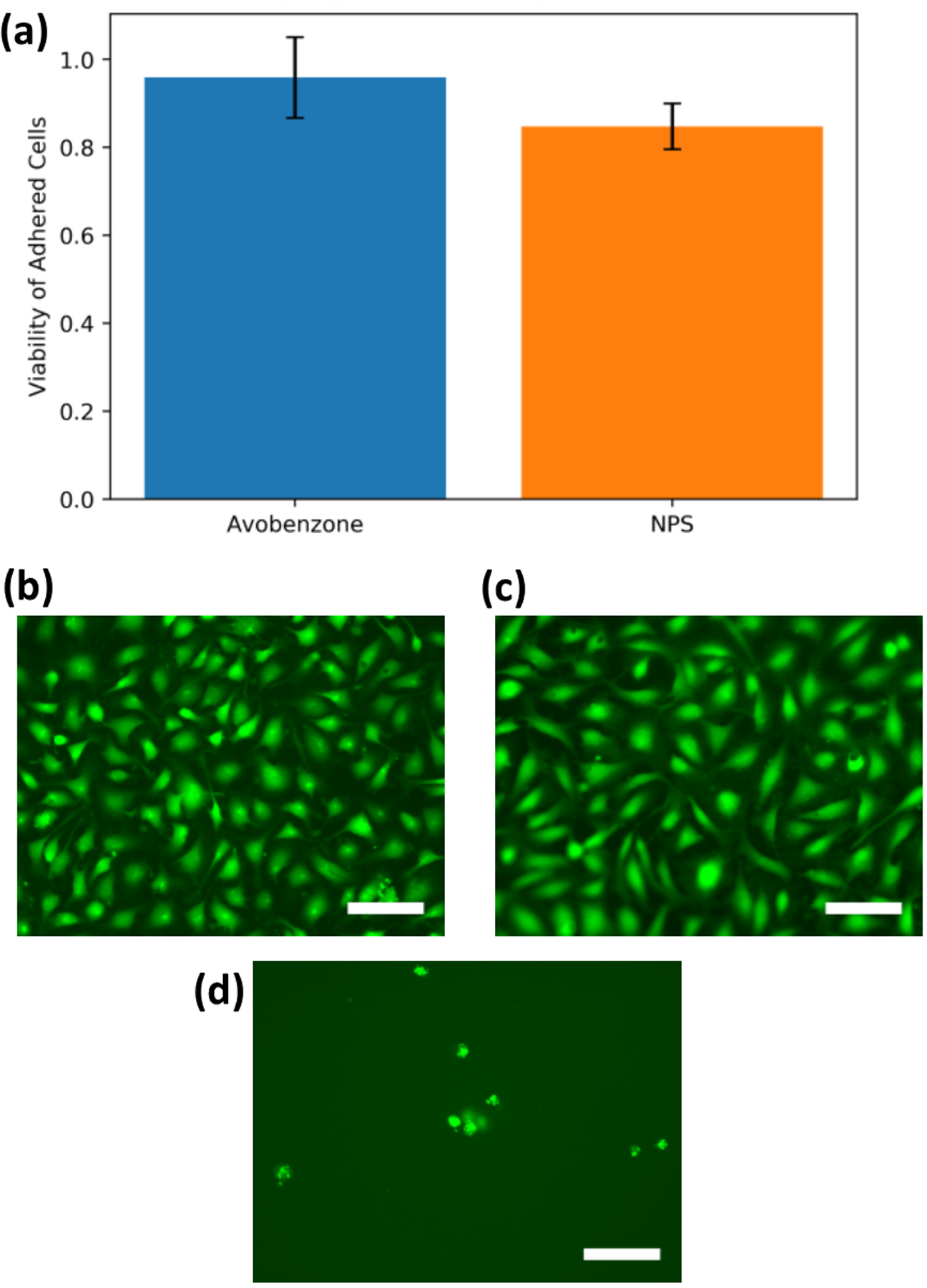
(a) Cell viability of EA.hy926 cells adhered to plasma-treated A-PEGDA and NPS-PEGDA. Data are shown as the fractional surface coverage of the cells normalized to a TCPS control. Error bars indicate standard deviation. The average and standard deviation are, respectively, 0.959 and 0.092 for A-PEGDA and 0.848 and 0.052 for NPS-PEGDA. Images of adhered and stained EA.hy926 cells for plasma-treated (b) unwashed A-PEGDA and (c) washed NPS-PEGDA, and (d) washed NPS-PEGDA with no plasma treatment. Scale bars are 100 μm.
