Abstract
Liver cancer is a malignant tumor that occurs in the liver and can be divided into primary and secondary liver cancer. Long non-coding RNA (lncRNA) breast cancer anti-estrogen resistance 4 (BCAR4) has been demonstrated to promote the development of various types of cancer. However, the function of lncRNA BCAR4 in liver cancer remains unclear. In the present study, the expression of lncRNA BCAR4 was notably elevated in liver cancer compared with adjacent non-tumor tissues. Functional in vitro assays demonstrated that knockdown of lncRNA BCAR4 inhibited the proliferation, migration and invasion of Huh-7 cells. In addition, lncRNA BCAR4 was demonstrated to directly bind to microRNA (miR)-1261, and miR-1261 expression negatively correlated with the expression of lncRNA BCAR4. Through bioinformatics analysis, lncRNA BCAR4 was predicted to target anaphase-promoting complex subunit 11 (ANAPC11) through miR-1261. In addition, the results demonstrated that lncRNA BCAR4 increased the expression of ANAPC11 by inhibiting miR-1261 expression. Consistently, overexpression of ANAPC11 or inhibition of miR-1261 significantly rescued liver cancer cell proliferation induced by knockdown of lncRNA BCAR4. Collectively, the results of the present study demonstrated that lncRNA BCAR4 may promote liver cancer development by directly binding to miR-1261 and targeting ANAPC11.
Keywords: BCAR4, liver cancer, miR-1261, ANAPC11, migration, invasion
Introduction
Hepatic malignant tumors can be divided into primary and secondary: Primary hepatic malignant tumors originate from the epithelium or mesenchymal tissue of the liver; the etiology and exact molecular mechanisms of primary liver cancer are not completely clear (1,2). The pathogenesis of liver cancer is considered to be a complex multifactor process of multiple steps, which is affected by both environmental and dietary factors (3).
Long non-coding RNAs (lncRNAs) have been reported to serve important roles in regulating a number of biological processes, especially cancer development (4-6). LncRNA breast cancer anti-estrogen resistance 4 (BCAR4) has been demonstrated to regulate the progression of various types of cancer, including colon, breast and non-small cell lung cancer. Ouyang et al (7) have reported that lncRNA BCAR4 can activate the Wnt/β-catenin signaling pathway to promote colon cancer progression. In addition, Xing et al (8) have demonstrated that lncRNA BCAR4 participates in the regulation of breast cancer development. Furthermore, Li et al (9) reported that lncRNA BCAR4 regulated the progression of non-small cell lung cancer through epithelial-mesenchymal transition. However, the function of lncRNA BCAR4 in regulating liver cancer remains unknown.
MicroRNAs (miRNAs) have been reported to participate in the regulation of multiple types of cancer by binding to lncRNAs (10-13). MicroRNA (miR)-1261 was first reported to promote invasion and migration of prostate cancer cells by binding to lncRNA prostate cancer antigen 3 (14). In addition, miR-1261 has been demonstrated to regulate the development of papillary thyroid cancer and glioma (15,16). However, the role of miR-1261 in regulating liver cancer progression remains unclear. Thus, the present study aimed to examine the role of miR-1261 in the regulation of liver cancer.
Anaphase-promoting complex subunit 11 (ANAPC11) has been reported to mediate the degradation of cell cycle (17). In addition, overexpression of ANAPC11 was associated with chromosomal instability in colorectal cancer, lymphovascular invasion and residual tumors (18). However, the role of ANAPC11 in the regulation of liver cancer remains unknown. This study aimed to determine the function of ANAPC11 in regulating liver cancer progression.
Materials and methods
Samples and cell lines
Human liver cancer samples and adjacent healthy liver tissues were obtained from 30 patients (14 male and 16 female; mean age, 54 years; age range, 47-72 years). Patients were diagnosed with liver cancer and underwent surgery at Taiyuan Second People's Hospital between July 2016 and September 2019. The matched adjacent non-cancer tissue was collected 2 cm away from the edge of cancer tissue. Samples were obtained from patients during surgery, and experienced pathologists confirmed the diagnosis. All procedures were performed in accordance with the Helsinki Declaration. Exclusion criteria included radiotherapy or chemotherapy prior to surgical treatment, prior history of cancer and a lack of the written informed consent. All samples were kept in liquid nitrogen before use. This study was approved by the Ethics Committee of Taiyuan Second People's Hospital. Written informed consent was obtained from all patients.
The expression levels of BCAR4 were examined by reverse transcription-quantitative PRC (RT-qPCR) and categorized as low or high according to the median value.
