Abstract
The pathologic mechanisms of pulmonary fibrosis (PF), one of the most common chronic pulmonary diseases, remain unclear. Napsin A is an aspartic proteinase that has been regarded as a hallmark of pulmonary adenocarcinoma. The present study aimed to investigate the specific function and molecular mechanisms of Napsin A in PF from the perspective of microRNA (miRNA or miR) regulation. In the present study, it was found that miR-1290 downregulated the expression of Napsin A by binding to its 3′-UTR. Cell viability was examined by MTT assay. The protein levels of α-smooth muscle actin (α-SMA), Collagen I and Napsin A were examined by western blot analysis. The predicted targeting of Napsin A by miR-1290 was validated by luciferase reporter assay. The protein content of α-SMA was examined by immunofluorescence staining. miR-1290 was found to be upregulated in blood samples from patients with PF and in TGF-β1-stimulated A549 cells. miR-1290 was found to directly target Napsin A. miR-1290 overexpression also significantly promoted A549 cell proliferation and increased the protein levels of markers of fibrosis. Napsin A knockdown exerted effects on A549 cell proliferation and TGF-β1-induced fibrosis that were similar to those induced by miR-1290 overexpression; more importantly, Napsin A knockdown significantly reversed the effects of miR-1290 inhibition, indicating that miR-1290 promotes TGF-β1-induced fibrosis by targeting Napsin A. Moreover, TGF-β1-induced CAMP responsive element binding protein 1 (CREB1) overexpression promoted the transcription of miR-1290 in A549 cells. On the whole, the findings of the present study demonstrate that TGF-β1-induced CREB1 over-expression induces the significant upregulation of miR-1290 expression, thus aggravating TGF-β1-induced fibrotic changes in A549 cells via the miR-1290 downstream target, Napsin A.
Keywords: pulmonary fibrosis, Napsin A, miR-1290, CAMP responsive element binding protein 1
Introduction
Pulmonary fibrosis (PF) is one of the most common chronic pulmonary diseases, and it is characterized by restrictive functional ventilation disorder, hypoxemia and chronic progressive diffuse lung fibrosis; it is associated with clinical features, such as wheezing, dyspnea and dry cough. It has a high incidence and mortality rate worldwide (1). To date, the pathological mechanisms of PF have not been fully elucidated (2,3); thus, no effective drug or treatment has yet been developed.
Napsin A is an aspartic proteinase expressed not only in type II lung cells, but also in alveolar macrophages and it may be expressed secondary to phagocytosis (4,5). The alveolar cavity contains active Napsin A at high levels, which are associated with the levels of surfactant protein B (SP-B), precursor protein proSP-B and SP-C (6). As a hallmark of the most significant number of pulmonary adenocarcinomas, immunohistochemical analysis for Napsin A yields negative results in the majority of squamous cell carcinomas and adenocarcinomas of other organs (7,8). Reportedly, its local expression not only aids in the classification of primary pulmonary tumors as adenocarcinomas, but also in the identification of the lung as the origin in the context of metastatic adenocarcinoma (7,8). In addition, elevated serum levels of Napsin A in patients with idiopathic PF (IPF) are related to the severity of the illness (9-11). Ueno et al (12) transfected the Napsin A gene into 293 cells and found that the cell proliferative and migratory ability were significantly inhibited. Consistently, in a previous study by the authors, it was also demonstrated that PF could be suppressed by the transfection of the Napsin A gene into type II alveolar epithelial cells, possibly through the inhibition of integrin signal transduction (13). However, the expression of Napsin A is significantly downregulated in the blood samples obtained from PF patients. Investigating the mechanism by which Napsin A expression is suppressed during PF might provide potent novel strategies for PF treatment.
MicroRNAs (miRNAs or miRs), a family of non-coding single-stranded small RNAs with lengths of 21-23 nucleotides, can modulate gene expression at the post-transcriptional level by base pairing to sequence motifs in the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs) (14-17). Numerous studies have screened and identified several deregulated miRNAs in lung fibrosis and other diseases. Reportedly, miR-1343 can directly bind to the 3′-UTR of TGF-β receptor 1 (TGFBR1) and TGFBR2 to inhibit the TGF-β signaling pathway, thus alleviating PF (18). miR-449a has been found to target autophagy-related Bcl2 mRNA to play an antifibrotic role in silica-induced PF (19). Moreover, miR-489 has been shown to bind to both MyD88 and Smad3 to alleviate inflammation and the development of fibrosis (19,20). In summary, miRNAs may be involved in the fibrotic development process by targeting various downstream transcripts. Therefore, it is reasonable to hypothesize that miRNAs may target Napsin A to exert an effect on PF development.
Herein, the online tool TargetScan was used to identify miRNAs that reduce the expression of Napsin A by targeting its 3′-UTR. Among 17 candidate miRNAs, the expression of miR-1290 was the most upregulated in blood samples obtained from patients with PF (Fig. S1); moreover, to the best of our knowledge, no study to date has reported a role for miR-1290 in PF. To investigate the cellular functions of miR-1290 and its interaction with Napsin A, A549 cells were stimulated with TGF-β1 and miR-1290 expression was examined. In addition, the effects of miR-1290 on A549 proliferation and markers of fibrosis, and the predicted binding of miR-1290 to Napsin A were examined. The dynamic effects of miR-1290 and Napsin A on TGF-β1-induced fibrosis were also evaluated. To determine the mechanisms through which miR-1290 is upregulated during PF, the TransmiR v2.0 database and ChIP-Atlas were used to identify transcription factors that are related to PF and may bind the miR-1290 promoter to activate its transcription. On the whole, the present study demonstrates a novel mechanism through which Napsin A can be downregulated in PF by regulating miRNA; miR-1290 may be a novel potential target for the treatment of PF.
