Abstract
Vertebrate segmentation is regulated by the segmentation clock, a biological oscillator that controls periodic formation of somites, or embryonic segments, which give rise to many mesodermal tissue types. This molecular oscillator generates cyclic gene expression with the same periodicity as somite formation in the presomitic mesoderm (PSM), an area of mesenchymal cells that give rise to mature somites. Molecular components of the clock include the Hes/her family of genes that encode transcriptional repressors, but additional genes cycle. Cyclic gene transcripts are cleared rapidly, and clearance depends upon the pnrc2 (proline-rich nuclear receptor co-activator 2) gene that encodes an mRNA decay adaptor. Previously, we showed that the her1 3’UTR confers instability to otherwise stable transcripts in a Pnrc2-dependent manner, however, the molecular mechanism(s) by which cyclic gene transcripts are cleared remained largely unknown. To identify features of the her1 3’UTR that are critical for Pnrc2-mediated decay, we developed an array of transgenic inducible reporter lines carrying different regions of the 3’UTR. We find that the terminal 179 nucleotides (nts) of the her1 3’UTR are necessary and sufficient to confer rapid instability. Additionally, we show that the 3’UTR of another cyclic gene, deltaC (dlc), also confers Pnrc2-dependent instability. Motif analysis reveals that both her1 and dlc 3’UTRs contain terminally-located Pumilio response elements (PREs) and AU-rich elements (AREs), and we show that the PRE and ARE in the last 179 nts of the her1 3’UTR drive rapid turnover of reporter mRNA. Finally, we show that mutation of Pnrc2 residues and domains that are known to facilitate interaction of human PNRC2 with decay factors DCP1A and UPF1 reduce the ability of Pnrc2 to restore normal cyclic gene expression in pnrc2 mutant embryos. Our findings suggest that Pnrc2 interacts with decay machinery components and cooperates with Pumilio (Pum) proteins and ARE-binding proteins to promote rapid turnover of cyclic gene transcripts during somitogenesis.
Keywords: Hes/her, deltaC, PRE, ARE, somitogenesis, 3’UTR
Summary statement:
We show that her1 and dlc 3’UTR regulatory sequences confer Pnrc2-mediated decay to reporter transcripts and that a Pumilio response element (PRE) and an AU-rich element (ARE) in the terminal her1 3’UTR drive transcript destabilization. Our work suggests that Pnrc2 interacts with decay machinery components and cooperates with Pumilio (Pum) proteins and ARE-binding proteins to promote rapid cyclic gene transcript turnover during somitogenesis.
INTRODUCTION
Genetic oscillations underlie many cellular events and function in the regulation of critical developmental processes. A well-studied example of genetic oscillation is the segmentation clock, a rapid ultradian oscillator that generates periodic expression in developing embryos (Hubaud and Pourquie, 2014; Oates et al., 2012; Pourquie, 2011). The segmentation clock controls vertebrate somitogenesis, the process by which the mesoderm is sequentially divided into segmental units called somites that later give rise to vertebrae, ribs, body musculature, and dermis. Molecular evidence for the segmentation clock was first uncovered with the characterization of the c-hairy gene, a chick homolog of the Drosophila pair rule gene hairy (Palmeirim et al., 1997). c-hairy encodes a member of the Hairy/Enhancer of split-related (Hes)/Hes-related (her) family of helix-loop-helix transcription factors that oscillate through a negative feedback loop in which the Hes/Her protein inhibits its own transcription (Bessho et al., 2003; Chen et al., 2005; Hirata et al., 2002; Lewis, 2003). Homologs of hairy have since been identified in vertebrate models such as mouse, fish, frog, and snake (Bessho et al., 2001; Gomez et al., 2008; Holley et al., 2000; Li et al., 2003), and in each species, members of the Hes/her family undergo oscillatory expression in the presomitic mesoderm (PSM). Genetic studies have shown that the mouse hairy1 homolog Hes7 and the zebrafish hairy1 homologs her1 and her7 are core segmentation genes that cycle in the PSM of each species with the same periodicity of segment formation (Bessho et al., 2001; Delaune et al., 2012; Gajewski et al., 2003; Harima et al., 2013; Henry et al., 2002; Hirata et al., 2002; Holley et al., 2000; Holley et al., 2002; Oates and Ho, 2002; Shih et al., 2015; Takashima et al., 2011; Takke and Campos-Ortega, 1999; Williams et al., 2016). Maintenance of oscillation periodicity requires many levels of regulation as a somite pair develops rapidly in developing embryos (30 minutes in zebrafish, 120 minutes in mouse) (Kageyama et al., 2012; Oates et al., 2012). Studies have explored transcriptional activation and the effect of negative feedback inhibition on oscillatory expression (Bessho et al., 2003; Giudicelli et al., 2007; Gonzalez et al., 2013; Hirata et al., 2002; Lewis, 2003; Schwendinger-Schreck et al., 2014), and have emphasized the importance of post-transcriptional regulation in maintaining proper oscillatory periodicity (Cibois et al., 2010; Fujimuro et al., 2014; Hanisch et al., 2013; Nitanda et al., 2014). For example, splicing (Harima et al., 2013; Takashima et al., 2011) and mRNA export (Hoyle and Ish-Horowicz, 2013) are rate-limiting steps of oscillatory expression. Additionally, cyclic gene transcript 3’UTRs can promote rapid decay of oscillating transcripts (Delaune et al., 2012; Fujimuro et al., 2014; Gallagher et al., 2017; Giudicelli et al., 2007). miRNAs regulate decay of some cyclic gene transcripts (Bonev et al., 2012; Riley et al., 2013; Tan et al., 2012; Wong et al., 2015), but not others (Gallagher et al., 2017; Zhang et al., 2011). Thus, specific mechanisms that govern cyclic gene transcript turnover are still not well-understood.
In a forward genetic screen, we discovered a zebrafish mutant, tortuga (tor), with disrupted cyclic gene expression (Dill and Amacher, 2005). Over 20 genes are deleted in the tor deficiency allele, but loss of a single gene in the deletion interval, pnrc2, is responsible for defects in cyclic gene expression (Gallagher et al., 2017). Recent work in cell culture systems has shown that human PNRC2 interacts with factors such as UPF1, DCP1A, and STAU1, revealing that Pnrc2 can function as a decay adaptor in nonsense-mediated mRNA decay (NMD) and STAU1-mediated mRNA decay (SMD) (Cho et al., 2013a; Cho et al., 2013b; Cho et al., 2012; Cho et al., 2009; Cho et al., 2015; Lai et al., 2012; Mugridge et al., 2016). Tethering of PNRC2 to reporter mRNAs confers instability (Cho et al., 2009; Lai et al., 2012; Nicholson et al., 2018), suggesting that PNRC2 can recruit decay machinery when directly associated with transcripts. However, recent work reported that destabilization of reporters containing NMD-inducing premature termination codons (PTCs) in HeLa cells is unaffected by PNRC2 knockdown, suggesting that PNRC2 is not necessary for NMD-induced decay in all cases (Nicholson et al., 2018).
We have shown that pnrc2 functions in clearance of cyclic gene transcripts such as her1, her7, and deltaC (dlc) during zebrafish segmentation (Gallagher et al., 2017). By in situ hybridization, cyclic gene transcript expression appears striped in the anterior PSM of wild-type embryos due to rapid oscillatory transcription followed by rapid mRNA decay. In pnrc2oz22 mutants, the accumulation of cyclic gene transcripts obscures this dynamic striped expression (Gallagher et al., 2017). We also previously showed that pnrc2 is maternally-provided and zygotically-expressed throughout somitogenesis (Gallagher et al., 2017) and show here that maternally-provided pnrc2 partially compensates for zygotic pnrc2 in regulating cyclic gene transcript turnover during somitogenesis. Using a series of inducible transgenic reporter lines driving expression of a reporter mRNA fused to various portions of the her1 3’UTR, we find that the last 179 nucleotides (nts) of the 725 nt her1 3’UTR is necessary and sufficient to confer Pnrc2-dependent instability to reporter transcripts. We find the dlc 3’UTR also confers Pnrc2-dependent instability, demonstrating 3’UTR instability elements are present in both cyclic gene transcripts. Motif analysis comparing the last 179 nts of the her1 3’UTR to the dlc 3’UTR uncovered two potential cis-regulatory motifs: a Pumilio response element (PRE) and an AU-rich element (ARE). Mutation of the PRE or ARE motif partially disrupts the destabilizing effect of the her1 3’UTR on reporter mRNA, and mutation of the PRE and ARE severely disrupts the destabilizing effect, suggesting that the PRE and ARE both contribute to rapid turnover of endogenous her1 transcripts. Finally, we show that mutation of Pnrc2 residues and domains that are known to facilitate interaction of human PNRC2 with decay factors DCP1A and UPF1 eliminate or severely reduce the ability of Pnrc2 to restore normal cyclic gene expression in pnrc2 mutant embryos. Together, this work suggests that Pumilio proteins, ARE-binding proteins (ARE-BPs), and/or Pnrc2 interact with decay machinery components to regulate 3’UTR-mediated turnover of cyclic gene transcripts during vertebrate segmentation.
METHODS
Animal stocks and husbandry
Adult zebrafish strains (Danio rerio) were kept at 28.5°C on a 14 hour (h) light/10h dark cycle and obtained by natural spawning or in vitro fertilization, and were staged according to Kimmel et al (1995). The pnrc2oz22 line has been described previously (Gallagher et al., 2017). The stable transgenic reporter lines generated and analyzed in this study are: Tg(hsp70l:Venus-her1 3’UTR-SV40 pA)oz44, oz45, oz46; Tg(hsp70l:Venus-her1 3’UTRΔ1–362-SV40 pA)oz47, oz48, oz50; Tg(hsp70l:Venus-her1 3’UTRΔ363–725-SV40 pA)oz51, oz54, oz96; Tg(hsp70l:Venus-her1 3’UTRΔ1–546-SV40 pA)oz55, oz57, oz58; Tg(hsp70l:Venus-her1 3’UTRΔ1–362;Δ547–725-SV40 pA)oz60, oz61; Tg(hsp70l:Venus-disrupted PRE her1 3’UTR-SV40 pA)oz69, oz70, oz71, oz72, oz73; Tg(hsp70l:Venus-disrupted ARE her1 3’UTR-SV40 pA)oz74, oz75, oz77, oz78, oz79; Tg(hsp70l:Venus-SV40 pA)oz64, oz65, oz66, oz67, oz68; Tg(hsp70l:Venus-dlc 3’UTR-SV40 pA)oz80, oz81, oz83; and Tg(hsp70l:Venus-disrupted PRE & ARE her1 3’UTR-SV40 pA)oz93, oz94, oz95. To control for potential locus-specific effects on transgene expression, all of the above independent lines were analyzed by in situ hybridization and showed consistent Venus reporter decay across all heat shock experiments except for Tg(hsp70l:Venus-disrupted ARE her1 3’UTR-SV40 pA)oz79 which had abnormally high Venus induction levels compared to the three other lines and was therefore excluded from the analysis. For the reporters hsp70l:Venus-her1 3’UTR-SV40 pA, hsp70l:Venus-disrupted PRE her1 3’UTR-SV40 pA, hsp70l:Venus-disrupted ARE her1 3’UTR-SV40 pA, and hsp70l:Venus-disrupted PRE & ARE her1 3’UTR-SV40 pA, three independent lines per reporter were analyzed by qPCR across three biological replicates and exhibited comparable Venus decay dynamics for each reporter (Figs. S2–S5). Animal experiments were performed according to institutional and national guidelines and regulations and were approved by the Ohio State University Animal Care and Use Committee.