Huh7 cell lines were obtained from the Japanese Cancer Research Resources Bank and cultured in DMEM supplied with 10% FBS (Thermo Fisher Scientific, Inc.) with 5% CO2 at 37°C (19,20).
RT-qPCR
Total RNA form tissues and cells was extracted using TRIzol® regent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse-transcribed into cDNA by PrimeScript RT Reagent kit (Promega Corporation) according to the manufacturer's instructions. QPCR was performed using SYBR® Green PCR Master Mix (Takara Bio, Inc.) on a 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 40 sec (21). The primers used were as follows: 18S forward, 5′-GTA ACC CGT TGA ACC CCA TT-3′ and reverse, 5′-CCA TCC AAT CGG TAG TAG CG-3′; lncRNA BCAR4 forward, 5′-GTG GGG ACA TTC AAG TGA AC-3′ and reverse, 5′-GAT GAG CAA CAA GCT GCT GTG-3′; ANAPC11 forward, 5′-GGA TGG CAT TTA ACG GAT G-3′ and reverse, 5′-GTC TGG GGA CCT AGA AGA CTC-3′; miR-1261 forward, 5′-TGC TAT GGA TAA GGC TTT G-3′ and reverse, 5′-GCT GCT ATG GAG AAA GTT TC-3′.
Bioinformatics analysis
The target miRNAs of lncRNA BCAR4 were predicted by bioinformatics analysis using the miRDB tool (http://mirdb.org). The potential target genes regulated by miR-1261 were predicted by TargetScan 7.2 (http://www.targetscan.org/vert_72/).
Plasmid construction and transfection
Full-length ANAPC11 coding region (ANAPC11 forward, 5′-ATG AAG GTG AAG ATT AAG TGC TGG AAC G-3′ and reverse, 5′-TCA GGA TGC CCC TCC AGC GAG AG-3′) was cloned into pMy vectors (Addgene, Inc.) to overexpress ANAPC11. An empty vector was used as a negative control. miR-1261 mimic (5′-ACU AUG UUG ACA CUU UUA UCC AA-3′), miR-1261 inhibitor (5′-UGA UAC AAC YGA AAA UAG GUU-3′), mimic control (5′-ACA UCU GCG UAA GAU UCG AGU CUA-3′), control inhibitor (5′-UAA CUA AUA CAU CGG AUU-3′), short hairpin (sh)RNA targeting lncRNA BCAR4 (1 mg; shBCAR4-1; 5′-GCU GCG AGG GUA GAC AUC U-3′ and shBCAR4-2; 5′-GUG AUU GCC AAA CGC UCC C-3′), shANAPC11 (5′-UCC CAG GAC AGG CAC AGG C-3′) and scramble control shRNA (1 mg; 5′-UAA GGC UAU GAA GAG AUA C-3′) were purchased from Shanghai GenePharma Co, Ltd. and cloned into a PLKO.1 puro vector (cat. no. 8453; Addgene, Inc.). Huh7 cells were counted and seeded in a 6-well plate at 1×106 cells/well. When the confluence reached 90%, the cells were transfected with 50 nM miRNA mimic, 50 nM miRNA inhibitor or 4.0 µg corresponding plasmids using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were cultured at 37°C in a 5% CO2 incubator for 48 h and harvested for subsequent experimentation.
Luciferase reporter assay
Luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's instructions. Cells (1×106 cells/well) were seeded into 24-well plates 1 day prior to transfection. Cells were transfected with a wild-type (wt)-BCAR4 luciferase reporter gene vector, a mutant (mut)-BCAR4 vector containing a 6-bp mutation on the predicted miR-1261 binding site within BCAR4, a wt-ANAPC11 3′UTR vector, or a mut-ANAPC11 3′UTR vector (all from Shanghai GenePharma Co., Ltd.) containing a mutation in the predicted miR-1261 binding site in the 3′UTR of ANAPC11, along with the miR-1261 mimic or mimic NC using Lipofectamine® 2000 into Huh7 cells. Cells were cultured at 37°C in a 5% CO2 incubator for 48 h. Then, luciferase activity was measured. Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell Counting Kit-8 (CCK8) assay
CCK8 assay was performed using CCK8 (7Sea Biotech Co., Ltd.) according to the manufacturer's instructions.