Materials and methods
Clinical patients and samples
PF was diagnosed according to the American Thoracic Society/European Respiratory Society consensus criteria (21). The patients with PF and healthy volunteers were enrolled at the First People's Hospital of Changzhou from March, 2018 to March, 2019. The plasma samples were isolated from patients (n=14; age, 58.5±10.94 years; sex: F/M ratio, 5/9) with PF and healthy volunteers (n=20; age, 51.2±9.64; sex: F/M, 8/12). All the experiments were approved by the Ethics Committee of The First People's Hospital of Changzhou (ethics number: 2018KY047). Written informed consent was also obtained from all study participants.
Cell lines and cell transfection
A human non-small cell lung cancer cell line (A549; ATCC® CCL-185) and a normal human lung epithelial cell line (BEAS-2B; ATCC® CRL-9609) were purchased from ATCC and cultured in F-12K medium (for A549 cells, cat. no. 30-2004, ATCC) or bronchial epithelial basal medium BEBM (for BEAS-2B cells, Lonza/Clonetics Corp.) supplemented with 10% FBS. The cells were cultured at 37°C with 5% v/v CO2. For TGF-β1 treatment, cells were stimulated with 10 ng/ml TGF-β1 for 48 h.
miR-1290 expression in target cells was assessed by transfection with 50 nM miR-1290 mimics or 100 nM miR-1290 inhibitor (RiboBio). Napsin A was knocked down by transfection with 20 nM si-Napsin A (RiboBio). CREB1 overexpression was achieved by transfection with 1 µg/ml CREB1 overexpression vector (CREB1 OE, RiboBio). All transfections were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the cells were harvested for further experiments. The sequences are listed in Table SI.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA extraction, reverse transcription, and PCR were performed following previously described methods (22,23) using the miScript Reverse Transcription kit (Qiagen, Inc.) and SYBR-Green PCR Master Mix (Qiagen, Inc.). RNU6B expression or GAPDH expression was used as an endogenous control for miRNA or mRNA determination. The 2−ΔΔCq method (24) was used to analyze relative fold changes. The primer sequences are listed in Table SI.
Determination of cell viability by MTT assay
Cell viability was examined by MTT assay following previously described methods (25). The OD values were measured at 490 nm using microplate reader (Bio-Rad Laboratories, Inc.). The cell viability of the untreated cells (control) was defined as 100%.
Western blot analysis
The protein levels of α-smooth muscle actin (α-SMA), Collagen I and Napsin A were examined by western blot analysis following previously described methods (25). A total of 50 µg protein samples were then separated by 10% SDS-PAGE and transferred onto nitrocellulose membrane (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% non-fat milk in TBST (0.1% Tween-20) for 2 h at room temperature and incubated with the following antibodies: Anti-α-SMA (1:1,000, ab5694, Abcam), anti-Collagen I (1:1,000, ab34710), anti-Napsin A (1:1,000, ab73021, Abcam), anti-GAPDH (1:1,000, ab8245, Abcam), anti-AKT (1:1,000, 10176-2-AP, Proteintech), ani-p-AKT (1:1,000, 66444-1-Ig, Proteintech) and goat-anti-rabbit or mouse HRP-conjugated secondary antibodies (SA00001-1 and SA00001-2, Proteintech). Signals were visualized using enhanced chemiluminescence (ECL) substrates (Merck KGaA) using GAPDH as an endogenous reference protein. ImageJ software version 1.8.0 (National Institute of Health) was used for densitometric analysis.
Luciferase reporter assay
The Napsin A 3′-UTR was amplified by PCR and was then cloned downstream of the Renilla psiCHECK2 vector (Promega Corp.); the product was named wt-Napsin A 3′-UTR. To generate a Napsin A 3′-UTR mutant reporter, the predicted miR-1290 binding site was mutated to remove the complementarity, and this type of reporter vector was named mut-Napsin A 3′-UTR. 293 cells (ATCC) were co-transfected with miR-1290 mimics or miR-1290 inhibitor and wt-Napsin A 3′UTR or mut-Napsin A 3′UTR, and the Dual-Luciferase Reporter Assay System (Promega Corp.) was employed to determine the luciferase activity. Renilla lucif-erase activity was normalized to Firefly luciferase activity for each transfected well of cells.
Transcription factors predicted to regulate miR-1290 were identified using the TransmiR v2.0 database (http://www.cuilab.cn/transmir) and ChIP-Atlas (http://chip-atlas.org/enrichment_analysis). miR-1290 with the mutant or wild-type seed region was synthesized and cloned into the Renilla psiCHECK2 vector (Promega Corp.). A total of 7 nucleotides in the seed region were mutated to obtain the mutant sequence. The CREB1 protein-coding sequence was cloned into the pcDNA3.1 vector (Applied Biosystems). 293 cells were co-transfected with pcDNA3.1/CREB1 and wild- or mutant-miR-1290 vectors. After 48 h, the cells were harvested and luciferase activity assays were performed in the following treatment with TGF-β1 or following no treatment.
Immunofluorescence (IF) staining
α-SMA protein expression was examined by IF staining using anti-α-SMA antibody (ab5694, Abcam) according to previously described methods (26). The cells were incubated with FITC-conjugated secondary antibody (Beyotime Institute of Biotechnology) for 1 h in the dark at room temperature. Nuclei were stained with DAPI (Beyotime Institute of Biotechnology) for 5 min at room temperature. Images were observed and acquired by a fluorescence microscope (Nikon Corp.). Green fluorescence indicates α-SMA expression and blue fluorescence indicates nuclei.