DNA extraction and pnrc2oz22 and Venus genotyping
Individual embryos and adult fin tissue were lysed in 50 ul 1X ThermoPol Buffer (NEB) at 95°C for 10 minutes, digested at 55°C for 1–4 hours using 25–50 ug Proteinase K (Thermo Fisher), followed by Proteinase K inactivation at 95°C for 10 minutes. 1 ul of DNA extract was used as template in a standard 25 ul PCR with Taq polymerase according to manufacturer’s protocol (NEB). To molecularly identify pnrc2oz22 carriers after PCR amplification, samples were digested with 20 units NsiI-HF (NEB) to distinguish cleavable wild-type from un-cleavable mutant amplicons. Reaction products were analyzed on a 2% agarose gel stained with Gel Red (Biotium). To identify carriers of the heat shock inducible reporter transgenes, embryos were either screened post-heat shock (pHS) for Venus fluorescence or molecularly identified by PCR amplification of Venus coding sequence. Genotyping was performed with 1 ul of DNA extract as template in a standard 25 ul reaction with Taq polymerase according to manufacturer’s protocol (NEB). Primers were designed to amplify presence of Venus coding sequence (Table S2) and reaction products were analyzed on a 2% agarose gel stained with Gel Red (Biotium).
Plasmid construction and Transgenesis
The heat shock reporter construct hsp70l:Venus-her1 3’UTR-SV40 pA was assembled using standard restriction digestion-based cloning and replacement of the 1.1 kilobase (kb) her1 3’ noncoding sequence present in construct hsp70l:Venus-her1 3’UTR (Gallagher et al., 2017) with a synthetic 725 nt her1 3’UTR sequence directly fused to an SV40 polyadenylation (pA) sequence synthesized by GeneArt® Gene Synthesis (Thermo Fisher). Derivative her1 3’UTR constructs, were generated by restriction digestion or PCR amplification of the hsp70l:Venus-her1 3’UTR-SV40 pA plasmid, in parallel with removal of the full-length her1 3’UTR from the hsp70l:Venus-her1 3’UTR-SV40 pA plasmid by restriction digestion, followed by ligation of the truncated her1 3’UTR sequence into the digested hsp70l:Venus-her1 3’UTR-SV40 pA plasmid. Modification of either the her1 3’UTR PRE or ARE sequence was performed by site-directed mutagenesis of the hsp70l:Venus-her1 3’UTR-SV40 pA reporter using KOD polymerase (EMD Millipore) with mutagenic primers (Table S2), followed by DpnI digestion and transformation into E. coli. To generate the construct with disruptions in both the PRE and ARE sequences, we performed site-directed mutagenesis of the reporter hsp70l:Venus-her1 3’ UTR with disrupted ARE-SV40 pA using KOD polymerase (EMD Millipore) with primers that mutate the PRE sequence without affecting the mutated ARE sequence (Table S2). The full-length 1327 nt dlc 3’UTR was cloned by extracting total RNA from wild-type embryos at mid-segmentation with Trizol Reagent according to manufacturer’s protocol (Thermo Fisher Scientific), followed by reverse-transcription with Superscript III (Thermo Fisher) using a dlc-specific reverse primer (Table S2) followed by PCR amplification of the dlc 3’UTR using gene-specific primers containing restriction enzyme sites for cloning. In parallel, construct hsp70l:Venus-her1 3’UTR-SV40 pA was digested to remove the her1 3’UTR followed by replacement with the dlc 3’UTR. All constructs were sequence confirmed. Transgenic lines were generated as previously described using I-SceI-based transgenesis (Thermes et al., 2002).
Heat shock assay
Adult fish carrying the stable reporter transgenes were crossed to AB wild-type fish. Progeny were raised to mid-segmentation, heat shocked at 37°C for 15 minutes, and fixed in 4% PFA at defined intervals post-heat shock. All transgenic lines were analyzed for Venus expression post-heat shock by colorimetric in situ hybridization or qPCR analysis in parallel (each method described below).
In situ hybridization
Whole mount in situ hybridization was performed as previously described (Broadbent and Read, 1999; Jowett, 1998) using DIG-labeled antisense probes. Riboprobes for her1, dlc, and Venus were made as previously described (Delaune et al., 2012; Dill and Amacher, 2005).
RNA extraction
Whole embryos at mid-segmentation (n=10 per time point or condition) were solubilized in Trizol for RNA extraction and purified following standard procedures (Thermo Fisher). 0.5 –1 ug total RNA was reverse transcribed using random primers or gene-specific reverse primers and Superscript III reverse transcriptase (RT) according to the manufacturer’s instructions (Thermo Fisher).
Quantitative PCR and half-life calculations
Quantitative PCR was performed using PowerUp SYBR Green Master Mix (Thermo Fisher) and 4.5 ul cDNA (diluted 1:50) in 10 ul reactions, following manufacturer’s procedures. Negative controls lacking template were included for each primer set. All reactions were subjected to thermal melting to confirm that each reaction gave single peaks. For each transgenic line and time point, three biological replicates were performed, and transcript levels were normalized to mobk13 (mob4) (Hu et al., 2016; Gangras et al, 2019). Cycle thresholds (Ct) were determined using Bio-Rad CFX manager software. Changes in mRNA expression were calculated by ΔΔCt = ΔCt target − ΔCt control. Relative changes in mRNA expression levels are represented graphically as fold change, where relative mRNA fold change = 2−ΔΔCt. All graphs were generated using Prism 8.1 (GraphPad). For half-life calculations, 30 minutes pHS was set as “time 0” because our experiments revealed that heat shock induction continues temporarily after the downshift from 37ºC to 28.5ºC. To determine half-lives, we fitted exponential decay equations to normalized Venus mRNA levels across time for each heat shock experiment in the form y = ae−(bx), with y representing fold change, a representing fold change at the y intercept, b representing a decay constant, and x representing time. We determined the value for x when y = 0.5 using the online algebraic tool MathPapa at http://www.mathpapa.com. Three biological replicates per reporter line were used to calculate average half-lives and standard deviation.
Polyadenylation site determination
Polyadenylation (pA) site use for each reporter was determined using 3’-rapid amplification of cDNA ends. Briefly, 0.5 ug total RNA was reverse transcribed using an oligo-dT adapter primer and Superscript III reverse transcriptase (Thermo Fisher), followed by amplification using Taq polymerase (NEB) and a Venus-specific forward primer and universal reverse adapter primer (Table S2). Amplified products were gel purified and TOPO cloned according to manufacturer’s instructions (Thermo Fisher). pA site use was determined by plasmid restriction digestion with BamHI-HF (New England Biolabs) which generates an additional cleaved product of unique size when the SV40 pA rather than the endogenous her1 pA site is used. A subset of clones utilizing the SV40 and her1 pA sites were sequence analyzed to confirm that the digestion strategy accurately distinguishes between clones derived from use of the her1 or SV40 pA sites.
Microscopy and Imaging
In situ hybridized embryos were mounted in Permount and imaged using an Axiocam HRc digital camera with AxioPlan2 microscope (Zeiss). Immunofluorescent embryos were mounted in 80% glycerol and imaged at 20x magnification using MetaMorph software (Molecular Devices) on an Andor™ SpinningDisc Confocal Microscope (Oxford Instruments) with a Nikon Neo camera. Laser wavelength and intensity were set at 488 nm and 50%, respectively, and bit depth at 16-bit. Maximum intensity projections are shown (Fig. S1).
Immunohistochemistry
Standard immunohistochemistry protocols were followed using 4% PFA fixation, dehydration and rehydration in a methanol series, and incubation in blocking solution for 1 hour. Mid-segmentation embryos from wild-type and pnrc2oz22 crosses were incubated in 2% BSA/5% goat serum/0.1% Tween-20/PBS blocking solution containing 1:200 anti-zdc2 that recognizes Dlc protein (ab73336, Abcam) according to previously published methods (Giudicelli et al., 2007), followed by goat anti-mouse Alexa-Fluor-488 (1:800) (Thermo Fisher).
Plasmid construction and mRNA injection
Full-length pnrc2 cDNA was generated as previously described (Gallagher et al., 2017) to create plasmid SP6-pnrc2-cDNA (pTLG109). The Cerulean coding sequence was translationally fused to pnrc2 by overlap extension PCR (Horton et al., 2013) using Phusion polymerase (NEB), pTLG109 and pCS-H2B-Cerulean (Megason, 2009) as template in separate reactions, and primers that incorporate a flexible linker between Cerulean and pnrc2 coding sequences. Stitching of fragments was performed using Phusion polymerase (NEB), Cerulean and pnrc2 overlap fragments as templates, and outer primers complementary to the 5’ and 3’ ends of the Cerulean and pnrc2 coding sequences, respectively, containing restriction sites for subcloning into expression vector pCS2+ (Rupp et al., 1994; Turner and Weintraub, 1994) to generate SP6-Cerulean-pnrc2-cDNA (pTLG149). To generate mutant pnrc2 versions, site-directed mutagenesis (Hutchison et al., 1978) of pTLG109 was performed using KOD polymerase (EMD Millipore) and mutagenic primers, followed by DpnI (New England Biolabs) digestion and transformation into E. coli, creating SP6-pnrc2F138→stop-cDNA (pTLG137), SP6-pnrc2 K119→A-cDNA (pTLG138), and SP6-pnrc2 W124→A-cDNA (pTLG139). Mutant pnrc2 versions from pTLG137–139 were subcloned into pTLG149 to replace the wild-type pnrc2 coding sequence, creating SP6-Cerulean-pnrc2F138→stop-cDNA (pTLG151), SP6-Cerulean-pnrc2 K119→A-cDNA (pTLG152), and SP6-Cerulean-pnrc2 W124→A-cDNA (pTLG153). All plasmids were sequence validated. Primer sequences are listed in Table S2.
For rescue experiments, wild-type and mutant Cerulean-pnrc2 mRNAs were synthesized using the SP6 mMessage Machine Kit (Thermo Fisher), diluted in 0.2M KCl with 0.1% phenol red, and injected into 1-cell stage embryos (40 pg mRNA per embryo). To determine the minimal dose required for consistent rescue, we performed dose response experiments with wild-type Cerulean-pnrc2 mRNA and found that 40–100 pg doses rescued almost all MZpnrc2 mutants, whereas doses between 5–20 pg doses were less penetrant.