Colony formation assay
Huh7 cells were cultured to the logarithmic growth phase and digested with 0.25% trypsin into single cells. The wells of the control or shBACR4 group were inoculated with 1,000 cells in DMEM with 10% FBS for about two weeks. When the clones were visible, the supernatant was discarded, and the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After removing the fixative solution, cells were stained with Giemsa solution for 30 min at room temperature. Subsequently, the staining solution was washed out with running water, and the cells were dried. Finally, images were acquired under an optical microscope with x10 magnification (Olympus Corporation).
Transwell assay
Matrigel was used on the upper chamber of Transwell chamber for the invasion assay (37°C for 30 min). Huh7 cells were seeded (4×104 cells/well) into Transwell chambers with 8-µm pores (BD Biosciences). Serum-free DMEM was added to the upper chamber, and DMEM containing 10% FBS was added to the lower chamber. After 24 h, the cells on the upper side of membrane were removed using a cotton swab; the cells on the lower side of the membrane were fixed with 4% paraformaldehyde for 30 min at room temperature and stained with 0.1% crystal violet for 30 min at room temperature. Finally, images were acquired using a light microscope (Olympus Corporation) at x100 magnification. Each sample was analyzed in three random fields.
Statistical analysis
Data are presented as the mean ± SD. GraphPad Prism 6 software (GraphPad Software, Inc.) was used to analyze the data. Unpaired Student's t-test was used to analyze the differences between two groups. One-way ANOVA followed by a Tukey's post hoc test was used for multiple comparisons. The overall survival was analyzed by Kaplan-Meier analysis and a log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA BCAR4 is expressed in liver cancer cells and associated with liver cancer progression
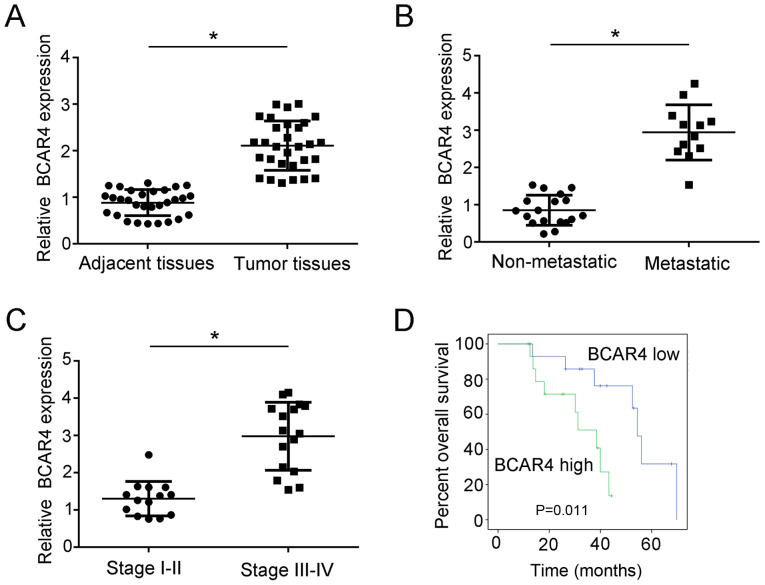
To explore the expression of lncRNA BCAR4 in liver cancer tissues, RT-qPCR assay was performed. The results demonstrated that the expression of lncRNA BCAR4 in liver cancer tissues was significantly higher compared with that in paired healthy liver tissues (Fig. 1A). In addition, the association between liver cancer metastasis and lncRNA BCAR4 expression was analyzed; lncRNA BCAR4 expression was higher in the metastatic liver cancer group compared with that in the non-metastatic group (Fig. 1B). The expression level of lncRNA BCAR4 was also higher in liver cancer stages III and IV compared with that in stages I and II (Fig. 1C). To examine the overall survival rate of patients with liver cancer with high lncRNA BCAR4 expression, Kaplan-Meier analysis was performed. Patients with liver cancer were divided into two groups based on the median expression of lncRNA BCAR4, and the results demonstrated that the high lncRNA BCAR4 expression group exhibited a lower survival rate compared with the low expression group (Fig. 1D). Taken together, these data demonstrated that the expression of lncRNA BCAR4 was associated with liver cancer progression.
Figure 1.