Statistical analysis
Data from at least 3 independent experiments were processed using SPSS 17.0 (IBM, Inc.) and are presented as the means ± SD. Student's t-tests were used for statistical comparisons between 2 groups. One-way ANOVA followed by Tukey's multiple comparison test was used to estimate the differences among >2 groups. Pearson's correlation analysis was also used to determine the correlation between the expression of miR-1290 and Napsin A in the plasma samples. Values of P<0.05 and P<0.01 were considered to indicate statistically significant and highly statistically significant differences, respectively.
Results
miR-1290 expression is upregulated in plasma samples from patients with PF and TGF-β1-stimulated A549 cells
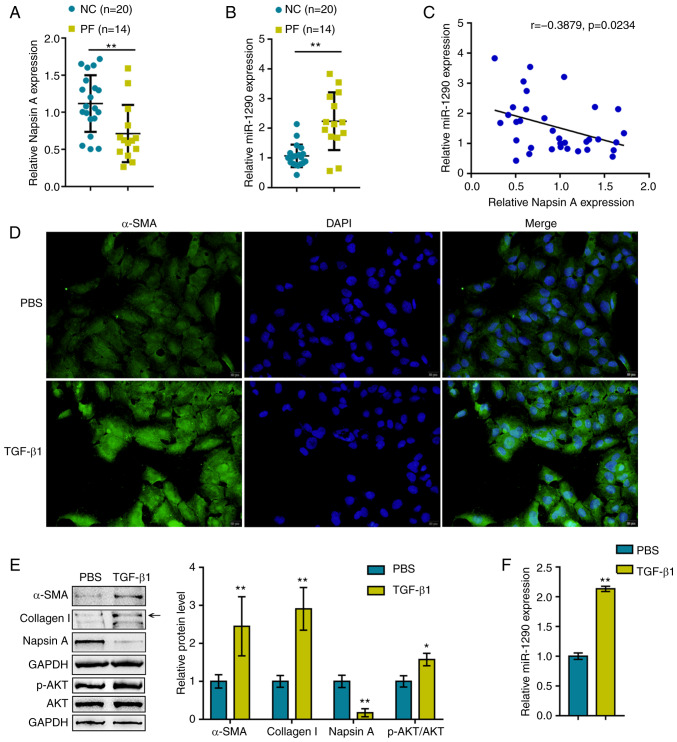
Before investigating the cellular functions of miR-1290, the first step of the present study was to evaluate Napsin A and miR-1290 expression within blood samples obtained from healthy donors and patients with PF. As shown in Fig. 1A and B, compared to the healthy donors, the expression of Napsin A was significantly decreased, whereas the expression of miR-1290 was increased within the plasma samples obtained from patients with PF. Moreover, the expression levels of miR-1290 negatively correlated with Napsin A expression levels in the samples (Fig. 1C).
Figure 1.
miR-1290 expression is upregulated in blood samples from patients with pulmonary fibrosis and in TGF-β1-stimulated A549 cells. (A and B) miR-1290 and Napsin A expression levels in blood samples obtained from healthy volunteers (negative control) and patients with pulmonary fibrosis. (C) The correlation of miR-1290 and Napsin A expression in blood samples was analyzed by Pearson's correlation analysis. A549 cells were treated with TGF-β1 and examined for (D) α-SMA protein content (green) by immunofluorescence staining. (E) Protein levels of Napsin A, α-SMA, Collagen I, AKT and p-AKT were assessed by western blot analysis. (F) Expression of miR-1290 was assessed by RT-qPCR. *P<0.05 and **P<0.01 compared to the control. PF, pulmonary fibrosis.
Previous studies have demonstrated that TGF-β1 is related to the AKT pathway in PF (27), lung cancer (28), live fibrosis (29). Based on this, the present study examined the association between TGF-β1 and the AKT pathway in A549 cells under TGF-β1 stimulation. The A549 cells were treated with TGF-β1 to generate a cell model of TGF-β1-induced fibrosis, which was validated by IF staining and western blot analysis for α-SMA, Collagen I, Napsin A and AKT (total AKT and phosphorylated AKT) protein expression levels. As shown in Fig. 1D and E, the α-SMA, Collagen I and p-AKT protein levels were significantly upregulated by TGF-β1 stimulation, indicating fibrotic changes in the A549 cells. The protein levels of Napsin A were decreased upon TGF-β1 stimulation. In addition, the expression of miR-1290 was significantly upregulated by TGF-β1 stimulation (Fig. 1F), suggesting that miR-1290 participates in TGF-β1-induced fibrosis.