RESULTS
Zygotically-expressed and maternally-provided pnrc2 promotes cyclic gene transcript decay
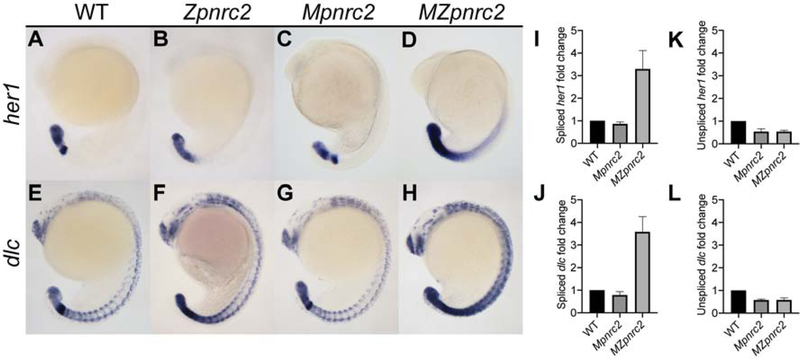
We previously showed that zygotically-expressed pnrc2 promotes decay of cyclic gene transcripts, including her1 and dlc, during somitogenesis (Gallagher et al., 2017). Because pnrc2 transcript is detected in wild-type (WT) embryos at early time points including the 8-cell stage (Gallagher et al., 2017), we hypothesized that maternally-provided pnrc2 transcript and/or Pnrc2 protein may partially compensate for loss of zygotic pnrc2 function during somitogenesis. To assess maternal contribution, we examined cyclic gene transcript expression in maternal-zygotic pnrc2oz22 (MZpnrc2) mutant embryos that lack both maternal and zygotic pnrc2 function, maternal pnrc2oz22 (Mpnrc2) mutant embryos that have only zygotic pnrc2 function, and zygotic pnrc2oz22 (Zpnrc2) mutant embryos that have only maternal pnrc2 function (Fig. 1). As shown previously (Gallagher et al., 2017), her1 and dlc transcripts are misexpressed in Zpnrc2 embryos (Fig. 1A vs B, E vs F). Although her1 and dlc expression appear normal in Mpnrc2 embryos (Fig. 1A vs C, E vs G), MZpnrc2 embryos show an enhanced phenotype compared to Zpnrc2 embryos (Fig. 1B vs D, F vs H). To confirm that Pnrc2 functions post-transcriptionally to negatively regulate cyclic gene expression, we quantitatively assessed her1 and dlc transcript levels in Mpnrc2 and MZpnrc2 mutant and wild-type sibling embryos using quantitative PCR (qPCR). To distinguish unspliced from spliced transcripts, we used primers that selectively amplify one or the other form of transcript, with the expectation that only spliced transcripts would be elevated in MZpnrc2 mutants. Indeed, spliced her1 and dlc transcript levels are elevated almost 4-fold in MZpnrc2 mutant embryos compared to Mpnrc2 mutant and wild-type sibling embryos (Fig. 1I and J). In contrast, unspliced her1 and dlc transcripts are down-regulated ~2-fold in MZpnrc2 compared to wild-type embryos (Fig. 1K and L), suggesting that cyclic expression might be transcriptionally reduced despite accumulation of cyclic gene transcripts or that splicing efficiency is increased in pnrc2-deficient embryos. Surprisingly, Mpnrc2 mutant embryos also show reduced unspliced cyclic gene transcript levels compared to wild-type embryos despite normal levels of spliced cyclic gene transcript, demonstrating that post-transcriptional accumulation of cyclic gene transcripts in MZpnrc2 mutant embryos is unrelated to transcriptional downregulation. Together, these data strongly support that Pnrc2 regulates cyclic gene expression at the post-transcriptional level, and extend our previous qualitative work (Dill and Amacher, 2005; Gallagher et al., 2017). Overall, these results suggest maternal and zygotic pnrc2 function promotes rapid turnover of her1 and dlc transcripts during somitogenesis.
Figure 1. Maternal and zygotic pnrc2 promotes proper her1 and dlc expression.
Wild-type (WT), zygotic pnrc2oz22 (Zpnrc2), maternal pnrc2oz22 (Mpnrc2), and maternal-zygotic pnrc2oz22 (MZpnrc2) mutant embryos were raised to mid-segmentation stage (16–18 hpf) and probed for her1 (A–D) and dlc expression (E–F) by in situ hybridization (n ≥ 7 each). WT, Mpnrc2 mutant, and MZpnrc2 mutant embryos (n = 10 per biological replicate) were analyzed by qPCR using primers to amplify across exon-exon boundaries to detect spliced her1 (I) and dlc (J) transcripts or primers to amplify across intron-exon boundaries to detect her1 (K) and dlc (L) unspliced transcripts. MZpnrc2 mutant embryos have ~4-fold higher levels of spliced her1 and dlc mRNA than wild-type or Mpnrc2 mutant embryos, which have comparable levels (I and J). Both MZpnrc2 and Mpnrc2 mutant embryos have ~2-fold less unspliced her1 and dlc transcripts compared to WT embryos (K and L). hpf = hours post-fertilization.
Unlike cyclic gene transcript, cyclic gene protein does not accumulate in MZpnrc2 mutant embryos
We previously showed that expression of the cyclic gene protein Dlc is unaffected in zygotic pnrc2 mutants (Gallagher et al., 2017). To assess whether the same is true for MZpnrc2 mutants, we examined endogenous Dlc expression by immunofluorescence. While dlc transcript levels are almost 4-fold higher in MZpnrc2 mutants than in wild-type embryos (Fig. 1J), there is no obvious corresponding increase in Dlc protein expression (Fig. S1). Thus, despite accumulation of cyclic gene transcripts in MZpnrc2 mutants, cyclic gene protein expression appears normal, which may explain why mutants lack a morphological segmentation phenotype.
The terminal 179 nucleotides of the her1 3’UTR are necessary and sufficient for Pnrc2-mediated decay of reporter transcripts
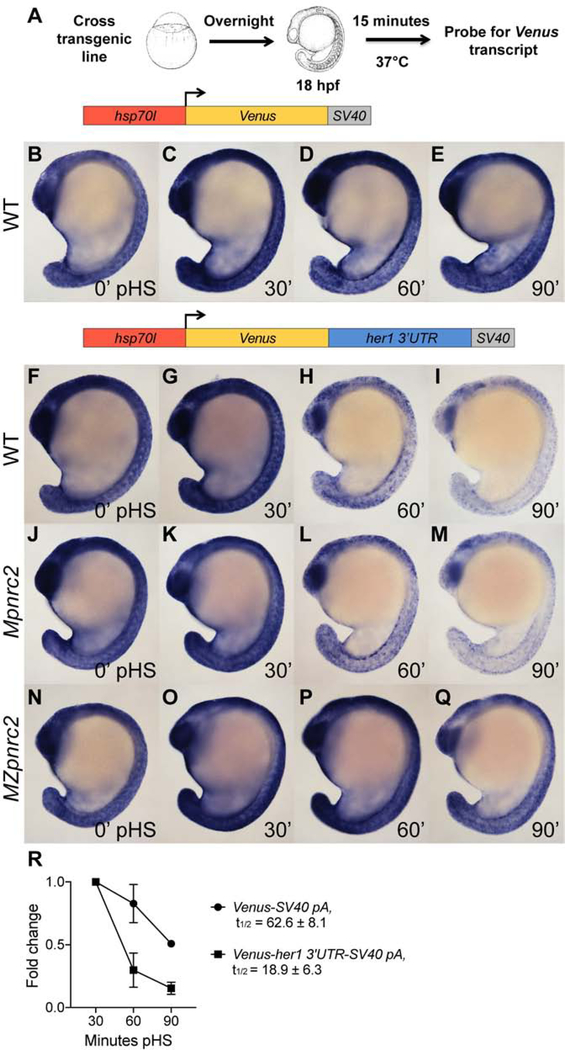
Previous studies have shown that reporter transcripts containing the her1 3′UTR are rapidly degraded (Gallagher et al., 2017; Giudicelli et al., 2007), and that rapid decay requires Pnrc2 function (Gallagher et al., 2017). To identify regulatory elements within the her1 3’UTR that are required for Pnrc2-mediated decay, we modified our transgenic heat shock-inducible reporter (Gallagher et al., 2017), generated stable transgenic lines driving expression of Venus transcript followed by various portions of the her1 3’UTR, and performed heat shock induction assays (Fig. 2A). Because a subset of reporter lines lacks the endogenous her1 polyadenylation (pA) signal, we included an SV40 pA signal on all reporters, including the full-length her1 3’UTR, to ensure that all reporters contained at least one bona fide pA signal. As a control for Venus transcript stability, a transgenic line driving inducible expression of Venus transcript with an SV40 pA signal (Venus-SV40 pA) was tested in parallel. Reporter mRNA containing the full-length her1 3’UTR (Venus-her1 3’UTR-SV40 pA) is rapidly decayed between 30 to 90 minutes post-heat shock (pHS) when compared to reporter mRNA lacking the her1 3’UTR sequence (Venus-SV40 pA) by in situ hybridization (Fig. 2B–I), confirming that the full-length her1 3’UTR is sufficient to confer rapid instability. Consistent with in situ hybridization data, qPCR analysis reveals that reporter half-life between 30 to 90 minutes pHS is >3-fold lower when the her1 3’UTR is present (Fig. 2R) and that this effect is consistent across three independent lines (Fig. S2). To test whether Pnrc2 is required for rapid reporter decay, we performed the same heat shock experiments in Mpnrc2 and MZpnrc2 mutant embryos carrying transgenic reporters. Wild-type and Mpnrc2 mutant embryos expressing Venus-her1 3’UTR-SV40 pA reporter mRNA show comparable expression patterns across all pHS time points (Fig. 2F–M). In contrast, reporter mRNA in MZpnrc2 mutant embryos perdures (Fig. 2N–Q), indicating that Pnrc2-mediated decay of her1 transcripts occurs through destabilizing features of the her1 3’UTR.
Figure 2. The her1 3’UTR confers Pnrc2-mediated instability to reporter transcripts.
(A) Diagram illustrating the heat shock protocol used for transgenic lines in this study. (B–I) Transgenic embryos carrying the hsp70l:Venus-her1 3’UTR-SV40 pA reporter (line oz44) or hsp70l:Venus-SV40 pA reporter (line oz68) were raised to mid-segmentation stage, heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 7 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 per reporter line) (data not shown). (J–Q) Mid-segmentation stage Mpnrc2 mutant embryos and MZpnrc2 mutant embryos carrying the hsp70l:Venus-her1 3’UTR-SV40 pA reporter (line oz44) were heat shocked and processed by Venus in situ hybridization as above (n ≥ 8 embryos per time point). Representative embryos were genotyped post-imaging to confirm genotype. (R) qPCR analysis comparing Venus transcript fold change from 30 minutes pHS to 60 and 90 minutes pHS for the Tg(hsp70l:Venus-her1 3’UTR-SV40 pA)oz44 and Tg(hsp70l:Venus-SV40 pA)oz68 reporter lines (n = 10 embryos per time point across three biological replicates). Three independent lines carrying the hsp70l:Venus-her1 3’UTR-SV40 pA reporter and five independent lines carrying the hsp70l:Venus-SV40 pA reporter were analyzed in wild-type embryos by in situ hybridization and exhibited comparable Venus decay across all lines carrying the same reporter (data not shown); one representative line for each is shown (see Methods for details). For the hsp70l:Venus-her1 3’UTR-SV40 pA reporter, three independent lines were analyzed by qPCR and exhibited comparable decay (Fig. S2). pHS = post-heat shock; hpf = hours post-fertilization; t1/2 = half-life; ± = standard deviation; pA = polyadenylation sequence.