LncRNA BCAR4 is associated with liver cancer progression. (A) The relative expression of lncRNA BCAR4 in liver cancer tissues and normal healthy tissues was measured by RT-qPCR. The expression was normalized to that of 18S RNA. (B) RT-qPCR was performed to determine the expression of lncRNA BCAR4 in non-metastatic (n=18) and metastatic (n=12) liver cancer samples. The expression was normalized to that of 18S RNA. (C) RT-PCR was used to examine the expression of lncRNA BCAR4 in stage I and II (n=14) and stage III and IV (n=16) liver cancer samples. The expression was normalized to that of 18S. (D) Overall survival rate of high and low lncRNA BCAR4 expression groups was analyzed by Kaplan-Meier analysis and the long rank test. All experiments were repeated independently three times. *P<0.05. LncRNA, long non-coding RNA; BCAR4, breast cancer anti-estrogen resistance 4; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of BCAR4 significantly decreases the proliferative, migratory and invasive abilities of liver cancer cells
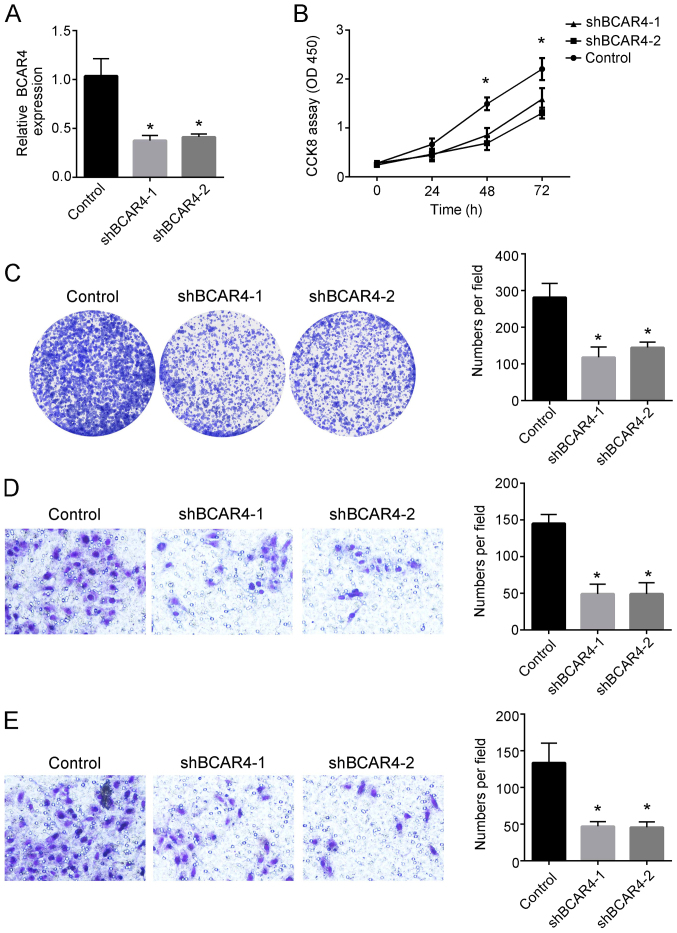
To analyze the functions of lncRNA BCAR4 in regulating liver cancer, shRNAs targeting the lncRNA BCAR4 sequence were constructed and transfected into Huh7 liver cancer cells (Fig. 2A). CCK8 assay results demonstrated that knockdown of lncRNA BCAR4 decreased the proliferative ability of liver cancer cells compared with that of the control group (Fig. 2B). In addition, colony formation assay was performed to further examine the proliferative abilities of Huh7 cells. The results revealed that the colony numbers were decreased after knockdown of lncRNA BCAR4 in Huh7 cells compared with those in the control group (Fig. 2C). Transwell assay results also demonstrated that knockdown of lncRNA BCAR4 significantly decreased the migration and invasion of Huh7 cells compared with the control group (Fig. 2D and E). Collectively, these results suggested that knockdown of lncRNA BCAR4 inhibited liver cancer cell abilities associated with cancer progression.
Figure 2.
Knockdown of BCAR4 inhibits liver cancer proliferation, migration and invasion. (A) Relative expression of lncRNA BCAR4 was determined by reverse transcription-quantitative PCR after transfecting shBCAR4 plasmids and control plasmid in Huh7 cells. The expression was normalized to that of 18S. (B) Cell Counting Kit-8 assay was performed to examine the proliferative abilities of Huh7 cells after BCAR4 knockdown. (C) Colony formation assay was used to detect the colony-forming ability of Huh7 cells transfected with shBCAR4 or control shRNA. (D and E) The (D) migratory and (E) invasive abilities of Huh7 cells were examined using Transwell assay following transfection with shBCAR4 or control shRNA. Plates were imaged at ×4 magnification. All experiments were repeated independently three times. *P<0.05. LncRNA, long non-coding RNA; BCAR4, breast cancer anti-estrogen resistance 4; sh, short hairpin.