miR-1290 directly targets Napsin A to modulate A549 cell proliferation and TGF-β1-induced fibrosis
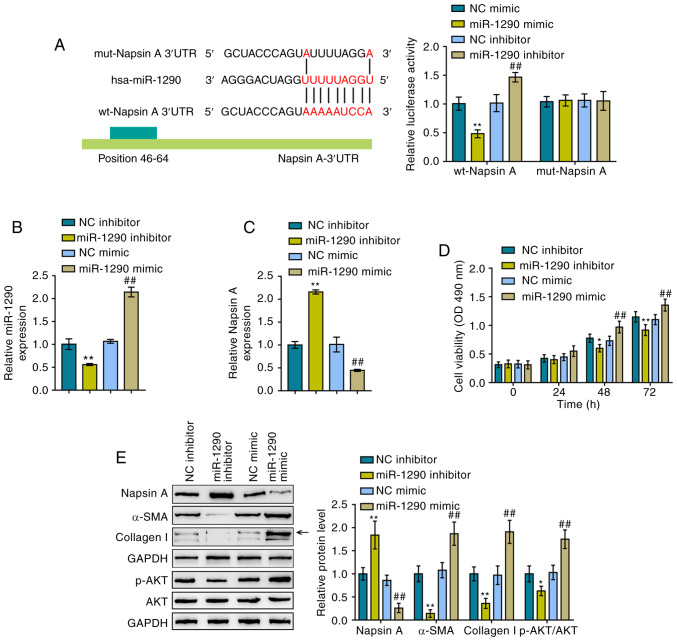
To further determine the interaction between miR-1290 and Naspin A, luciferase reporter assays were performed. As described in the Materials and methods section, two different types of Napsin A 3′-UTR luciferase reporter vectors were constructed, the wild-type and mutant-type, and these were named wt-Napsin A 3′-UTR and mut-Napsin A 3′-UTR, respectively (Fig. 2A). These vectors were co-transfected into 293 cells with miR-1290 mimics or a miR-1290 inhibitor and luciferase activity examined. miR-1290 overexpression induced a significant decrease in the luciferase activity of wild-type Napsin A 3′-UTR, which was increased by the inhibition of miR-1290; mutating the putative miR-1290 binding site abolished the changes in luciferase activity (Fig. 2A). Based on these data, miR-1290 directly targeted the Napsin A 3′-UTR. Subsequently, miR-1290 mimics/inhibitor were transfected into the cells to achieve miR-1290 overexpression/inhibition in A549 cells and RT-qPCR was performed to verify the transfection efficiency (Fig. 2B). Consistently, miR-1290 overexpression inhibited the expression of Napsin A, while miR-1290 inhibition promoted it (Fig. 2C).
Figure 2.
miR-1290 directly targets Napsin A to modulate A549 cell proliferation and TGF-β1-induced fibrosis. (A) Schematic diagram showing the predicted binding site between miR-1290 and Napsin A 3′UTR. Wild- and mutant-type Napsin A 3′UTR luciferase reporter vectors were constructed. The mutant-type Napsin A 3′UTR vector contained a 7-bp mutation in the predicted miR-1290 binding site. These vectors were co-transfected into 293 cells with miR-1290 mimics or inhibitor and the luciferase activity was determined. (B) miR-1290 overexpression and inhibition in A549 cells were achieved by transfection with miR-1290 mimics or an inhibitor, and results were confirmed by RT-qPCR. (C) Napsin A mRNA expression in response to miR-1290 overexpression or inhibition was determined by RT-qPCR. (D) Viability of A549 cells in which miR-1290 was overexpressed or inhibited was determined by MTT assays. (E) Protein levels of Napsin A, α-SMA, Collagen I, AKT and p-AKT in A549 cells in which miR-1290 was overexpressed or inhibited were determined by western blot analysis. *P<0.05 and **P<0.01, compared to the NC (negative control) inhibitor group; ##P<0.01, compared to the NC (negative control) mimics group.
The cellular functions of miR-1290 were then evaluated under TGF-β1 stimulation. As shown in Fig. 2D, miR-1290 overexpression significantly promoted the proliferation of the A549 cells, whereas miR-1290 inhibition suppressed it; in other words, miR-1290 overexpression enhanced TGF-β1-stimulated A549 cell growth, while miR-1290 inhibition decreased the growth. To evaluate the cytotoxicity of miR-1290 against normal cells, the viability of the human normal lung epithelial cell line, BEAS-2B, was examined by MTT assay (Fig. S2). The transfection efficiency of miR-1290 mimics and miR-1290 inhibitor in BEAS-2B cells is illustrated in Fig. S2A. According to the results of MTT assay (Fig. S2B), miR-1290 overexpression markedly promoted the proliferation of BEAS-2B cells, whereas miR-1290 inhibition restrained it. In addition, miR-1290 overexpression decreased the Napsin A protein levels and increased the protein levels of α-SMA, Collagen I and p-AKT in A549 cells. However, miR-1290 inhibition exerted opposing effects on the protein levels of Napsin A, α-SMA, Collagen I and p-AKT under TGF-β1 stimulation in A549 cells (Fig. 2E). In summary, miR-1290 may directly bind to the 3′-UTR of Napsin A, and miR-1290 inhibition improves TGF-β1-induced fibrosis.
Dynamic effects of miR-1290 and Napsin A on A549 cell proliferation and TGF-β1-induced fibrotic changes
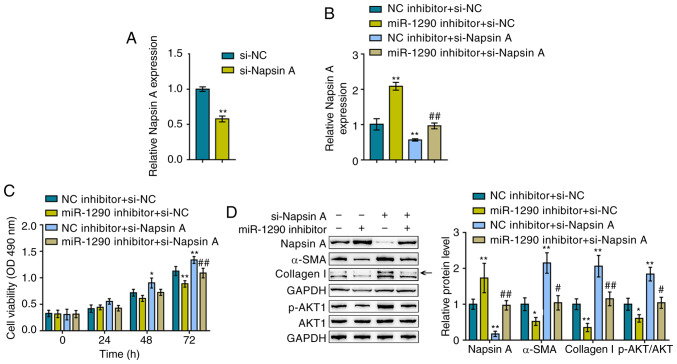
After confirming the effects of miR-1290 on TGF-β1-induced fibrotic changes, the dynamic effects of miR-1290 and its downstream target, Napsin A, on A549 proliferation and TGF-β1-induced fibrotic changes were evaluated. The A549 cells were trans-fected with si-Napsin A to knock down its expression and RT-qPCR was then performed to verify the transfection efficiency (Fig. 3A). The A549 cells were co-transfected with miR-1290 inhibitor/si-Napsin A, and Napsin A expression, cell viability, and Napsin A, α-SMA and Collagen I protein levels were then evaluated. The TGF-β1-induced suppression of Napsin A expression was significantly reversed by transfection with miR-1290 inhibitor, whereas it was further suppressed by Napsin A knockdown; Napsin A knockdown significantly attenuated the effects of the miR-1290 inhibitor (Fig. 3B).