Several surface modifications were attempted to allow for adherence of proteins to the PEGDA surface including a topographical modification and oxygen plasma treatment. Topo-graphical modification and micropatterning have been shown to be effective in helping cells that adhere weakly to PEG have access to increased surface area and thus have a higher chance of adhering to the surface.30 A simple 3D printed topographical modification of small parallel channels (width: 20 μm, depth:10 μm, spacing: 20 μm) was attempted with little improvement over the unmodified surface.
Oxygen plasma treatment has also been shown to increase cellular adherence on PEGDA.27 As shown in Fig. 4, plasma-treated 3D printed surfaces resulted in cell adherence at an average of 96 ± 9% for A-PEGDA and 85 ± 5% for NPS-PEGDA in comparison to a TCPS control. The p-value for A-PEGDA compared to the TCPS control is 0.26, indicating that their difference is not statistically significant. The corresponding p-value for NPS-PEGDA and TCPS is 0.001, indicating that there is a difference, which may be due to residual cytotoxic NPS not fully leached out by the wash process. In either case, our results show that a simple plasma treatment is sufficient to modify the PEGDA surface such that proteins adhere to the surface and allow cell attachment. Moreover, cell attachment to A-PEGDA was statistically indistinguishable from the TCPS control, which bodes well for its future use in microfluidic cell culture.
As a note for future work, we observe that direct addition of membrane proteins to the PEGDA resin may be useful as an alternate route to achieve cellular adherence.31
3D Growth and Cell Migration
Having established the cytocompatibility of 3D printed A-PEGDA and the ability to provide a non-adherent or adherent surface depending on the absence or presence of plasma treatment, we then evaluated the feasibility of using the material to initiate, maintain, and promote adherence of 3D cell cultures (spheroids). We first characterized 3D printed A-PEGDA’s compatibility with initiating 3D growth since its surface has very low cell adhesion. As shown in Fig. 5e, we observed that seeded A549 cancer cells formed 3D structures, although not as compacted as those made using force-aggregation (Fig. 5a–b). Nonetheless, 3D structure growth was still observed. We then looked at the ability of pre-aggregated spheroids to survive and for cells to migrate onto the A-PEGDA surfaces with and without plasma treatment. We observed virtually no cell migration when using the native PEGDA devices (Fig. 5b–d). In contrast, a short plasma treatment of the device allowed for significantly increased adherence and subsequent migration of cells outward from the spheroids as expected on the treated surface (Fig. 5b–d).