miR-1261 directly binds to lncRNA BCAR4 and affects its expression
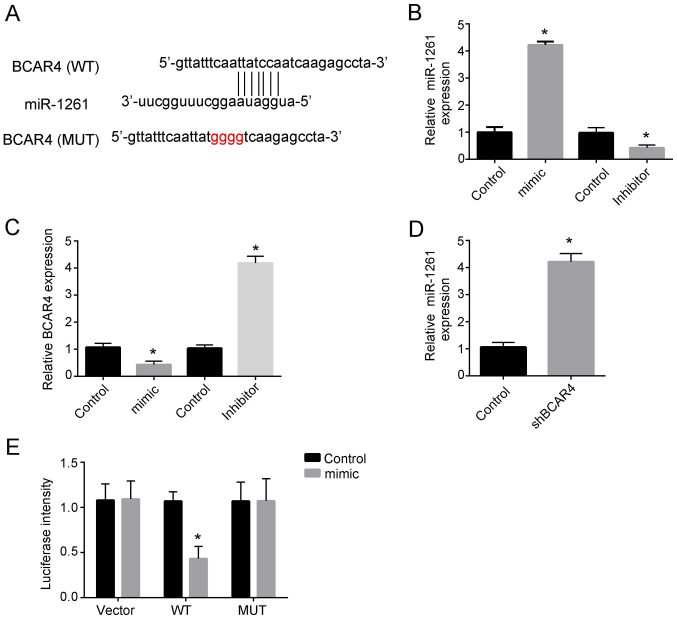
As the aforementioned results demonstrated that lncRNA BCAR4 was associated with liver cancer proliferation, migration and invasion, bioinformatics analysis was performed to identify the potential mechanism by which lncRNA BCAR4 regulated liver cancer progression. The results revealed that lncRNA BCAR4 could directly bind miR-1261 (Fig. 3A). To explore the relationship between lncRNA BCAR4 and miR-1261, miR-1261 mimic plasmid and inhibitor were used (Fig. 3B). miR-1261 mimics significantly inhibited the expression of lncRNA BCAR4, whereas inhibition of miR-1261 promoted BCAR4 expression compared with the corresponding control groups (Fig. 3C). Additionally, knockdown of lncRNA BCAR4 decreased the expression of miR-1261 compared with the control group (Fig. 3D). Luciferase assays were also performed, and the results confirmed that BCAR4 bound to miR-1261 directly (Fig. 3E). Collectively, these results demonstrated that miR-1261 directly bound to BCAR4 and affected the expression of lncRNA BCAR4.
Figure 3.
miR-1261 binds to lncRNA BCAR4 and affects its expression. (A) The binding sites between miR-1261 and BCAR4. (B) Relative expression of miR-1261 was examined by RT-qPCR after transfection with the miR-1261 mimic, miR-1261 inhibitor or control plasmids in Huh7 cells. The expression was normalized to that of 18S. (C) Relative expression of BCAR4 was examined by RT-qPCR following transfection with the miR-1261 mimic, miR-1261 inhibitor or control plasmids in Huh7 cells. The expression was normalized to that of 18S. (D) Relative expression of miR-1261 was examined by RT-qPCR after transfection with the shBCAR4 plasmid in Huh7 cells. The expression was normalized to that of 18S. (E) Luciferase assay in Huh7 cells was performed to detect the luciferase intensity after co-transfecting the cells with miR-1261 plasmid and WT or MUT pMIR-lncBCAR4. All experiments were repeated independently three times. *P<0.05. miR, microRNA; lncRNA, long non-coding RNA; BCAR4, breast cancer anti-estrogen resistance 4; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; WT, wild-type; MUT, mutant.