Figure 3.
Dynamic effects of miR-1290 and Napsin A on A549 cell proliferation and TGF-β1-induced fibrosis (A) Napsin A knockdown was conducted by the transfection of si-Napsin A, as confirmed by RT-qPCR. A549 cells were co-transfected with miR-1290 inhibitor and si-Napsin A upon TGF-β1 stimulation and examined for (B) the expression of Napsin A, as determined by RT-qPCR; (C) cell viability, as determined by MTT assays; and (D) the protein levels of Napsin A, α-SMA, Collagen I, AKT and p-AKT as determined by western blot analysis. *P<0.05 and **P<0.01, compared to NC (negative control) inhibitor + si-NC group; #P<0.05 and ##P<0.01, compared to NC (negative control) inhibitor + si-Napsin A group.
As regards cellular functions, miR-1290 inhibition reduced, whereas Napsin A knockdown increased TGF-β1-induced A549 cell proliferation; the effect of miR-1290 inhibitor was also reversed by Napsin A knockdown (Fig. 3C). Consistently, TGF-β1 decreased the Napsin A levels, whereas it increased the α-SMA, Collagen I and p-AKT levels, and this effect was attenuated by treatment with the miR-1290 inhibitor, but enhanced by Napsin A knockdown; Napsin A knockdown significantly attenuated the effect of the miR-1290 inhibitor (Fig. 3D). In summary, these findings demonstrate that miR-1290 regulates TGF-β1-induced fibrotic changes via Napsin A.
TGF-β1-induced CREB1 expression promotes the transcription of miR-1290
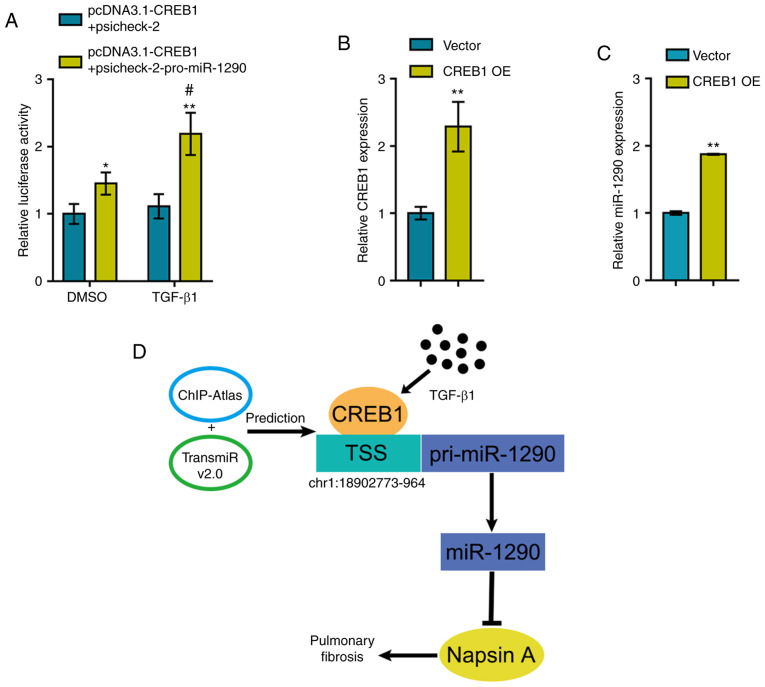
To further investigate the mechanisms through which TGF-β1 induces the expression of miR-1290, the online tools, TransmiR v2.0 database and ChIP-Atlas, were used to identify transcription factors that may regulate miR-1290 expression and can be induced by TGF-β1 stimulation; CREB1 was identified (30,31). The miR-1290 luciferase reporter vector was constructed as described in the Materials and methods section and was co-transfected into 293 cells with pcDNA3.1/CREB1. Luciferase activity was determined in the presence or absence of TGF-β1 treatment. As shown in Fig. 4A, CREB1 overexpression significantly enhanced the luciferase activity of the miR-1290 reporter vector and TGF-β1 treatment further enhanced the CREB1 overexpression-induced increase in luciferase activity.
Figure 4.
TGF-β1-induced CREB1 promotes the transcription of miR-1290. (A) A luciferase reporter vector with the miR-1290 promoter was constructed and co-transfected into 293 cells with pcDNA3.1/CREB1. Luciferase activity was determined in the presence or absence of TGF-β1. (B) CREB1 overexpression was achieved in A549 cells by transfection of the CREB1 overexpression vector (CREB1 OE), as confirmed by RT-qPCR. (C) miR-1290 expression was determined in CREB1-overexpressing A549 cells by RT-qPCR. (D) Schematic diagram showing that TGF-β1-induced CREB1 promotes the transcription of miR-1290, therefore inhibiting Napsin A expression and promoting pulmonary fibrosis. *P<0.05 and **P<0.01, compared to pcDNA3.1-CREB1 + psicheck-2 group or vector group; #P<0.05, compared to DMSO group.