Figure 5:
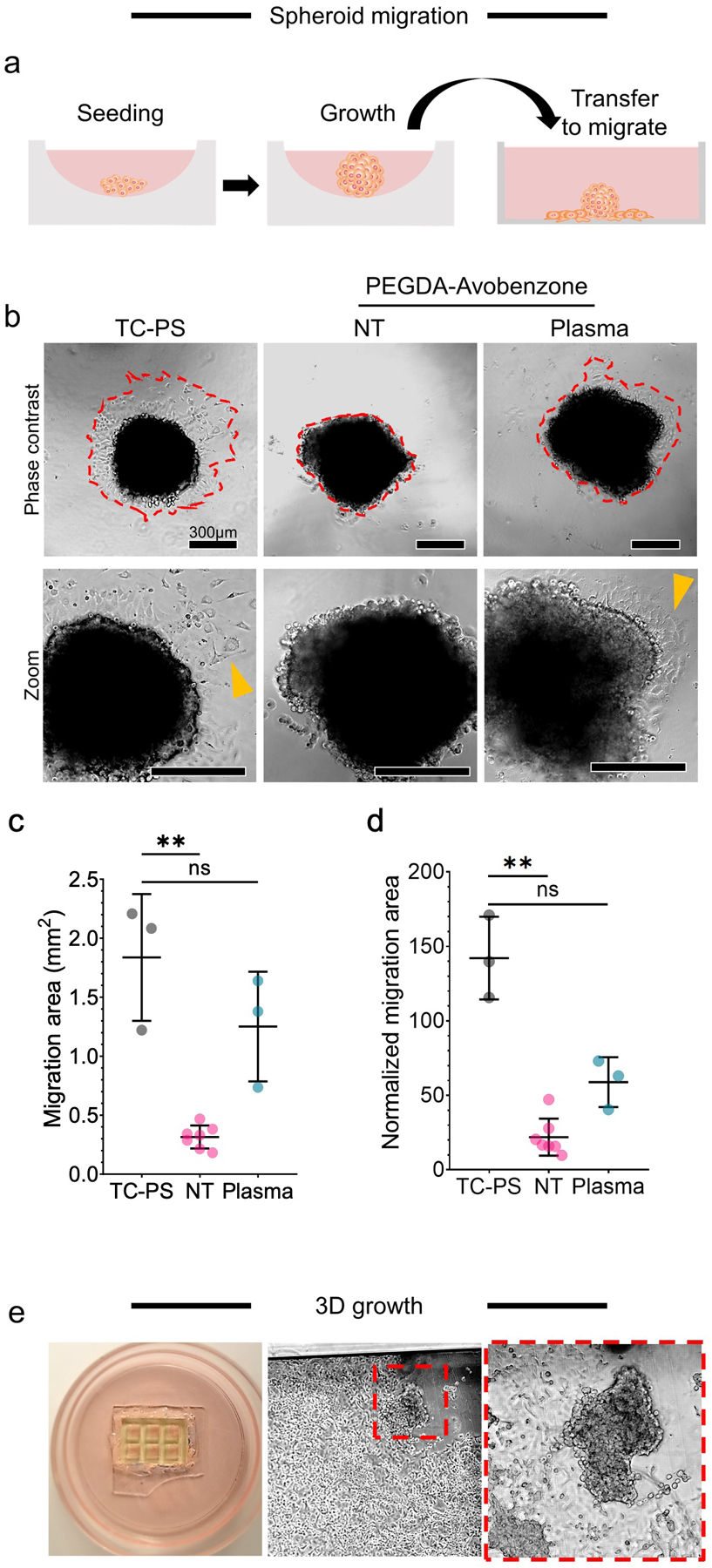
Characterization of 3D growth and migration. a-d) migration of cells from spheroids in flat printed plates, where: a) workflow that includes transfer from low-adhesion seeding and growth well to a 3D printed A-PEGDA well; b) phase-contrast images of spheroids 24hr post-transfer to the plates (NT = no plasma treatment) where zoom images show migrated cells out of the spheroid (yellow arrow); and quantifications of cell migration according to Fig. 2 where c) is the migration area and d) is the migration area normalized to the spheroid area. Significance is based on the Kruskal-Wallis non-parametric test with sample size of 3 (TC-PS), 7 (PEGDA NT), and 3 (PEGDA + Plasma); bars indicate mean and standard deviation; points are observed individual values. e) 3D growth in printed A-PEGDA micro 6-well device. The red box shows 3D spheroid growth in epithelial cell (A549) culture.
In addition to epithelial (A549) tumoroid induction, we also looked into endothelial 3D formation on the device through a fibroblast (HFL1) co-culture system. Initially, monocultured endothelial (EAHy) cells showed similar 3D formation (Fig. 6) to epithelial cells (Fig. 5e). However, regardless of surface area, endothelial cells also formed 3D networks on the device, although, similar to the epithelial cells, they did not form compacted spheroids. We posit that 3D compaction could not be induced due to partial adherence on the surface. By incorporation of fibroblasts with endothelial cells, we observed significantly enhanced 3D formation as seen in Fig. 7.