ANAPC11 is the downstream target of miR-1261 and lncRNA BCAR4
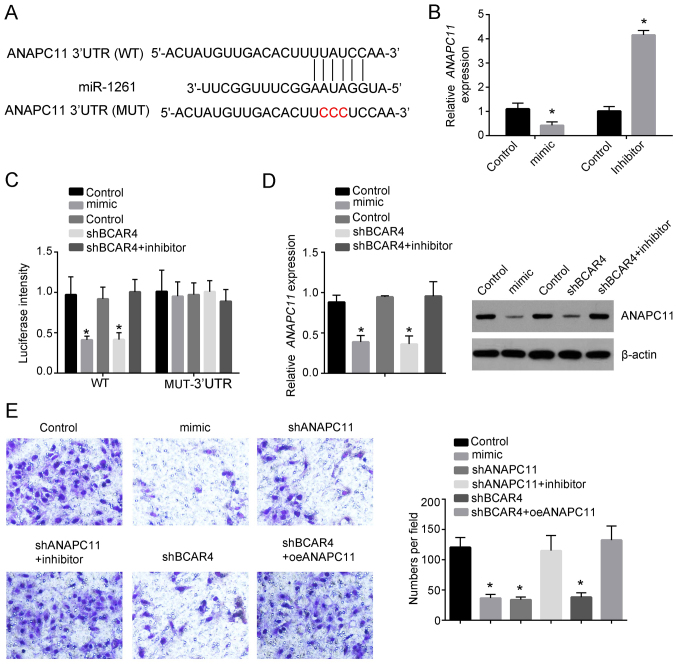
To identify the potential target gene for lncRNA BCAR4 and miR-1261, bioinformatics analysis was performed. The results revealed that ANAPC11 was the potential target of miR-1261 (Fig. 4A). miR-1261 mimics notably inhibited ANAPC11 expression, whereas inhibition of miR-1261 increased the expression of ANAPC11 compared with the corresponding control groups (Fig. 4B). Luciferase assay results revealed that knockdown of BCAR4 resulted in a significant decrease the luciferase activity of wild-type ANPC11 (Fig. 4C). In addition, knockdown of lncRNA BCAR4 decreased the expression of ANAPC11 compared with that in the control group, whereas inhibition of miR-1261 rescued the decreased expression of ANAPC11 caused by knockdown of lncRNA BCAR4 (Fig. 4D). Consistently, overexpression of miR-1261, knockdown of lncRNA BCAR4 or knockdown of ANAPC11 significantly decreased the migration of Huh7 cells compared with that in the control group (Fig. 4E). Additionally, inhibition of miR-1261 rescued the reduced migration of Huh7 cells caused by knockdown of ANAPC11, whereas overexpression of ANAPC11 rescued the decreased migration cells caused by knockdown of lncRNA BCAR4 (Fig. 4E). These results suggested that ANAPC11 may be a potential target of lncRNA BCAR4 and miR-1261.
Figure 4.
ANAPC11 is targeted by lncRNA BCAR4 and miR-1261. (A) The binding sites between miR-1261 and ANAPC11. (B) Relative expression of ANAPC11 was examined by RT-qPCR after transfecting miR-1261 overexpression plasmid or miR-1261 inhibitor in Huh7 cells. The expression was normalized to that of 18S. (C) Luciferase assay was performed to detect the luciferase intensity after transfection with miR-1261 mimic, miR-1261 inhibitor, shBCAR4 or shBCAR4 together with the miR-1261 inhibitor or control plasmids in Huh7 cells. (D) Relative expression of ANAPC11 was examined by RT-qPCR after transfection with the miR-1261 mimic, miR-1261 inhibitor, shBCAR4 or shBCAR4 together with the miR-1261 inhibitor or control in Huh7 cells. The expression was normalized to that of 18S. (E) Transwell assay was performed to determine the migratory ability of Huh7 cells following transfection with the miR-1261 the mimic, shANAPC11 or shANAPC11 together with the miR-1261 inhibitor, shBCAR4, shBCAR4, oeANAPC11 or control plasmids. Plates were imaged at x4 magnification. All experiments were repeated independently three times. *P<0.05. ANAPC11, anaphase-promoting complex subunit 11; miR, microRNA; lncRNA, long non-coding RNA; BCAR4, breast cancer anti-estrogen resistance 4; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; WT, wild-type; MUT, mutant; oe, overexpression.
Discussion
As a malignant tumor with high incidence, liver cancer is a serious threat to human health (22,23). The incidence of liver cancer is associated with hepatitis B and C virus infection, aflatoxin, drinking water pollution, alcohol, liver cirrhosis, sex hormones, nitrosamines and trace elements (24-26). However, the pathogenesis and treatment of liver cancer is still unclear (27,28). Therefore, it is of great significance to study the regulatory mechanism of liver cancer tumorigenesis.
A number of lncRNAs have been reported to regulate liver cancer progression. Mao et al (29) have reported that lncRNA LALR1 upregulates small nucleolar RNA C/D box 72 to promote the proliferation and invasion of liver cancer cells. In addition, lncRNA HAND2 antisense RNA 1 has been demonstrated to inhibit the proliferation and migration of liver cancer cells (30). According to previous studies, lncRNAs can promote or inhibit the progression of liver cancer; therefore, it is important to explore the roles of different lncRNAs in the regulation of liver cancer. LncRNA BCAR4 has been reported to participate in the regulation of the progression of multiple types of cancer (31-33). However, the role of lncRNA BCAR4 in liver cancer remains unknown. The results of the present study demonstrated that the expression of lncRNA BCAR4 was significantly higher in liver cancer compared with adjacent non-tumor tissues. In addition, the proliferation, migration and invasion of Huh7 cells were significantly decreased after knockdown of lncRNA BCAR4 compared with those in the control group.