To validate the effects of CREB1 on miR-1290, the CREB1-overexpressing vector was transfected into the A549 cells to induce CREB1 overexpression and RT-qPCR was then performed to verify the transfection efficiency (Fig. 4B). As predicted, CREB1 overexpression significantly promoted the expression of miR-1290 (Fig. 4C). Therefore, a novel mechanism is demonstrated through which TGF-β1-induced CREB1 upregulation promotes the transcription of miR-1290, thus inhibiting Napsin A expression and promoting fibrotic changes in A549 cells (Fig. 4D).
Discussion
Herein, miR-1290 was regarded as an upstream regulatory miRNA, that reduces the expression of Napsin A by binding to its 3′-UTR. miR-1290 was upregulated in PF blood samples and in TGF-β1-stimulated A549 cells. miR-1290 can directly target Napsin A, significantly promote A549 proliferation and increase the protein levels of markers of fibrosis. Napsin A knockdown exerted effects on A549 proliferation and TGF-β1-induced fibrosis that were similar to those observed following miR-1290 overexpression; more importantly, Napsin A knockdown significantly reversed the effects of miR-1290 inhibition, indicating that miR-1290 promotes TGF-β1-induced fibrosis by targeting Napsin A. Moreover, TGF-β1-induced CREB1 overexpression promoted the transcription of miR-1290 in A549 cells.
Over the past decade, emerging evidence has revealed potential biomarkers for the prediction of PF. According to microarray profiles, multiple miRNAs, including miR-29 (32,33), miR-326 (34), miR-98 (35) and miR-let-7d (36), may participate in the pathogenesis of PF. Herein, the expression of miR-1290 was significantly upregulated in blood samples obtained from patients with PF and in A549 cells upon TGF-β1 stimulation, and the overexpression of miR-1290 promoted A549 cell proliferation and fibrosis marker proteins levels under TGF-β1 stimulation conditions, indicating the potential of miR-1290 as a novel biomarker for PF. Previously, miR-1290 expression has been reported to be significantly upregulated by Matrigel and may thus be cancer-related. Matrigel is considered to be a medium that is rich in extracellular matrix (ECM) components and capable of altering cellular cell phenotypes and gene expression (37). miR-1290 expression is abnormally upregulated in non-small cell lung cancer (NSCLC); thus, serum levels of miR-1290 may serve as an underlying prognostic biomarker for NSCLC (38). However, to the best of our knowledge, this is the first study to report the upregulation of miR-1290 in blood samples from patients with PF. miR-1290 may thus play a critical role in PF progression.
As has already been mentioned, miR-1290 expression is significantly upregulated by TGF-β1 stimulation. Upon TGF-β1 treatment, miR-1290 overexpression further enhanced the promotive effects of TGF-β1 on A549 cell growth and α-SMA and Collagen I protein levels. In other words, miR-1290 may antagonize TGF-β1-induced fibrotic changes in A549 cells. More importantly, as predicted by an online tool, miR-1290 overexpression significantly inhibited the protein levels of Napsin A. Previously, it was demonstrated that Napsin A overexpression significantly reversed or attenuated TGF-β1-induced fibrotic changes in A549 cells (39) Herein, it was demonstrated that miR-1290 targets the 3′-UTR of Napsin A to exert a negative regulatory effect on the expression of Napsin A upon TGF-β1 stimulation. The effects of miR-1290 inhibition on TGF-β1-induced fibrotic changes in A549 cells were reversed by Napsin A silencing, indicating that miR-1290 promotes TGF-β1-induced fibrosis by targeting Napsin A.
Since the expression of miR-1290 was significantly increased in the serum obtained from patients with PF, the mechanisms of the abnormal miR-1290 upregulation in serum from patients with PF were further investigated. According to bioinformatics analyses, CREB1 may activate the transcription of miR-1290 via targeting its promoter region. Inflammation, cell growth, differentiation, adaptation and survival, as well as other cell functions, have been shown to be modulated by CREB (40). It has been revealed that an increase in CREB activity can be related to the pathologic mechanism of asthma, changes in cognitive memory, and the process of chronic obstructive pulmonary disease (COPD) (41). Herein, the predicted binding of CREB1 to miR-1290 was validated by revealing that TGF-β1-induced CREB1 overexpression promoted the expression of miR-1290 by targeting the promoter region.