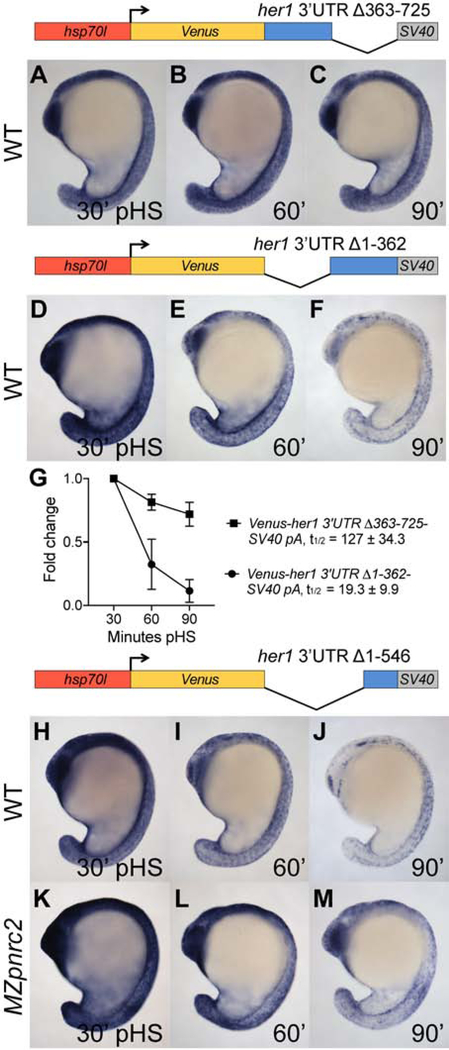
To define regions of the her1 3’UTR that are necessary and sufficient for Pnrc2-dependent decay, we conducted a deletion analysis of the her1 3’UTR. Transgenic lines were generated with hsp70l promoter sequence driving expression of Venus coding sequence fused to the first 362 nts or the last 363 nts of the her1 3’UTR followed by an SV40 pA. Reporter induction was performed and analyzed as in Fig. 2A. By in situ hybridization, reporter transcripts containing the first 362 nts of the her1 3’UTR display minimal destabilization by 90 minutes pHS, whereas reporter transcripts containing the last 363 nts of the her1 3’UTR are rapidly degraded (Fig. 3A–F). Correspondingly, qPCR analysis reveals that the reporter containing the terminal half of the her1 3’UTR has a short half-life that is very similar to that of the reporter containing the full-length her1 3’UTR and that the reporter containing the first half of the her1 3’UTR is >6-fold more stable (Figs. 2R and 3G). These data indicate that the terminal half of the her1 3’UTR is necessary and sufficient to confer instability to reporter transcripts.
Figure 3. The terminal 179 nucleotides of the her1 3’UTR is sufficient for Pnrc2-mediated decay of reporter transcripts.
(A–F) Transgenic embryos carrying the hsp70l:Venus-her1 3’UTRΔ363–725-SV40 pA reporter (line oz54) or hsp70l:Venus-her1 3’UTRΔ1–362-SV40 pA reporter (line oz47) were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 6 per time point). Venus transcript is not detected in the absence of heat shock (n = 10 per reporter line) (data not shown). (G) qPCR analysis comparing Venus transcript fold change from 30 minutes pHS to 60 and 90 minutes pHS for the Tg(hsp70l:Venus-her1 3’UTRΔ363–725-SV40 pA)oz54 or Tg(hsp70l:Venus-her1 3’UTRΔ1–362-SV40 pA)oz47 reporter lines (n = 10 embryos per time point across three biological replicates). (H–M) Mid-segmentation stage wild-type (WT) and MZpnrc2 mutant embryos carrying the hsp70l:Venus-her1 3’UTRΔ1–546-SV40 pA reporter (line oz50) were heat shocked and processed by Venus in situ hybridization (n ≥ 7 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 wild-type embryos) (data not shown). Representative embryos were genotyped post-imaging to confirm genotype. For each reporter, three independent lines were analyzed in wild-type embryos by in situ hybridization and exhibited comparable Venus decay across all lines carrying the same reporter (data not shown); one representative line for each is shown (see Methods for details). pHS = post-heat shock; t1/2 = half-life; ± = standard deviation; pA = polyadenylation sequence.
To elucidate specific destabilizing features of the her1 3’UTR, we generated heat shock inducible transgenic lines containing the last quarter of the her1 3’UTR. Reporter transcripts containing the last 179 nts of the her1 3’UTR are rapidly degraded, similar to reporters containing the full-length or the last half of the her1 3’UTR (Fig. 2F–I and Fig. 3D–F vs Fig. 3H–J). Collectively, all of our her1 3’UTR deletion reporters indicated that instability elements are located near the end of the her1 3’UTR (Fig. 4). To investigate whether the terminal 179 nt her1 3’UTR is Pnrc2-dependent, we examined reporter decay in MZpnrc2 mutant embryos and observed that the reporter persists longer than in wild-type embryos (Fig. 3H–M) or Mpnrc2 mutant controls (data not shown). Even though rapid decay of the reporter containing the terminal 179 nts is similar to that containing the full-length her1 3’UTR (Figs. 2F–I and 3H–J), the two reporters are differentially affected upon loss of Pnrc2 function (Figs. 2N–Q and 3K–M). Loss of Pnrc2 function stabilizes full-length reporter transcripts to a greater extent than that observed for reporters containing the terminal 179 nts, suggesting that additional pnrc2-dependent destabilizing elements lie upstream of the terminal 179 nts. Because the terminal 179 nts are necessary and sufficient to trigger reporter decay in wild-type embryos (Fig. 4), such upstream pnrc2-dependent elements could only elicit decay when the terminal 179-nt region is present.
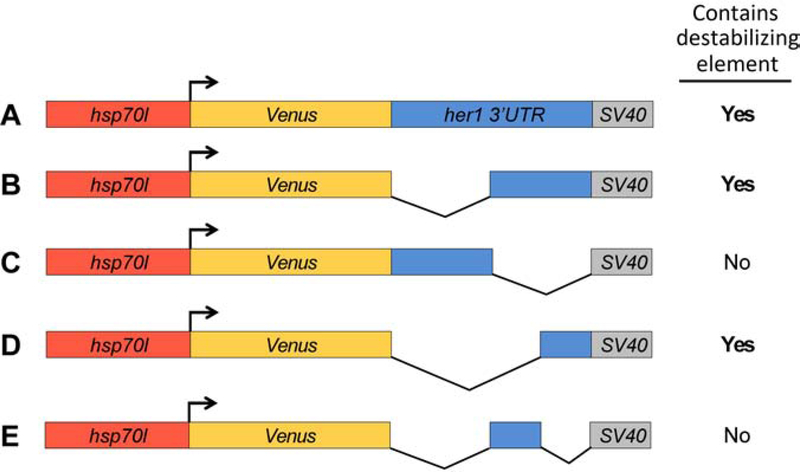
Figure 4. Transgenic reporters reveal that the terminal her1 3’UTR is necessary and sufficient to confer transcript destabilization.
Summary of reporter destabilization in transgenic embryos carrying various derivatives of the her1 3’UTR (A–E). All lines were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected and processed by Venus in situ hybridization at 0, 30, 60, and 90 minutes pHS. (A) hsp70l:Venus-her1 3’UTR-SV40 pA reporter line (see Figs. 2 and 6). (B) hsp70l:Venus-her1 3’UTRΔ1–362-SV40 pA reporter line (see Fig. 3). (C) hsp70l:Venus-her1 3’UTRΔ363–725-SV40 pA reporter line (see Fig. 3). (D) hsp70l:Venus-her1 3’UTRΔ1–546-SV40 pA reporter line (see Fig. 3). (E) hsp70l:Venus-her1 3’UTRΔ1–362; Δ547–725-SV40 pA reporter line (data not shown). Three independent lines were analyzed in wild-type embryos by in situ hybridization for each reporter, except for the hsp70l:Venus-her1 3’UTRΔ1–362; Δ547–725-SV40 pA reporter (E) for which two independent lines were analyzed. Each reporter exhibited comparable Venus decay across all independent lines (data not shown); see Methods for details. pHS = post-heat shock.
An alternative possibility is that Pnrc2-independent stabilizing features are present in the first 556 nts of the her1 3’UTR that are suppressed or overcome by a Pnrc2-dependent destabilizing element in the terminal 179 nts. The activity of such stabilizing features could only be unmasked when Pnrc2 is absent, leading to greater stabilization of the full-length versus the terminal 179 nt reporter in MZpnrc2 mutant embryos.
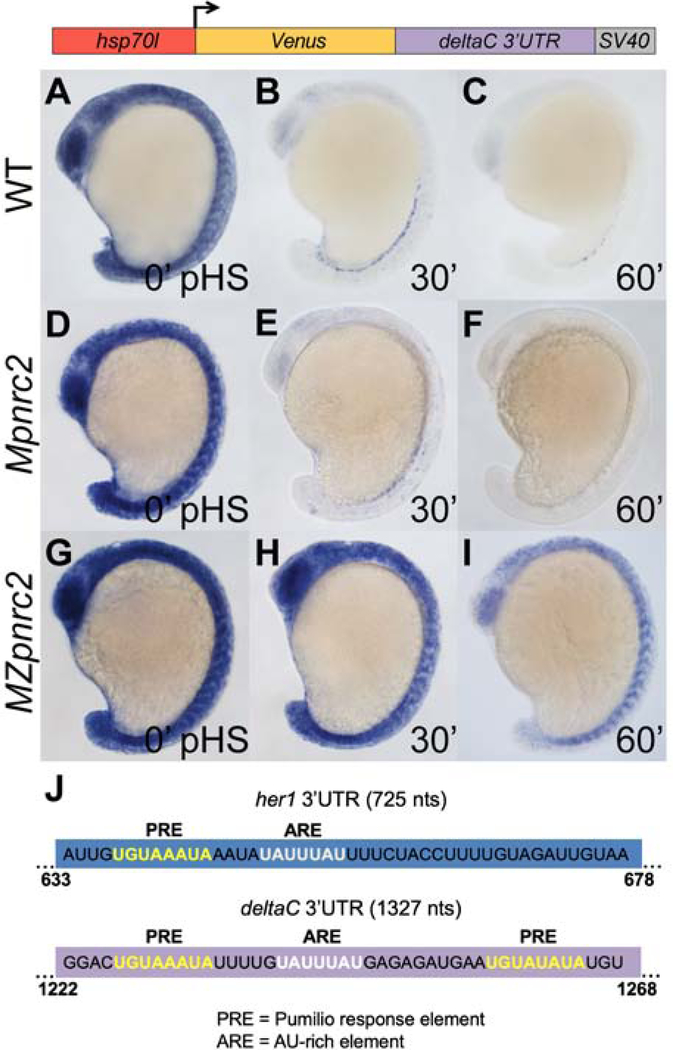
The dlc 3’UTR confers instability to reporter transcripts in a Pnrc2-dependent manner
Pnrc2 function is important for proper expression of cyclic genes her1 and dlc (Dill and Amacher, 2005; Gallagher et al., 2017). To determine if the dlc 3’UTR, like the her1 3’UTR, contains features that promote Pnrc2-dependent decay, we used our heat shock-inducible system to drive expression of Venus transcripts containing the full-length 1327 nt dlc 3’UTR and SV40 pA sequence (Venus-dlc 3’UTR-SV40 pA) and compared reporter decay among wild-type, Mpnrc2 mutant, and MZpnrc2 mutant embryos at 0, 30, and 60 minutes pHS (Fig. 5). We find that reporters in wild-type and Mpnrc2 mutant embryos are rapidly decayed (Fig. 5A–C vs D–F). In contrast, reporter expression in MZpnrc2 mutants is stabilized relative to wild-type and Mpnrc2 mutant embryos (Fig. 5A–F vs G–I). These data reveal that the dlc 3’UTR promotes Pnrc2-dependent transcript decay and suggests cyclic gene transcripts may share common Pnrc2-dependent 3’UTR decay features.