Figure 6:
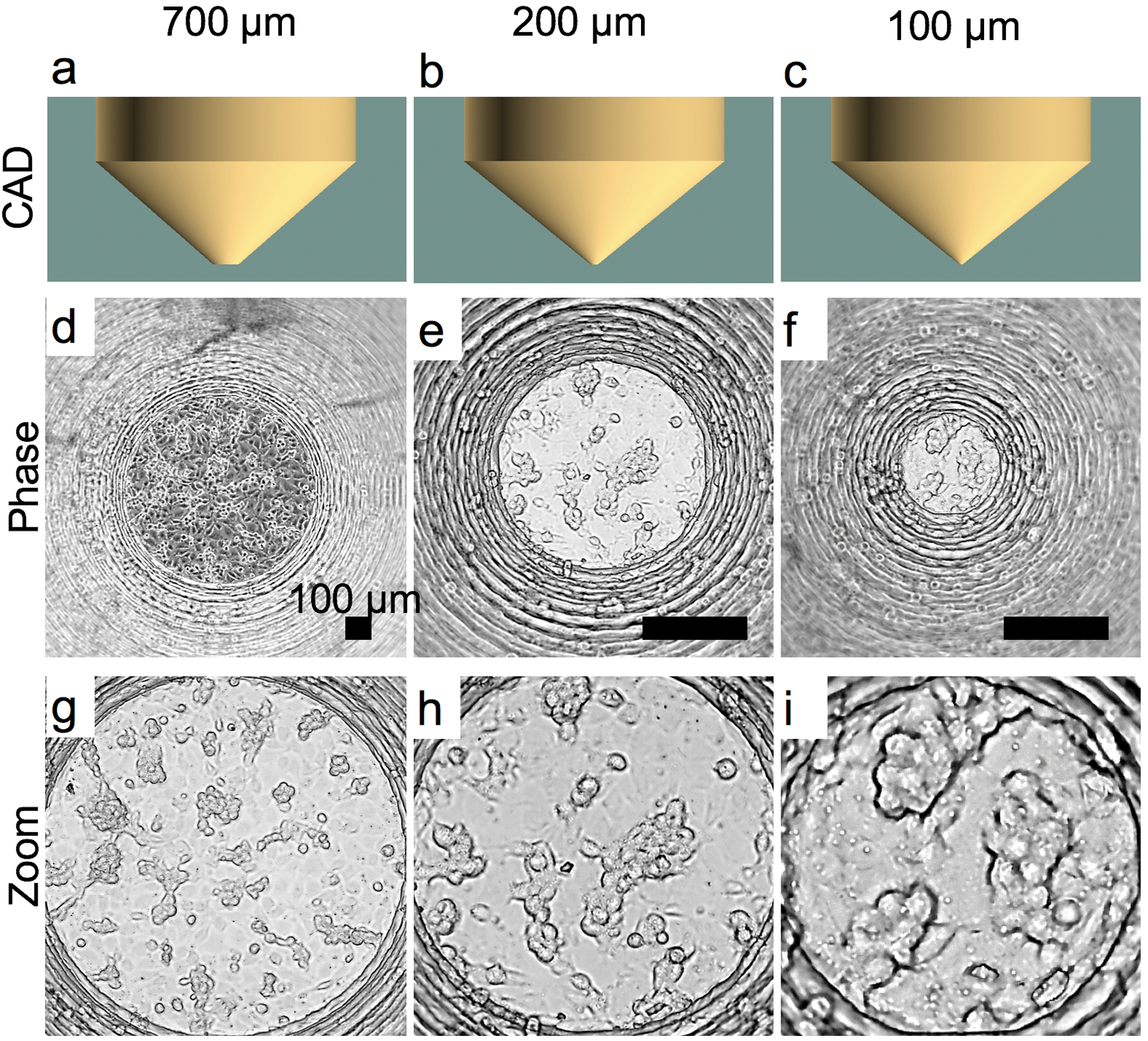
Endothelial (EA.hy-926) 3D cell growth in V-slope flat-bottom A-PEGDA printed devices with varying bottom diameters of 700 μm (a,d,g), 200 μm (c,e,h), and 100 μm (c,f,i). CAD designs (a-c) and representative phase microscopy images (d-i). Zoom images are shown to illustrate 3D networks that formed.