In conclusion, the present study demonstrates that TGF-β1-induced CREB1 overexpression significantly upregulates miR-1290 expression, therefore antagonizing TGF-β1-induced fibrotic changes in A549 cells through the miR-1290 downstream target, Napsin A. The CREB1/miR- 1290/Napsin A axis may thus be a potent target in the treatment of TGF-β1-induced PF. However, further in vivo studies and clinical investigations are warranted to confirm these findings.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by the 2018 Applied Basic Research Program of Changzhou Science and Technology Bureau (CJ20189021).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
SG and JZ made substantial contributions to the conception and design of the study. SG and YW contributed to the experimental design and study implementation. QZ analyzed and interpreted the data. SG and QZ drafted the manuscript. JZ revised the study critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in experiments involving human participants were in accordance with the ethical standards of the Ethics Committee of The First People's Hospital of Changzhou. All participants signed informed consent forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lepparanta O, Sens C, Salmenkivi K, Kinnula VL, Keski-Oja J, Myllärniemi M, Koli K. Regulation of TGF-β storage and activation in the human idiopathic pulmonary fibrosis lung. Cell Tissue Res. 2012;348:491–503. doi: 10.1007/s00441-012-1385-9. [DOI] [PubMed] [Google Scholar]
- 2.Ni S, Wang D, Qiu X, Pang L, Song Z, Guo K. Bone marrow mesenchymal stem cells protect against bleomycin-induced pulmonary fibrosis in rat by activating Nrf2 signaling. Int J Clin Exp Pathol. 2015;8:7752–7761. [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniou KM, Margaritopoulos GA, Siafakas NM. Pharmacological treatment of idiopathic pulmonary fibrosis: From the past to the future. Eur Respir Rev. 2013;22:281–291. doi: 10.1183/09059180.00002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uchida A, Samukawa T, Kumamoto T, Ohshige M, Hatanaka K, Nakamura Y, Mizuno K, Higashimoto I, Sato M, Inoue H. Napsin A levels in epithelial lining fluid as a diagnostic biomarker of primary lung adenocarcinoma. BMC Pulm Med. 2017;17:195. doi: 10.1186/s12890-017-0534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Zhang Y, Ding T, Cheng R, Gong W, Guo Y, Luo Y, Pan Y, Zhai Q, Sun W, et al. Napsin A expression in subtypes of thyroid tumors: Comparison with lung adenocarcinomas. Endocr Pathol. 2020;31:39–45. doi: 10.1007/s12022-019-09600-6. [DOI] [PubMed] [Google Scholar]
- 6.Beck J, Miller MA, Frank C, DuSold D, Ramos-Vara JA. Surfactant Protein A and Napsin A in the immunohistochemical characterization of canine pulmonary carcinomas: Comparison with thyroid transcription factor-1. Vet Pathol. 2017;54:767–774. doi: 10.1177/0300985817712559. [DOI] [PubMed] [Google Scholar]
- 7.Hirano T, Gong Y, Yoshida K, Kato Y, Yashima K, Maeda M, Nakagawa A, Fujioka K, Ohira T, Ikeda N, et al. Usefulness of TA02 (napsin A) to distinguish primary lung adenocarcinoma from metastatic lung adenocarcinoma. Lung Cancer. 2003;41:155–162. doi: 10.1016/S0169-5002(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 8.Bishop JA, Sharma R, Illei PB. Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol. 2010;41:20–25. doi: 10.1016/j.humpath.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Samukawa T, Hamada T, Uto H, Yanagi M, Tsukuya G, Nosaki T, Maeda M, Hirano T, Tsubouchi H, Inoue H. The elevation of serum napsin A in idiopathic pulmonary fibrosis, compared with KL-6, surfactant protein-A and surfactant protein-D. BMC Pulm Med. 2012;12:55. doi: 10.1186/1471-2466-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.du Bois RM, Weycker D, Albera C, Bradford WZ, Costabel U, Kartashov A, Lancaster L, Noble PW, Raghu G, Sahn SA, et al. Ascertainment of individual risk of mortality for patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;184:459–466. doi: 10.1164/rccm.201011-1790OC. [DOI] [PubMed] [Google Scholar]
- 11.du Bois RM, Weycker D, Albera C, Bradford WZ, Costabel U, Kartashov A, King TE, Jr, Lancaster L, Noble PW, Sahn SA, et al. Forced vital capacity in patients with idiopathic pulmonary fibrosis: Test properties and minimal clinically important difference. Am J Respir Crit Care Med. 2011;184:1382–1389. doi: 10.1164/rccm.201105-0840OC. [DOI] [PubMed] [Google Scholar]
- 12.Ueno T, Linder S, Elmberger G. Aspartic proteinase napsin is a useful marker for diagnosis of primary lung adenocarcinoma. Br J Cancer. 2003;88:1229–1233. doi: 10.1038/sj.bjc.6600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng JX, Guan SH, Xu Q, Tang Y, Liu JZ, Lu XT. Effect of Napsin A transfection into type II alveolar epithelial cells on pulmonary fibrosis. Zhonghua Yi Xue Za Zhi. 2010;90:3294–3299. In Chinese. [PubMed] [Google Scholar]
- 14.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 15.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 16.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 17.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolzenburg LR, Wachtel S, Dang H, Harris A. miR-1343 attenuates pathways of fibrosis by targeting the TGF-β receptors. Biochem J. 2016;473:245–256. doi: 10.1042/BJ20150821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han R, Ji X, Rong R, Li Y, Yao W, Yuan J, Wu Q, Yang J, Yan W, Han L, et al. miR-449a regulates autophagy to inhibit silica-induced pulmonary fibrosis through targeting Bcl2. J Mol Med (Berl) 2016;94:1267–1279. doi: 10.1007/s00109-016-1441-0. [DOI] [PubMed] [Google Scholar]