Figure 5. The dlc 3’UTR confers Pnrc2-mediated decay to reporter transcripts.
(A–C) Transgenic embryos carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter (line oz81) were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 6 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 embryos) (data not shown). (D–I) Mpnrc2 (D–F) and MZpnrc2 mutant embryos (G–I) carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter (line oz81) were also heat shocked and processed for Venus transcript (n ≥ 10 embryos per time point). Representative embryos were genotyped post-imaging to confirm genotype. (J) RBPmap motif analysis (Paz et al., 2014) identifies a canonical PRE (5’UGUAAAUA; yellow) and a canonical ARE (5’UAUUUAU; white) near the end of the her1 3’UTR and two PREs and one ARE near the end of the dlc 3’UTR. The indicated PRE and ARE are the only such motifs in the 725 nt full-length her1 3’UTR, whereas the 1327 nt dlc 3’UTR contains an additional PRE and two additional AREs. Three independent lines carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter were analyzed in wild-type embryos by in situ hybridization and exhibited comparable Venus decay across all three lines (data not shown); one representative line is shown (see Methods for details). pHS = post-heat shock; nts = nucleotides; PRE = Pumilio response element; ARE = AU-rich element; pA = polyadenylation sequence.
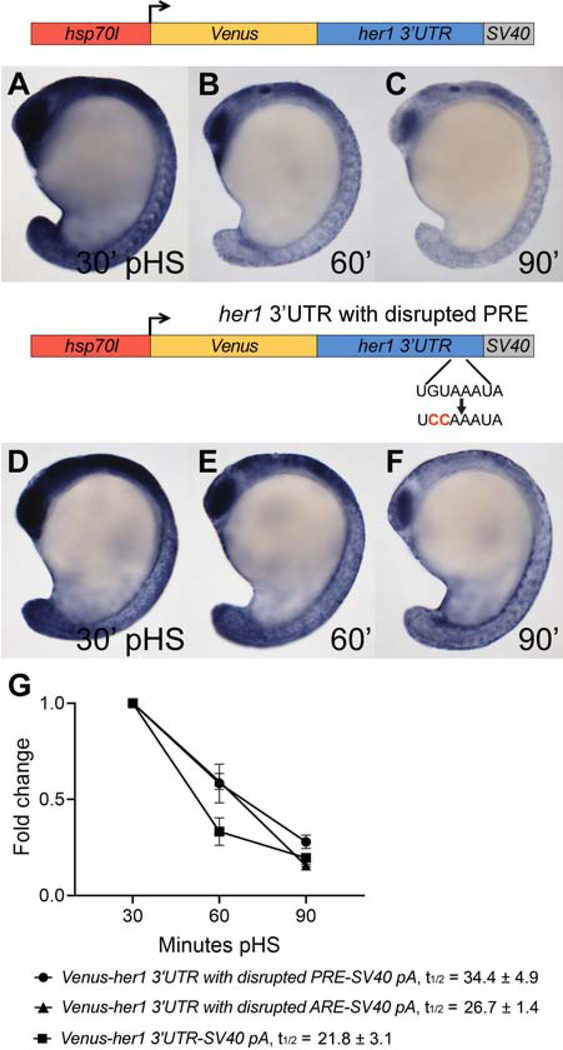
Pumilio response and AU-rich elements in the her1 3’UTR promote transcript decay
To determine if her1 and dlc 3’UTRs contain shared motifs, we compared the terminal 179 nt her1 3’UTR with the dlc 3’UTR using RBPmap motif analysis (Paz et al., 2014) and manual inspection. Both share two perfect matches to motifs that are not present in the first 75% (1–546 nts) of the her1 3’UTR: a Pumilio response element (PRE) and an AU-rich element (ARE) (Fig. 5J). The terminal her1 3’UTR contains a single 5’UGUAHAUA PRE and a single 5’UAUUUAU ARE while the full-length dlc 3’UTR contains three 5’UGUAHAUA PREs and three 5’UAUUUAU AREs. Because PREs and AREs are associated with mRNA instability (Goldstrohm et al., 2018; Schoenberg and Maquat, 2012), we determined whether they contribute to cyclic gene transcript turnover by introducing mutations within the context of the full-length her1 3’UTR reporter and comparing transcript stability to that of the unmodified reporter. The PRE was disrupted by changing PRE base positions 2 and 3 from GU to CC, a replacement that disrupts the ability of human PUMILIO proteins to bind target mRNAs (Miles et al., 2015). By in situ hybridization, reporter mRNA with the PRE mutation appears partially stabilized relative to unmodified reporter mRNA (Fig. 6A–F). Correspondingly, qPCR analysis reveals that the PRE mutation increases reporter half-life by ~1.6-fold when compared to the full-length, unmodified reporter (Fig. 6G) and that these results are consistent across three independent lines (Fig. S3). To determine if the her1 ARE promotes decay of reporter transcript, ARE positions 3–5 were changed from UUU to CCC, a replacement that disrupts ARE-binding protein association with target mRNAs (Lai et al., 2005). qPCR analysis reveals that the ARE mutation increases reporter half-life, although this affect is modest relative to the PRE mutation (Fig. 6G). Reporter stabilization due to ARE mutation is consistent across three independent lines (Fig. S4).
Figure 6. The Pumilio response and AU-rich elements in the her1 3’UTR contribute to reporter transcript turnover.
(A–F) Transgenic embryos carrying the hsp70l:Venus-her1 3’UTR-SV40 pA reporter (line oz44) or the hsp70l:Venus-her1 3’UTR with disrupted PRE-SV40 pA reporter (line oz71) with a 2 nt mutation in the PRE sequence were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 11 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 per reporter line) (data not shown). (G) qPCR analysis comparing Venus transcript fold change from 30 minutes pHS to 60 and 90 minutes pHS for the reporter lines Tg(hsp70l:Venus-her1 3’UTR-SV40 pA)oz44, Tg(hsp70l:Venus-her1 3’UTR with disrupted PRE-SV40 pA)oz71, and Tg(hsp70l:Venus-her1 3’UTR with disrupted ARE-SV40 pA)oz75 (n = 10 embryos per time point across three biological replicates). The PRE mutation changes the 5’UGUAAAUA site to 5’UCCAAAUA and the ARE mutation changes the 5’UAUUUAU site to 5’UACCCAU. Both mutations extend reporter half-life; the PRE-mutated reporter half-life is increased 1.7-fold and the ARE-mutated reporter half-life is increased 1.6-fold. Five independent lines carrying the hsp70l:Venus-her1 3’UTR with disrupted PRE-SV40 pA reporter and four independent lines carrying the hsp70l:Venus-her1 3’UTR with disrupted ARE-SV40 pA reporter were analyzed in wild-type embryos by in situ hybridization and exhibited comparable Venus decay across all lines carrying the same reporter (data not shown); one representative line for each is shown (see Methods for details). For each reporter, three independent lines were chosen for qPCR analysis and exhibited comparable Venus decay (Figs. S3–S4). pHS = post-heat shock; PRE = Pumilio response element; ARE = AU-rich element; t1/2 = half-life; ± = standard deviation; pA = polyadenylation sequence.
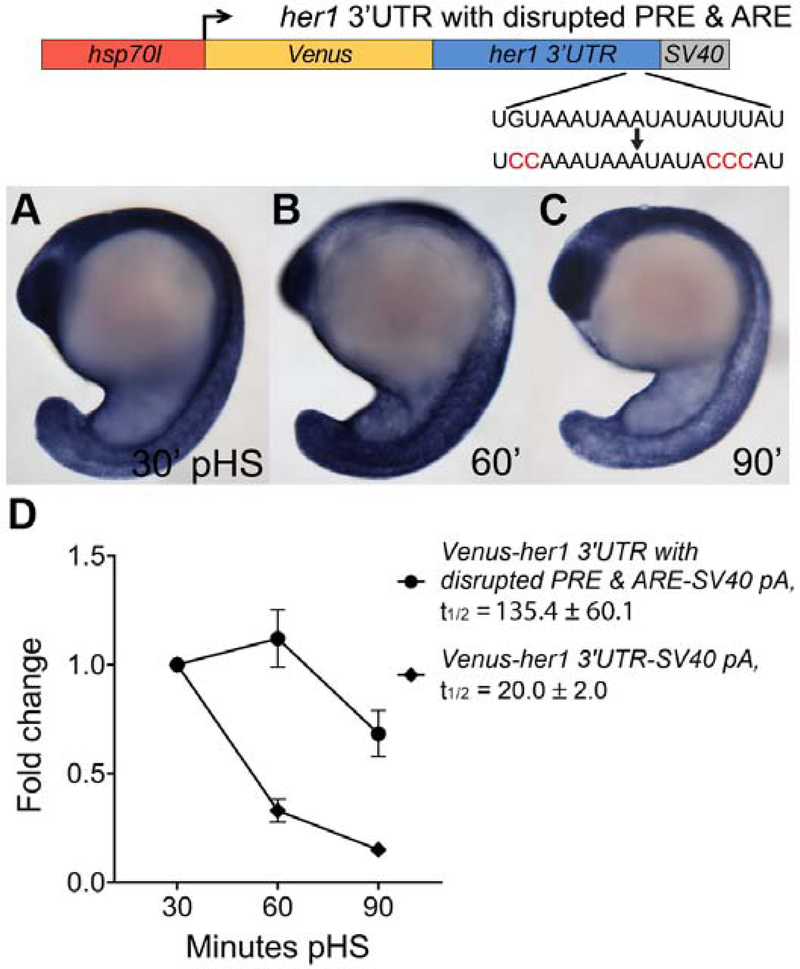
We next investigated the combinatorial activity of the PRE and ARE motifs on reporter decay by introducing both PRE and ARE mutations in the full-length her1 3’UTR reporter and comparing transcript stability to that of the unmodified reporter. By in situ hybridization, reporter mRNA with both PRE and ARE mutations appear dramatically stabilized (Fig. 7A–C). Correspondingly, qPCR analysis reveals that combined PRE and ARE mutations increase reporter half-life ~7-fold when compared to the full-length, unmodified reporter (Fig. 7D), and that these results are consistent across three independent lines (Fig. S5).To address the possibility that differences in pA site use between reporters might influence mRNA stability, we analyzed the 3’ ends of reporter transcripts and find that the unmodified and PRE-mutated her1 3’UTR reporters primarily use the SV40 pA, whereas reporters with disrupted ARE and disrupted PRE & ARE use either the SV40 or natural her1 pA with similar frequency (Table S1). Because reporters with a disrupted ARE motif exhibit a shift in pA site preference, we quantified Venus transcripts that use the SV40 pA and find no differences in decay when compared to quantification of all Venus transcripts regardless of pA site use for each reporter (Fig. S6). Taken together, our analysis of single- and double-mutated reporters suggests that the PRE and ARE in the her1 3’UTR both contribute to transcript decay and together these motifs dramatically destabilize reporter mRNA.