Figure 7:
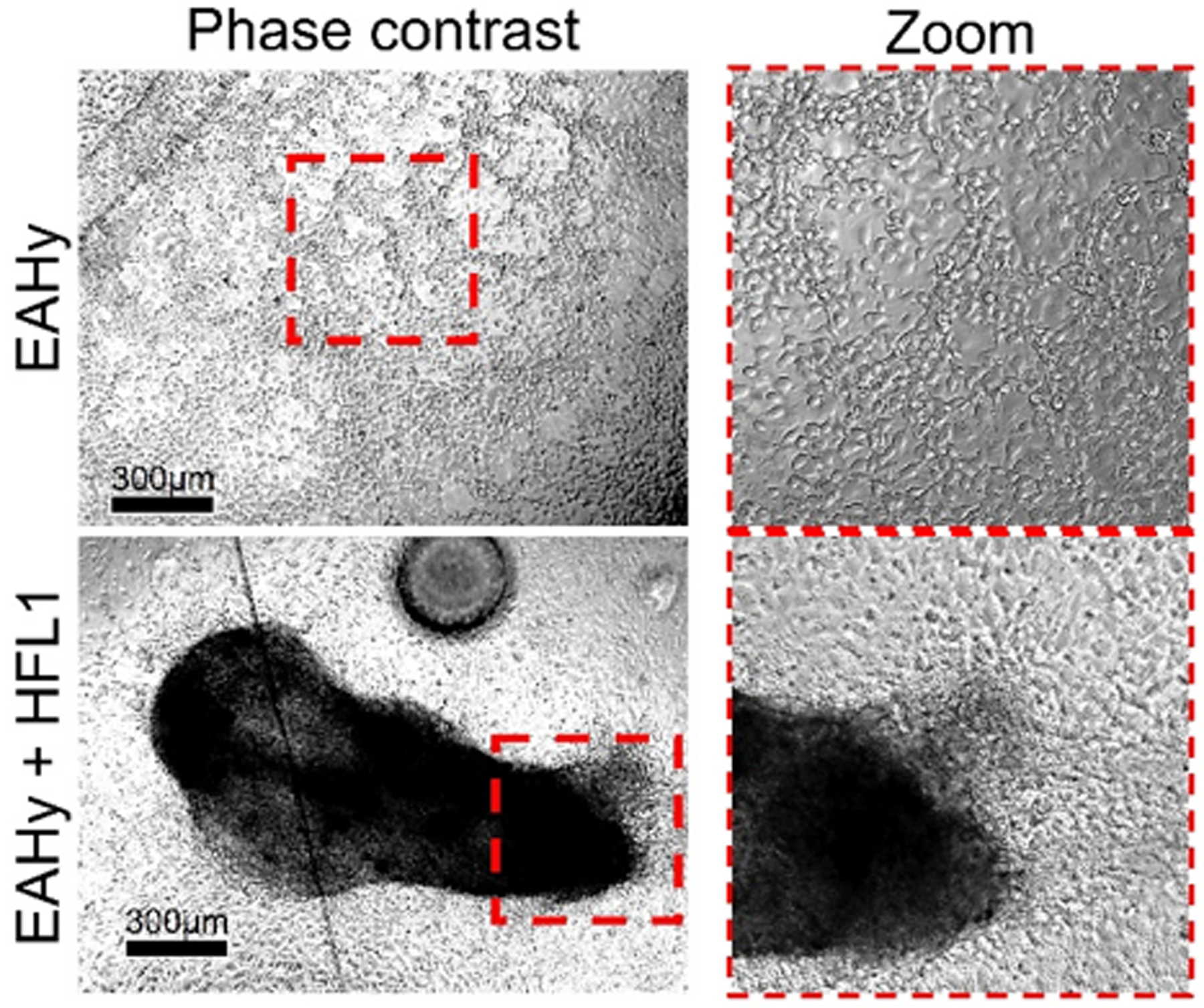
Endothelial (EAhy)-fibroblast(HFL1) coculture shows enhanced 3D formation on a low-adherence A-PEGDA device.