- 20.Wu Q, Han L, Yan W, Ji X, Han R, Yang J, Yuan J, Ni C. miR-489 inhibits silica-induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci Rep. 2016;6:30921. doi: 10.1038/srep30921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghu G, Rochwerg B, Zhang Y, Garcia CA, Azuma A, Behr J, Brozek JL, Collard HR, Cunningham W, Homma S, et al. An official ATS/ERS/JRS/ALAT clinical practice guideline: Treatment of idiopathic pulmonary fibrosis. An update of the 2011 clinical practice guideline. Am J Respir Crit Care Med. 2015;192:e3–19. doi: 10.1164/rccm.201506-1063ST. [DOI] [PubMed] [Google Scholar]
- 22.Aslani S, Mahmoudi M, Garshasbi M, Jamshidi AR, Karami J, Nicknam MH. Evaluation of DNMT1 gene expression profile and methylation of its promoter region in patients with ankylosing spondylitis. Clin Rheumatol. 2016;35:2723–2731. doi: 10.1007/s10067-016-3403-x. [DOI] [PubMed] [Google Scholar]
- 23.Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miR-34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2019;50:304–307. doi: 10.1007/s12029-018-0060-0. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:279. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang TH, Liang LZ, Liu XL, Wu JN, Su K, Chen JY, Zheng QY. LncRNA UCA1/miR-124 axis modulates TGFβ1-induced epithelial-mesenchymal transition and invasion of tongue cancer cells through JAG1/Notch signaling. J Cell Biochem. 2019;120:10495–10504. doi: 10.1002/jcb.28334. [DOI] [PubMed] [Google Scholar]
- 27.Gimenez A, Duch P, Puig M, Gabasa M, Xaubet A, Alcaraz J. Dysregulated collagen homeostasis by matrix stiffening and TGF-β1 in fibroblasts from idiopathic pulmonary fibrosis patients: Role of FAK/Akt. Int J Mol Sci. 2017;18:E2431. doi: 10.3390/ijms18112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Xu Z, Yu L, Che J, Zhang J, Yang J. MicroRNA-144-3p suppressed TGF-β1-induced lung cancer cell invasion and adhesion by regulating the Src-Akt-Erk pathway. Cell Biol Int. 2019 Apr 30; doi: 10.1002/cbin.11158. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 29.Mi XJ, Hou JG, Jiang S, Liu Z, Tang S, Liu XX, Wang YP, Chen C, Wang Z, Li W. Maltol mitigates thioacetamide-induced liver fibrosis through TGF-β1-mediated activation of PI3K/Akt signaling pathway. J Agric Food Chem. 2019;67:1392–1401. doi: 10.1021/acs.jafc.8b05943. [DOI] [PubMed] [Google Scholar]
- 30.Jang YS, Kim JH, Seo GY, Kim PH. TGF-β1 stimulates mouse macrophages to express APRIL through Smad and p38MAPK/CREB pathways. Mol Cells. 2011;32:251–255. doi: 10.1007/s10059-011-1040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Shankar BS, Sainis KB. TGF-β1-ROS-ATM-CREB signaling axis in macrophage mediated migration of human breast cancer MCF7 cells. Cell Signal. 2014;26:1604–1615. doi: 10.1016/j.cellsig.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Herrera J, Beisang DJ, Peterson M, Forster C, Gilbertsen A, Benyumov A, Smith K, Korenczuk CE, Barocas VH, Guenther K, et al. Dicer1 deficiency in the idiopathic pulmonary fibrosis fibroblastic focus promotes fibrosis by suppressing MicroRNA biogenesis. Am J Respir Crit Care Med. 2018;198:486–496. doi: 10.1164/rccm.201709-1823OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Xu K, Yang XY, Liu J, Zeng Q, Wang FS. Upregulated miR-29c suppresses silica-induced lung fibrosis through the Wnt/beta-catenin pathway in mice. Hum Exp Toxicol. 2018;37:944–952. doi: 10.1177/0960327117741750. [DOI] [PubMed] [Google Scholar]
- 34.Xu T, Yan W, Wu Q, Xu Q, Yuan J, Li Y, Li P, Pan H, Ni C. miR-326 inhibits inflammation and promotes autophagy in silica-induced pulmonary fibrosis through targeting TNFSF14 and PTBP1. Chem Res Toxicol. 2019;32:2192–2203. doi: 10.1021/acs.chemrestox.9b00194. [DOI] [PubMed] [Google Scholar]
- 35.Gao SY, Zhou X, Li YJ, Liu WL, Wang PY, Pang M, Xie SY, Lv CJ. Arsenic trioxide prevents rat pulmonary fibrosis via miR-98 overexpression. Life Sci. 2014;114:20–28. doi: 10.1016/j.lfs.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 36.Huleihel L, Ben-Yehudah A, Milosevic J, Yu G, Pandit K, Sakamoto K, Yousef H, LeJeune M, Coon TA, Redinger CJ, et al. Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2014;306:L534–L542. doi: 10.1152/ajplung.00149.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price KJ, Tsykin A, Giles KM, Sladic RT, Epis MR, Ganss R, Goodall GJ, Leedman PJ. Matrigel basement membrane matrix influences expression of microRNAs in cancer cell lines. Biochem Biophys Res Commun. 2012;427:343–348. doi: 10.1016/j.bbrc.2012.09.059. [DOI] [PubMed] [Google Scholar]
- 38.Jin JJ, Liu YH, Si JM, Ni R, Wang J. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int J Biochem Cell Biol. 2018;95:113–120. doi: 10.1016/j.biocel.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Zheng JX, Guan SH, Xu Q, Liu JZ, Song P. Inhibition of epithelial-mesenchymal transition in A549 cell by transfected Napsin A. Chin Med J (Engl) 2012;125:2734–2740. [PubMed] [Google Scholar]
- 40.Sirotkin AV, Benco A, Mlyncek M, Harrath AH, Alwasel S, Kotwica J. The involvement of the phosphorylatable and nonphosphorylatable transcription factor CREB-1 in the control of human ovarian cell functions. C R Biol. 2019;342:90–96. doi: 10.1016/j.crvi.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Mroz RM, Holownia A, Chyczewska E, Drost EM, Braszko JJ, Noparlik J, Donaldson K, Macnee W. Cytoplasm-nuclear trafficking of CREB and CREB phosphorylation at Ser133 during therapy of chronic obstructive pulmonary disease. J Physiol Pharmacol. 2007;58(Suppl 5):S437–S444. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.