Figure 7. The Pumilio response and AU-rich elements in the her1 3’UTR are both required for rapid reporter transcript turnover.
(A–C) Transgenic embryos carrying the hsp70l:Venus-her1 3’UTR with disrupted PRE & ARE-SV40 pA reporter (line oz93) with a 2 nt mutation in the PRE sequence and 3 nt mutation in the ARE sequence were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 11 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 embryos) (data not shown). (D) qPCR analysis comparing Venus transcript fold change from 30 minutes pHS to 60 and 90 minutes pHS for the reporter lines Tg(hsp70l:Venus-her1 3’UTR-SV40 pA)oz44 and Tg(hsp70l:Venus-her1 3’UTR with disrupted PRE & ARE-SV40 pA)oz93 (n = 10 embryos per time point across three biological replicates). The presence of both mutations extends reporter half-life by ~7-fold compared to the unmutated control. Three independent lines carrying the hsp70l:Venus-her1 3’UTR with disrupted PRE & ARE-SV40 pA reporter were analyzed by in situ hybridization and qPCR and each line exhibited comparable Venus decay dynamics (data not shown and Fig. S5); one representative line is shown (see Methods for details). pHS = post-heat shock; PRE = Pumilio response element; ARE = AU-rich element; t1/2 = half-life; ± = standard deviation; pA = polyadenylation sequence.
Pnrc2 may promote cyclic gene transcript decay by interacting with known decay pathway components
While our analysis has identified cis-elements and potential trans-factors that promote cyclic gene transcript decay, it is still unclear how they interact with transcript decay machinery. Pnrc2 lacks an obvious RNA-binding domain, but does contain C-terminal SRC-Homology 3 (SH3) and Nuclear Receptor-box (NR-box) domains that are 100% identical to human PNRC2 (Gallagher et al., 2017). Human PNRC2 interacts with the mRNA decapping factor DCP1A through the SH3 domain and with the nonsense-mediated mRNA decay (NMD) effector UPF1 through the NR-box domain (Albers et al., 2005; Cho et al., 2009; Lai et al., 2012; Loh et al., 2013; Mugridge et al., 2016). Mutation of W114A in the SH3 domain abrogates PNRC2 binding to DCP1A and deletion of the NR-box abrogates PNRC2 binding to UPF1, and these interactions are required to elicit decay of reporter mRNA (Lai et al., 2012; Nicholson et al., 2018). We hypothesized the highly conserved SH3 and/or NR-box domains of Pnrc2 might also be essential for Pnrc2-mediated decay of cyclic gene transcripts. We injected mRNA encoding wild-type or mutant Pnrc2 translationally fused to Cerulean fluorescent protein into MZpnrc2 mutants and assayed rescue (Table 1). Whereas injection of Cer-pnrc2 mRNA restores proper her1 expression in MZpnrc2 mutants, injection of Cer-pnrc2 mRNA containing a W124A mutation (orthologous to the human W114A mutation) into MZpnrc2 mutants did not rescue her1 expression (Table 1). Cer-pnrc2 lacking the NR-box retains partial function and restores her1 expression in 63% of injected MZpnrc2 mutant embryos (Table 1). Overall, these results suggest that interaction of Pnrc2 with known decay machinery is necessary for turnover of cyclic gene transcripts.
Table 1.
Pnrc2 SH3 domain and NR-box are important for cyclic gene transcript decay
| Condition | her1 expression a | ||
|---|---|---|---|
| Normal b | Accumulated b | Percent affected | |
| Uninjected | 14/29 | 15/29 | 51.7% |
| Cerulean-pnrc2 | 48/49 | ¼9 | 2.0% |
| Cerulean-pnrc2ΔNR (c) | 22/27 | 5/27 | 18.5% |
| Cerulean-pnrc2W->A (d) | 26/50 | 24/50 | 52.0% |
At 16–18 hpf, pnrc2 mRNA-injected embryos from a cross of a pnrc2 homozygous female to a pnrc2 heterozygous male were processed by her1 in situ hybridization. The cross yields 50% Mpnrc2 mutants with normal her1 expression and 50% MZpnrc2 mutants with accumulated her1 expression unless there is phenotypic rescue by the injected pnrc2 mRNA.
Chi-square analysis indicates a significant difference in her1 expression between Cerulean-pnrc2 mRNA-injected and uninjected MZpnrc2 mutant embryos (p < 0.0001) and between Cerulean-pnrc2ΔNR mRNA-injected and uninjected MZpnrc2 mutant embryos (p = 0.001) and no significant difference between Cerulean-pnrc2W->A mRNA-injected and uninjected MZpnrc2 mutant embryos (p = 0.7773).
ΔNR = deletion of NR box
W->A = W124A mutation
DISCUSSION
In this work, we show that Pnrc2 promotes 3’UTR-mediated mRNA decay of cyclic gene transcripts her1 and dlc. This work builds upon previous studies demonstrating that Pnrc2 promotes decay of PTC-containing transcripts (Cho et al., 2013a; Cho et al., 2009; Lai et al., 2012) and non-PTC-containing transcripts (Gallagher et al., 2017; Nicholson et al., 2018) and identifies mRNA elements that promote Pnrc2-mediated decay. We show here that the terminal 179 nts of the her1 3’UTR are necessary and sufficient for Pnrc2-dependent decay of non-PTC-containing reporter mRNA in vivo. We also show that the full-length dlc 3’UTR confers Pnrc2-mediated decay to reporter transcripts, demonstrating that elements within cyclic gene transcript 3’UTRs are sufficient for Pnrc2-mediated decay. Both the her1 and dlc 3’UTRs contain at least one PRE and ARE and disruption of either or both motifs in the her1 3’UTR stabilizes reporter mRNA, raising the possibility that Pnrc2 promotes decay via Pumilio proteins and/or ARE-BPs. Human PNRC2 is implicated in mRNA turnover through interactions with decay factors SMG5, DCP1A, UPF1, and STAU1 (Cho et al., 2013a; Cho et al., 2013b; Cho et al., 2012; Cho et al., 2009; Cho et al., 2015; Lai et al., 2012; Mugridge et al., 2016; Nicholson et al., 2018), and our work suggests Pnrc2 interacts with Dcp1a and Upf1 to promote cyclic gene transcript turnover.
Maternal pnrc2 contributes to cyclic gene transcript turnover
Zebrafish utilize a large contribution of maternally-provided mRNA and protein (Tadros and Lipshitz, 2009). We show here that maternal deposition of pnrc2 partially compensates for loss of zygotic pnrc2 during somitogenesis. Embryos without maternal and zygotic pnrc2 function display enhanced accumulation of her1 and dlc transcripts compared to embryos lacking only zygotic pnrc2 function (Fig. 1). Interestingly, loss of maternal pnrc2 function alone has no consequence on her1 and dlc transcript levels, showing that zygotic pnrc2 expression is sufficient to clear cyclic gene transcripts during somitogenesis (Figs. 1, 2, and 5). Incomplete compensation of Zpnrc2 mutants by maternal pnrc2 function is likely caused by eventual depletion of maternal stores during somitogenesis, but differences in post-transcriptional processing of zygotic pnrc2 and maternal pnrc2 transcripts, such as splicing and/or translation, may also contribute.
Other cyclic gene transcript 3’UTRs contain Pnrc2-dependent instability elements
Our previous work demonstrated that the full-length her1 3’UTR is sufficient to confer Pnrc2-mediated decay (Gallagher et al., 2017), but it was unknown if other cyclic gene transcript 3’UTRs similarly confer Pnrc2-mediated decay. We show here that the full-length dlc 3’UTR is also capable of destabilizing transcripts in a Pnrc2-dependent manner (Fig. 5), suggesting a conserved mechanism of Pnrc2-dependent destabilization may regulate cyclic gene transcripts through 3’UTR interactions. The 1327 nt dlc 3’UTR is almost twice the length of the 725 nt her1 3’UTR, and more rapid destabilization of reporter transcripts by the dlc 3’UTR may be attributed to decay factors such as UPF1 that display preference for longer 3’UTRs (Hogg and Goff, 2010), or to the presence of additional decay elements. Identification of additional Pnrc2-regulated destabilizing 3’UTRs will facilitate the identification and analysis of conserved cyclic gene 3’UTR decay features and determine if 3’UTR length is a factor in Pnrc2-mediated decay.
PRE and ARE motifs are found in multiple cyclic gene transcripts
Transcriptome-wide analyses show that the PRE is among the features most strongly correlated with mRNA instability (Schwanhausser et al., 2011; Sharova et al., 2009; Yang et al., 2003), and a recent study found global enrichment of PREs and AREs in the 3’UTRs of maternal transcripts rapidly degraded during zebrafish embryogenesis (Rabani et al., 2017). Our work shows that disruption of the PRE and ARE in the her1 3’UTR affects destabilization of reporter transcripts (Fig. 6 and Fig. 7), suggesting these motifs may regulate instability of endogenous her1 transcripts. Cyclic gene transcripts such as dlc and her7 and the clock-associated transcript dld also contain PRE and ARE motifs (Fig. 5J and data not shown), suggesting PRE and ARE motifs may function in the regulation of multiple cyclic gene transcripts during somitogenesis. The dlc 3’UTR appears to confer more rapid destabilization of reporter transcripts than the her1 3’UTR, and this may be due to the presence of multiple PREs and AREs in the dlc 3’UTR, allowing for more efficient recruitment of decay factors. Additionally, unidentified decay-promoting elements, such as sequence-specific RNA binding protein motifs or decay-inducing secondary structures, may be present. The terminal 179 nt her1 3’UTR and the dlc 3’UTR both contain a canonical 5’UGCUGU Muscleblind-like 1 (MBNL1) binding motif, and MBNL1 is known to promote mRNA turnover through 3’UTR interactions (Masuda et al., 2012; Wang et al., 2015). Additionally, human PNRC2 interacts with STAU1, a well-known RNA binding protein that recognizes RNA hairpin structures, rather than sequence-specific elements, to promote STAU1-mediated decay (Cho et al., 2012). Further her1 3’UTR mutagenesis may identify additional features, and precise mutagenesis of such 3’UTR regulatory features within the endogenous gene will reveal their respective contributions to cyclic gene expression.