Our results show that through the modulation of the surface adherence properties of A-PEGDA 3D printed wells, we are able to either maintain a spheroid or allow cellular migration. Both of these properties are often used separately in different spheroid formation processes. Our 3D printed devices, therefore, have the potential to be applied to multiple steps of 3D culture and not just 3D growth. Since A-PEGDA can be 3D printed with high resolution, this opens the possibility of creating complete organ-on-chip systems to study complex multi-organ processes for physiology and disease pathology.
Conclusion
We have shown that A-PEGDA resin is non-cytotoxic in its as-printed state, and that NPS-PEGDA can be rendered non-cytotoxic using a 12-hour post-print ethanol wash to leach out the cytotoxic NPS UV absorber. We have also shown that 3D printing with either PEGDA resin results in non-adherent surfaces for cells, but that a short plasma treatment creates adherent surfaces. Furthermore, we have used the non-cytotoxic properties of 3D printed A-PEGDA to illustrate spheroid formation in non-adherent 3D printed wells, and spheroid migration on plasma-treated adherent surfaces. Given that A-PEGDA can be 3D printed with very high resolution, it has significant promise for a wide variety of cell-based studies using 3D printed microfluidic structures, which opens the door to being able to rapidly explore systems as complex as coupled heterogeneous organs-on-a-chip.
Acknowledgments
We are grateful to the National Institutes of Health (R15GM123405) for partial support of this work.
References
- (1).Anderson KB; Lockwood SY; Martin RS; Spence DM A 3D printed fluidic device that enables integrated features. Analytical chemistry 2013, 85, 5622–5626. [DOI] [PubMed] [Google Scholar]
- (2).Shallan AI; Smejkal P; Corban M; Guijt RM; Breadmore MC Cost-effective three-dimensional printing of visibly transparent microchips within minutes. Analytical chemistry 2014, 86, 3124–3130. [DOI] [PubMed] [Google Scholar]
- (3).Bhargava KC; Thompson B; Malmstadt N Discrete elements for 3D microfluidics. Proceedings of the National Academy of Sciences 2014, 111, 15013–15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ho CMB; Ng SH; Li KHH; Yoon Y-J 3D printed microfluidics for biological applications. Lab on a Chip 2015, 15, 3627–3637. [DOI] [PubMed] [Google Scholar]
- (5).Bhattacharjee N; Urrios A; Kang S; Folch A The upcoming 3D-printing revolution in microfluidics. Lab on a chip 2016, 16, 1720–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Waheed S; Cabot JM; Macdonald NP; Lewis T; Guijt RM; Paull B; Breadmore MC 3D printed microfluidic devices: enablers and barriers. Lab on a Chip 2016, 16, 1993–2013. [DOI] [PubMed] [Google Scholar]
- (7).Chen C; Mehl BT; Munshi AS; Townsend AD; Spence DM; Martin RS 3D-printed microfluidic devices: fabrication, advantages and limitations-a mini review. Anal. Methods 2016, 8, 6005–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yazdi AA; Popma A; Wong W; Nguyen T; Pan Y; Xu J 3D printing: an emerging tool for novel microfluidics and lab-on-a-chip applications. Microfluidics and Nanofluidics 2016, 20, 50. [Google Scholar]
- (9).Macdonald NP; Cabot JM; Smejkal P; Guijt RM; Paull B; Breadmore MC Comparing microfluidic performance of three-dimensional (3D) printing platforms. Analytical chemistry 2017, 89, 3858–3866. [DOI] [PubMed] [Google Scholar]