Pumilio proteins and ARE-binding proteins may regulate cyclic gene transcript expression during somitogenesis
We show that disruption of either the her1 3’UTR PRE or ARE extends reporter transcript half-life (Fig. 6G) and that disruption of both PRE and ARE motifs dramatically extends reporter transcript half-life (Fig. 7D). Because mutation of the PRE or ARE alone is not sufficient to fully stabilize reporter mRNA, these motifs likely promote decay in parallel. PREs are well-studied binding sites for Pumilio proteins across many species (Filipovska et al., 2011; Morris et al., 2008; Van Etten et al., 2012; Wang et al., 2002). Pumilio proteins function in diverse biological processes (Chen et al., 2012; Gennarino et al., 2018; Gennarino et al., 2015; Mak et al., 2016; Siemen et al., 2011; Xu et al., 2007; Zahr et al., 2018; Zhang et al., 2017), and Pumilio dysfunction has been linked to diseases such as neurodegeneration and cancer (Gennarino et al., 2018; Gennarino et al., 2015; Kopp et al., 2019; Miles et al., 2016; Naudin et al., 2017). With rare exceptions, Pumilio-regulated transcripts are destabilized by Pumilio (Goldstrohm et al., 2018). In contrast, of the many characterized ARE-BPs, some function to stabilize target transcripts and others to destabilize transcripts. Examples of well-studied destabilizing ARE-BPs are the ARE/poly(U)-binding/degradation factor 1 (AUF1), tristetrapolin (TTP), and KH-type splicing regulatory protein (KSRP) (Briata et al., 2005; Gratacos and Brewer, 2010; Lykke-Andersen and Wagner, 2005; Petryszak et al., 2016; Sanduja et al., 2011). Zebrafish pumilio orthologs, pum1 and pum2, and the ARE-BP orthologs auf1, ksrp, ttp, and tia-1 are expressed throughout somitogenesis (Petryszak et al., 2016), and are thus candidate regulators of cyclic gene transcript decay.
One way that Pumilio proteins promote transcript decay is by recruiting the major deadenylation machine, the Ccr4-Not (CNOT) complex, to transcript 3’UTRs (Goldstrohm et al., 2006; Joly et al., 2013; Lau et al., 2009; Van Etten et al., 2012; Weidmann et al., 2014; Arvola et al, 2019). Similar to Pumilio-mediated repression, deadenylation is also the first and rate-limiting step in degradation of many ARE-containing mRNAs (Brewer and Ross, 1988; Laird-Offringa et al., 1990; Lieberman et al., 1992; Peppel and Baglioni, 1991; Shyu et al., 1991; Wilson and Treisman, 1988). A recent zebrafish study found that inhibiting the CNOT complex causes increased her1 and her7 transcripts, suggesting that her1 and her7 3’UTRs may recruit the CNOT deadenylase complex (Fujino et al., 2018). Together, our work and the work of others suggests that Pumilio proteins and/or ARE-BPs recruit the CNOT deadenylase complex to the her1 3’UTR and other cyclic gene transcript 3’UTRs.
In addition to promoting transcript decay, Pumilio proteins can also repress translation by antagonizing poly(A)-binding protein (PABP) function (Chritton and Wickens, 2011; Van Etten et al., 2012; Weidmann et al., 2014). ARE-BPs such as TIA-1 and TIAR have also been found to inhibit translation (Dixon et al., 2003; Gueydan et al., 1999; Piecyk et al., 2000). Translational repression conferred by PRE- and/or ARE-binding proteins may explain why cyclic gene protein levels are normal in Zpnrc2 and MZpnrc2 mutant embryos (Fig. S1) (Gallagher et al., 2017).
Cyclic gene transcript 3’UTR elements may confer Pnrc2-mediated decay by influencing decapping
In addition to deadenylation, removal of the 5’ cap facilitates mRNA destabilization by providing access to 5’ to 3’ exonucleases (Schoenberg and Maquat, 2012). PNRC2 has previously been shown to interact with DCP1A in human cells (Cho et al., 2009; Lai et al., 2012; Mugridge et al., 2016), which interacts directly with DCP2 to remove the 5’ cap (She et al., 2008). Our work shows that the orthologous residue in the SH3 domain, which was shown to promote binding to DCP1A in human cells, is important for promoting Pnrc2-mediated turnover of her1 mRNA. This suggests that zebrafish Pnrc2 may recruit decapping factors to cyclic gene transcripts to initiate degradation. Studies also suggest that Pumilio proteins and ARE-BPs can regulate gene expression through interactions with the 5’ cap. In Xenopus, Pum2 has been shown to regulate translation by directly interacting with the 5’ cap (Cao et al., 2010). In Drosophila cells, knockdown of the decapping factor Dcp2 abrogates Pum-mediated mRNA decay and translational repression (Arvola, 2019). In HeLa cells, the ARE-binding protein TTP associates with decapping factors, and the presence of an ARE in mRNA stimulates decapping (Gao et al., 2001; Lykke-Andersen and Wagner, 2005). Our data suggests interaction of Pnrc2 with Dcp1a is necessary for decay of cyclic gene transcripts and that PREs and AREs in cyclic gene transcript 3’UTRs may also regulate cyclic gene transcript expression through recruitment of decapping factors such as Dcp1a.
CONCLUSIONS
We propose Pnrc2 regulates cyclic gene transcript expression through 3’UTR-mediated mRNA decay. The last 179 nucleotides (nts) of the her1 3’UTR, as well as the full-length dlc 3’UTR, are necessary and sufficient to confer Pnrc2-mediated decay to reporter transcripts. We have shown that the PRE and ARE motifs in the last 179 nts of the her1 3’UTR each contribute to reporter transcript destabilization and that together, these motifs are potent drivers of decay. Future biochemical, molecular, and genetic studies of Pnrc2, Pum1, Pum2, and ARE-BPs, and further investigation of how these factors interact with decay machinery, will provide a deeper understanding of regulators of the segmentation clock and post-transcriptional mechanisms that regulate cyclic gene expression.
Supplementary Material
Maternally deposited pnrc2 contributes to cyclic gene transcript turnover
The terminal her1 3’UTR confers Pnrc2-mediated instability to reporter transcripts
The deltaC 3’UTR also confers Pnrc2-dependent instability
PRE and ARE motifs in the her1 3’UTR drive reporter transcript destabilization
Pnrc2 residues required for interaction with UPF1 and DCP1A contribute to activity
ACKNOWLEDGEMENTS
We thank the Ohio State Zebrafish Facilities staff for excellent zebrafish care, Paula Monsma and the Neurobiology Imaging Core for microscopy assistance and advice, and the OSU Center for RNA Biology. We thank Wayne Miles, Guramrit Singh, and Susan Cole for advice and sharing equipment. We thank Danielle Pvirre, Lauren Woodword, Kara Braunreiter, Zhongxia Yi, and Pooja Gangras for technical assistance and advice.
FUNDING
This work was supported by NIH grants R01GM117964 and R01NS098780 (to S.L.A.), an OSU Center for RNA Biology Predoctoral Fellowship (K.T.T.), and an OSU Dean’s Enrichment Fellowship (M.C.M). The OSU Neuroscience Imaging Core is funded by NIH grants P30-NS045758, P30-NS104177 and S10-OD010383.
Footnotes
COMPETING INTERESTS
The authors declare no competing or financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M, 2005. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Molecular & cellular proteomics: MCP 4, 205–213. [DOI] [PubMed] [Google Scholar]
- Arvola RM, Chang CT, Buytendorp JP, Levdansky Y, Valkov E, Freddolino PL, Goldstrohm AC, 2019. Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs. Nucleic Acids Research: Dec 21 pii: gkz1187. doi: 10.1093/nar/gkz1187. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R, 2003. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes & development 17, 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R, 2001. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes & development 15, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Stanley P, Papalopulu N, 2012. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell reports 2, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G, Ross J, 1988. Poly(A) shortening and degradation of the 3’ A+U-rich sequences of human c-myc mRNA in a cell-free system. Molecular and cellular biology 8, 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R, 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Molecular cell 20, 891–903. [DOI] [PubMed] [Google Scholar]
- Broadbent J, Read EM, 1999. Wholemount in situ hybridization of Xenopus and zebrafish embryos. Methods in molecular biology 127, 57–67. [DOI] [PubMed] [Google Scholar]
- Cao Q, Padmanabhan K, Richter JD, 2010. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 16, 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H, 2012. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Current biology : CB 22, 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kang L, Zhang N, 2005. Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis 43, 196–204. [DOI] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK, 2013a. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic acids research 41, 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Han S, Park OH, Kim YK, 2013b. SMG1 regulates adipogenesis via targeting of staufen1-mediated mRNA decay. Biochimica et biophysica acta 1829, 1276–1287. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK, 2012. Staufen1-mediated mRNA decay functions in adipogenesis. Molecular cell 46, 495–506. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK, 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Molecular cell 33, 75–86. [DOI] [PubMed] [Google Scholar]
- Cho H, Park OH, Park J, Ryu I, Kim J, Ko J, Kim YK, 2015. Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proceedings of the National Academy of Sciences of the United States of America 112, E1540–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chritton JJ, Wickens M, 2011. A role for the poly(A)-binding protein Pab1p in PUF protein-mediated repression. The Journal of biological chemistry 286, 33268–33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois M, Gautier-Courteille C, Legagneux V, Paillard L, 2010. Post-transcriptional controls - adding a new layer of regulation to clock gene expression. Trends in cell biology 20, 533–541. [DOI] [PubMed] [Google Scholar]
- Delaune EA, Francois P, Shih NP, Amacher SL, 2012. Single-cell-resolution imaging of the impact of Notch signaling and mitosis on segmentation clock dynamics. Developmental cell 23, 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KK, Amacher SL, 2005. tortuga refines Notch pathway gene expression in the zebrafish presomitic mesoderm at the post-transcriptional level. Developmental biology 287, 225–236. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM, 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. The Journal of experimental medicine 198, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A, Razif MF, Nygard KK, Rackham O, 2011. A universal code for RNA recognition by PUF proteins. Nature chemical biology 7, 425–427. [DOI] [PubMed] [Google Scholar]
- Fujimuro T, Matsui T, Nitanda Y, Matta T, Sakumura Y, Saito M, Kohno K, Nakahata Y, Bessho Y, 2014. Hes7 3’UTR is required for somite segmentation function. Scientific reports 4, 6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino Y, Yamada K, Sugaya C, Ooka Y, Ovara H, Ban H, Akama K, Otosaka S, Kinoshita H, Yamasu K, Mishima Y, Kawamura A, 2018. Deadenylation by the CCR4-NOT complex contributes to the turnover of hairy-related mRNAs in the zebrafish segmentation clock. FEBS letters 592, 3388–3398. [DOI] [PubMed] [Google Scholar]
- Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D, 2003. Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269–4278. [DOI] [PubMed] [Google Scholar]
- Gallagher TL, Tietz KT, Morrow ZT, McCammon JM, Goldrich ML, Derr NL, Amacher SL, 2017. Pnrc2 regulates 3’UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation. Developmental biology 429, 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangras P, Gallagher TL, Patton RD, Yi Z, Parthun MA, Tietz KT, Deans NC, tBundschuh R, Amacher SL, Singh G, 2019. Stop codon proximal 3’UTR introns in vertebrates can elicit EJC-dependent nonsense-mediated mRNA decay. Preprint on BioRxiv doi: 10.1101/677666. [DOI] [Google Scholar]
- Gao M, Wilusz CJ, Peltz SW, Wilusz J, 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. The EMBO journal 20, 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Palmer EE, McDonell LM, Wang L, Adamski CJ, Koire A, See L, Chen CA, Schaaf CP, Rosenfeld JA, Panzer JA, Moog U, Hao S, Bye A, Kirk EP, Stankiewicz P, Breman AM, McBride A, Kandula T, Dubbs HA, Macintosh R, Cardamone M, Zhu Y, Ying K, Dias KR, Cho MT, Henderson LB, Baskin B, Morris P, Tao J, Cowley MJ, Dinger ME, Roscioli T, Caluseriu O, Suchowersky O, Sachdev RK, Lichtarge O, Tang J, Boycott KM, Holder JL Jr., Zoghbi HY, 2018. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell 172, 924–936.e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, Cooper TA, Orr HT, Sillitoe RV, Zoghbi HY, 2015. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell 160, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli F, Ozbudak EM, Wright GJ, Lewis J, 2007. Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS biology 5, e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hall TMT, McKenney KM, 2018. Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins. Trends in genetics : TIG 34, 972–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M, 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nature structural & molecular biology 13, 533–539. [DOI] [PubMed] [Google Scholar]
- Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquie O, 2008. Control of segment number in vertebrate embryos. Nature 454, 335–339. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Manosalva I, Liu T, Kageyama R, 2013. Control of Hes7 expression by Tbx6, the Wnt pathway and the chemical Gsk3 inhibitor LiCl in the mouse segmentation clock. PloS one 8, e53323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos FM, Brewer G, 2010. The role of AUF1 in regulated mRNA decay. Wiley interdisciplinary reviews. RNA 1, 457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V, 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. The Journal of biological chemistry 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Hanisch A, Holder MV, Choorapoikayil S, Gajewski M, Ozbudak EM, Lewis J, 2013. The elongation rate of RNA polymerase II in zebrafish and its significance in the somite segmentation clock. Development 140, 444–453. [DOI] [PubMed] [Google Scholar]
- Harima Y, Takashima Y, Ueda Y, Ohtsuka T, Kageyama R, 2013. Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell reports 3, 1–7. [DOI] [PubMed] [Google Scholar]
- Henry CA, Urban MK, Dill KK, Merlie JP, Page MF, Kimmel CB, Amacher SL, 2002. Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693–3704. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R, 2002. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP, 2010. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell 143, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley SA, Geisler R, Nusslein-Volhard C, 2000. Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes & development 14, 1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Holley SA, Julich D, Rauch GJ, Geisler R, Nusslein-Volhard C, 2002. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175–1183. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai Z, Ho SM, Pease LR, 2013. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 54, 129–133. [DOI] [PubMed] [Google Scholar]
- Hoyle NP, Ish-Horowicz D, 2013. Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proceedings of the National Academy of Sciences of the United States of America 110, E4316–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xie S, Yao J, 2016. Identification of Novel Reference Genes Suitable for qRT-PCR Normalization with Respect to the Zebrafish Developmental Stage. PloS one 11, e0149277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaud A, Pourquie O, 2014. Signalling dynamics in vertebrate segmentation. Nature reviews. Molecular cell biology 15, 709–721. [DOI] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M, 1978. Mutagenesis at a specific position in a DNA sequence. The Journal of biological chemistry 253, 6551–6560. [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M, 2013. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem cell reports 1, 411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, 1998. Analysis of protein and gene expression. Methods in Cell Biology 59, 63–85. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Niwa Y, Isomura A, Gonzalez A, Harima Y, 2012. Oscillatory gene expression and somitogenesis. Wiley interdisciplinary reviews. Developmental biology 1, 629–641. [DOI] [PubMed] [Google Scholar]
- Kopp F, Elguindy MM, Yalvac ME, Zhang H, Chen B, Gillett FA, Lee S, Sivakumar S, Yu H, Xie Y, Mishra P, Sahenk Z, Mendell JT, 2019. PUMILIO hyperactivity drives premature aging of Norad-deficient mice. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H, 2012. Structural basis of the PNRC2-mediated link between mrna surveillance and decapping. Structure 20, 2025–2037. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carrick DM, Blackshear PJ, 2005. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. The Journal of biological chemistry 280, 34365–34377. [DOI] [PubMed] [Google Scholar]
- Laird-Offringa IA, de Wit CL, Elfferich P, van der Eb AJ, 1990. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Molecular and cellular biology 10, 6132–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT, 2009. Human Ccr4-Not complexes contain variable deadenylase subunits. The Biochemical journal 422, 443–453. [DOI] [PubMed] [Google Scholar]
- Lewis J, 2003. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Current biology : CB 13, 1398–1408. [DOI] [PubMed] [Google Scholar]
- Li Y, Fenger U, Niehrs C, Pollet N, 2003. Cyclic expression of esr9 gene in Xenopus presomitic mesoderm. Differentiation; research in biological diversity 71, 83–89. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Pitha PM, Shin ML, 1992. Poly(A) removal is the kinase-regulated step in tumor necrosis factor mRNA decay. The Journal of biological chemistry 267, 2123–2126. [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E, 2013. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes & development 27, 2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E, 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & development 19, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W, Fang C, Holden T, Dratver MB, Lin H, 2016. An Important Role of Pumilio 1 in Regulating the Development of the Mammalian Female Germline. Biology of reproduction 94, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K, 2012. CUGBP1 and MBNL1 preferentially bind to 3’ UTRs and facilitate mRNA decay. Scientific reports 2, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, 2009. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods in molecular biology 546, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles WO, Lembo A, Volorio A, Brachtel E, Tian B, Sgroi D, Provero P, Dyson N, 2016. Alternative Polyadenylation in Triple-Negative Breast Tumors Allows NRAS and c-JUN to Bypass PUMILIO Posttranscriptional Regulation. Cancer research 76, 7231–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles WO, Lepesant JM, Bourdeaux J, Texier M, Kerenyi MA, Nakakido M, Hamamoto R, Orkin SH, Dyson NJ, Di Stefano L, 2015. The LSD1 Family of Histone Demethylases and the Pumilio Posttranscriptional Repressor Function in a Complex Regulatory Feedback Loop. Molecular and cellular biology 35, 4199–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD, 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Molecular and cellular biology 28, 4093–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge JS, Ziemniak M, Jemielity J, Gross JD, 2016. Structural basis of mRNA-cap recognition by Dcp1-Dcp2. Nature structural & molecular biology 23, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naudin C, Hattabi A, Michelet F, Miri-Nezhad A, Benyoucef A, Pflumio F, Guillonneau F, Fichelson S, Vigon I, Dusanter-Fourt I, Lauret E, 2017. PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells. Blood 129, 2493–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Gkratsou A, Josi C, Colombo M, Muhlemann O, 2018. Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells. RNA 24, 557–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitanda Y, Matsui T, Matta T, Higami A, Kohno K, Nakahata Y, Bessho Y, 2014. 3’-UTR-dependent regulation of mRNA turnover is critical for differential distribution patterns of cyclic gene mRNAs. The FEBS journal 281, 146–156. [DOI] [PubMed] [Google Scholar]
- Oates AC, Ho RK, 2002. Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129, 2929–2946. [DOI] [PubMed] [Google Scholar]
- Oates AC, Morelli LG, Ares S, 2012. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development 139, 625–639. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O, 1997. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. [DOI] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M Jr., Cline M, Mandel-Gutfreund Y, 2014. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic acids research 42, W361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppel K, Baglioni C, 1991. Deadenylation and turnover of interferon-beta mRNA. The Journal of biological chemistry 266, 6663–6666. [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Fullgrabe A, Fuentes AM, Jupp S, Koskinen S, Mannion O, Huerta L, Megy K, Snow C, Williams E, Barzine M, Hastings E, Weisser H, Wright J, Jaiswal P, Huber W, Choudhary J, Parkinson HE, Brazma A, 2016. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic acids research 44, D746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P, 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. The EMBO journal 19, 4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O, 2011. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Pieper L, Chew GL, Schier AF, 2017. A Massively Parallel Reporter Assay of 3’ UTR Sequences Identifies In Vivo Rules for mRNA Degradation. Molecular cell 68, 1083–1094.e1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MF, Bochter MS, Wahi K, Nuovo GJ, Cole SE, 2013. Mir-125a-5p-mediated regulation of Lfng is essential for the avian segmentation clock. Developmental cell 24, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H, 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes & development 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA, 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley interdisciplinary reviews. RNA 2, 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE, 2012. Regulation of cytoplasmic mRNA decay. Nature reviews. Genetics 13, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M, 2011. Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Schwendinger-Schreck J, Kang Y, Holley SA, 2014. Modeling the zebrafish segmentation clock’s gene regulatory network constrained by expression data suggests evolutionary transitions between oscillating and nonoscillating transcription. Genetics 197, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS, 2009. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA research : an international journal for rapid publication of reports on genes and genomes 16, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H, 2008. Structural basis of dcp2 recognition and activation by dcp1. Molecular cell 29, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih NP, Francois P, Delaune EA, Amacher SL, 2015. Dynamics of the slowing segmentation clock reveal alternating two-segment periodicity. Development 142, 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Belasco JG, Greenberg ME, 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & development 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Siemen H, Colas D, Heller HC, Brustle O, Pera RA, 2011. Pumilio-2 function in the mouse nervous system. PloS one 6, e25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD, 2009. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Ohtsuka T, Gonzalez A, Miyachi H, Kageyama R, 2011. Intronic delay is essential for oscillatory expression in the segmentation clock. Proceedings of the National Academy of Sciences of the United States of America 108, 3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C, Campos-Ortega JA, 1999. her1, a zebrafish pair-rule like gene, acts downstream of notch signalling to control somite development. Development 126, 3005–3014. [DOI] [PubMed] [Google Scholar]
- Tan SL, Ohtsuka T, Gonzalez A, Kageyama R, 2012. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes to cells : devoted to molecular & cellular mechanisms 17, 952–961. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS, 2002. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mechanisms of development 118, 91–98. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H, 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & development 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC, 2012. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. The Journal of biological chemistry 287, 36370–36383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, Lambert NJ, Freese P, Saxena T, Cooper TA, Burge CB, 2015. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome research 25, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM, 2002. Modular recognition of RNA by a human pumilio-homology domain. Cell 110, 501–512. [DOI] [PubMed] [Google Scholar]
- Weidmann CA, Raynard NA, Blewett NH, Van Etten J, Goldstrohm AC, 2014. The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA 20, 1298–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Shifley ET, Braunreiter KM, Cole SE, 2016. Disruption of somitogenesis by a novel dominant allele of Lfng suggests important roles for protein processing and secretion. Development 143, 822–830. [DOI] [PubMed] [Google Scholar]
- Wilson T, Treisman R, 1988. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3’ AU-rich sequences. Nature 336, 396–399. [DOI] [PubMed] [Google Scholar]
- Wong SF, Agarwal V, Mansfield JH, Denans N, Schwartz MG, Prosser HM, Pourquie O, Bartel DP, Tabin CJ, McGlinn E, 2015. Independent regulation of vertebral number and vertebral identity by microRNA-196 paralogs. Proceedings of the National Academy of Sciences of the United States of America 112, E4884–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Chang R, Salmon NA, Reijo Pera RA, 2007. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Molecular reproduction and development 74, 912–921. [DOI] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr., 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome research 13, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, Miller FD, 2018. A Translational Repression Complex in Developing Mammalian Neural Stem Cells that Regulates Neuronal Specification. Neuron 97, 520–537.e526. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen D, Xia J, Han W, Cui X, Neuenkirchen N, Hermes G, Sestan N, Lin H, 2017. Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes & development 31, 1354–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, O’Rourke JR, McManus MT, Lewandoski M, Harfe BD, Sun X, 2011. The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Developmental biology 351, 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.