- (10).Castiaux AD; Pinger CW; Hayter EA; Bunn ME; Martin RS; Spence DM PolyJet 3D-Printed Enclosed Microfluidic Channels without Photocurable Supports. Analytical chemistry 2019, 91, 6910–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rogers CI; Qaderi K; Woolley AT; Nordin GP 3D printed microfluidic devices with integrated valves. Biomicrofluidics 2015, 9, 016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Beauchamp MJ; Nordin GP; Woolley AT Moving from millifluidic to truly microfluidic sub-100-μm cross-section 3D printed devices. Analytical and bioanalytical chemistry 2017, 409, 4311–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Beauchamp MJ; Gong H; Woolley AT; Nordin GP 3D Printed Microfluidic Features Using Dose Control in X, Y, and Z Dimensions. Micromachines (Basel) 2018, 9, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gong H; Bickham BP; Woolley AT; Nordin GP Custom 3D printer and resin for 18 mum × 20 mum microfluidic flow channels. Lab Chip 2017, 17, 2899–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gong H; Woolley AT; Nordin GP 3D printed high density, reversible, chip-to-chip microfluidic interconnects. Lab Chip 2018, 18, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gong H; Woolley AT; Nordin GP 3D printed selectable dilution mixer pumps. Biomicrofluidics 2019, 13, 014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Castiaux AD; Spence DM; Martin RS Review of 3D cell culture with analysis in microfluidic systems. Analytical Methods 2019, 11, 4220–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Parker EK; Nielsen AV; Beauchamp MJ; Almughamsi HM; Nielsen JB; Sonker M; Gong H; Nordin GP; Woolley AT 3D printed microfluidic devices with immunoaffinity monoliths for extraction of preterm birth biomarkers. Anal Bioanal Chem 2018, 411, 5405–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Beauchamp MJ; Nielsen AV; Gong H; Nordin GP; Woolley AT 3D printed microfluidic devices for microchip electrophoresis of pre-term birth biomarkers. Analytical chemistry 2019, 91, 7418–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ertl P; Sticker D; Charwat V; Kasper C; Lepperdinger G Lab-on-a-chip technologies for stem cell analysis. Trends Biotechnol 2014, 32, 245–53. [DOI] [PubMed] [Google Scholar]
- (21).Tsui JH; Lee W; Pun SH; Kim J; Kim DH Microfluidics-assisted in vitro drug screening and carrier production. Adv Drug Deliv Rev 2013, 65, 1575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wu J; Chen Q; Liu W; He Z; Lin J-M Recent advances in microfluidic 3D cellular scaffolds for drug assays. TrAC Trends in Analytical Chemistry 2017, 87, 19–31. [Google Scholar]
- (23).Wu J; He Z; Chen Q; Lin J-M Biochemical analysis on microfluidic chips. TrAC Trends in Analytical Chemistry 2016, 80, 213–231. [Google Scholar]
- (24).Zheng XT; Yu L; Li P; Dong H; Wang Y; Liu Y; Li CM On-chip investigation of cell-drug interactions. Adv Drug Deliv Rev 2013, 65, 1556–74. [DOI] [PubMed] [Google Scholar]
- (25).Zhuang Q-C; Ning R-Z; Ma Y; Lin J-M Recent Developments in Microfluidic Chip for in vitro Cell-based Research. Chinese Journal of Analytical Chemistry 2016, 44, 522–532. [Google Scholar]
- (26).Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature methods 2012, 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Urrios A; Parra-Cabrera C; Bhattacharjee N; Gonzalez-Suarez AM; Rigat-Brugarolas LG; Nallapatti U; Samitier J; DeForest CA; Posas F; Garcia-Cordero JL; Folch A 3D-printing of transparent bio-microfluidic devices in PEGDA. Lab Chip 2016, 16, 2287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Rogers CI; Pagaduan JV; Nordin GP; Woolley AT Single-monomer formulation of polymerized polyethylene glycol diacrylate as a material for microfluidics. Analytical chemistry 2011, 83, 6418–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Nge PN; Pagaduan JV; Yu M; Woolley AT Microfluidic chips with reversed-phase monoliths for solid phase extraction and on-chip labeling. J Chromatogr A 2012, 1261, 129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lensen Marga C., D. M, Schulte Vera A. In Biomaterials; Pignatello R, Ed.; IntechOpen, 2011. [Google Scholar]
- (31).Missirlis D; Spatz JP Combined effects of PEG hydrogel elasticity and cell-adhesive coating on fibroblast adhesion and persistent migration. Biomacromolecules 2014, 15, 195–205. [DOI] [PubMed] [Google Scholar]