In previous studies, lncRNAs were demonstrated to regulate the progression of liver cancer cells through binding miRNAs. For example, Wei et al (34) demonstrated that lncRNA MFI2 antisense RNA 1 promoted HCC progression by binding miR-134. In the present study, lncRNA BCAR4 was demonstrated to bind miR-1261 and target ANAPC11. He et al (14) have reported that miR-1261 can promote the progression of prostate cancer. Wei et al (15) have demonstrated that miR-1261 was involved in the regulation of papillary thyroid cancer progression. In addition, Zhang et al (16) reported that miR-1261 was involved in glioma progression. However, the function of miR-1261 in the regulation of other types of cancer remain unclear. ANAPC11 has been reported to regulate the cell cycle distribution of 293T cells (17). In addition, Drouet et al (18) and Moyret-Lalle et al (35) have demonstrated that ANAPC11 is involved in the regulation of colorectal cancer. However, the role of ANAPC11 in the regulation of liver cancer remains unclear. The results of the present study demonstrated that lncRNA BCAR4 promoted the expression of ANAPC11 by inhibiting miR-1261. Additionally, knockdown of ANAPC11 significantly inhibited the migration of liver cancer cells compared with the control, whereas overexpression of ANAPC11 rescued the decreased migration of liver cancer cells caused by knockdown of BCAR4.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or used during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YZ and HZ conceived and designed the present study. YZ performed the experiments, analyzed and interpreted the results. HZ wrote the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Taiyuan Second People's Hospital. Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lazăr DC, Avram MF, Romoșan I, Văcariu V, Goldiș A, Cornianu M. Malignant hepatic vascular tumors in adults: Characteristics, diagnostic difficulties and current management. World J Clin Oncol. 2019;10:110–135. doi: 10.5306/wjco.v10.i3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: Epithelial-mesenchymal transition-does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008;5:280–290. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Tienderen GS, Groot Koerkamp B, IJzermans JNM, van der Laan LJW, Verstegen MMA. Recreating tumour complexity in a dish: Organoid models to study liver cancer cells and their extracellular environment. Cancers (Basel) 2019;11 doi: 10.3390/cancers11111706. pii: E1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang M, Sang Y, Gu H, Zheng L, Wang L, Liu C, Shi Y, Shao A, Ding G, Chen S, et al. Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumour Biol. 2015;36:6401–6408. doi: 10.1007/s13277-015-3328-z. [DOI] [PubMed] [Google Scholar]
- 5.Klec C, Gutschner T, Panzitt K, Pichler M. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. Expert Opin Ther Targets. 2019;23:177–186. doi: 10.1080/14728222.2019.1570499. [DOI] [PubMed] [Google Scholar]
- 6.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/β-catenin signaling. Oncotarget. 2017;8:92815–92826. doi: 10.18632/oncotarget.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12:681–689. doi: 10.1080/15476286.2015.1053687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21:2075–2086. [PubMed] [Google Scholar]
- 10.Yin XH, Jin YH, Cao Y, Wong Y, Weng H, Sun C, Deng JH, Zeng XT. Development of a 21-miRNA signature associated with the prognosis of patients with bladder cancer. Front Oncol. 2019;9:729. doi: 10.3389/fonc.2019.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Gao S, Lindberg D, Panja D, Wakabayashi Y, Li K, Kleinman JE, Zhu J, Li Z. Temporal dynamics of miRNAs in human DLPFC and its association with miRNA dysregulation in schizophrenia. Transl Psychiatry. 2019;9:196. doi: 10.1038/s41398-019-0538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angius A, Uva P, Pira G, Muroni MR, Sotgiu G, Saderi L, Uleri E, Caocci M, Ibba G, Cesaraccio MR, et al. Integrated analysis of miRNA and mRNA endorses a twenty miRNAs signature for colorectal carcinoma. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20164067. pii: E4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen MC, Yeh IJ, Liu KT, Jian SF, Lin CJ, Tsai MJ, Kuo PL. Next-generation sequencing predicts interaction network between miRNA and target genes in lipoteichoic acid-stimulated human neutrophils. Int J Mol Med. 2019;44:1436–1446. doi: 10.3892/ijmm.2019.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He JH, Li BX, Han ZP, Zou MX, Wang L, Lv YB, Zhou JB, Cao MR, Li YG, Zhang JZ. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cance cells. Tumour Biol. 2016 Oct 14; doi: 10.1007/s13277-016-5450-y. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61. doi: 10.1016/j.bbrc.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 16.Zhang F, Mai SR, Cao FP, Cao CX, Zhang L. MiR-1261/circ-PTPRZ1/PAK1 pathway regulates glioma cell growth and invasion. Human Cell. 2019;32:540–547. doi: 10.1007/s13577-019-00271-3. [DOI] [PubMed] [Google Scholar]
- 17.Shi YJ, Huo KK. Knockdown expression of Apc11 leads to cell-cycle distribution reduction in G2/M phase. Genet Mol Res. 2012;11:2814–2822. doi: 10.4238/2012.August.24.6. [DOI] [PubMed] [Google Scholar]
- 18.Drouet Y, Treilleux I, Viari A, Léon S, Devouassoux-Shisheboran M, Voirin N, de la Fouchardière C, Manship B, Puisieux A, Lasset C, Moyret-Lalle C. Integrated analysis highlights APC11 protein expression as a likely new independent predictive marker for colorectal cancer. Sci Rep. 2018;8:7386. doi: 10.1038/s41598-018-25631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Fang X, Han M, Wang X, Huang Q. MicroRNA-493-5p promotes apoptosis and suppresses proliferation and invasion in liver cancer cells by targeting VAMP2. Int J Mol Med. 2018;41:1740–1748. doi: 10.3892/ijmm.2018.3358. [DOI] [PubMed] [Google Scholar]
- 20.Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, Li X, Zheng Q, Pu H, Gui X, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17:94. doi: 10.1186/s12943-018-0843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Zhu H, Yang D, Xia J, Zheng Z. Long noncoding RNA lncBRM promotes proliferation and invasion of colorectal cancer by sponging miR-204-3p and upregulating TPT1. Biochem Biophys Res Commun. 2019;508:1259–1263. doi: 10.1016/j.bbrc.2018.12.053. [DOI] [PubMed] [Google Scholar]
- 22.Francica G, Borzio M. Status of, and strategies for improving, adherence to HCC screening and surveillance. J Hepatocell Carcinoma. 2019;6:131–141. doi: 10.2147/JHC.S159269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viveiros P, Riaz A, Lewandowski RJ, Mahalingam D. Current state of liver-directed therapies and combinatory approaches with systemic therapy in hepatocellular carcinoma (HCC) Cancers (Basel) 2019;11 doi: 10.3390/cancers11081085. pii: E1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Q, Zhang G, Zheng Y, Yang Y, Chen C, Xia J, Liang L, Lei C, Hu Y, Cai X, et al. SLC27A5 deficiency activates NRF2/TXNRD1 pathway by increased lipid peroxidation in HCC. Cell Death Differ. 2020;27:1086–1104. doi: 10.1038/s41418-019-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feld J. Update on the risk of primary and recurrent HCC With the use of DAA therapy for HCV infection. Gastroenterol Hepatol (N Y) 2019;15:303–306. [PMC free article] [PubMed] [Google Scholar]
- 26.Fang G, Zhang P, Liu J, Zhang X, Zhu X, Li R, Wang H. Inhibition of GSK-3β activity suppresses HCC malignant phenotype by inhibiting glycolysis via activating AMPK/mTOR signaling. Cancer Lett. 2019;463:11–26. doi: 10.1016/j.canlet.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Shigeta K, Datta M, Hato T, Kitahara S, Chen IX, Matsui A, Kikuchi H, Mamessier E, Aoki S, Ramjiawan RR, et al. Dual programmed death receptor-1 and vascular endothelial growth factor receptor-2 blockade promotes vascular normalization and enhances antitumor immune responses in hepatocellular carcinoma. Hepatology. 2019 Aug 5; doi: 10.1002/hep.30889. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju JX, Zeng QJ, Xu EJ, He XQ, Tan L, Huang QN, Li K, Zheng RQ. Intraprocedural contrast-enhanced ultrasound-CT/MR fusion imaging assessment in HCC thermal ablation to reduce local tumor progression: Compared with routine contrast-enhanced ultrasound. Int J Hyperthermia. 2019;36:785–793. doi: 10.1080/02656736.2019.1640899. [DOI] [PubMed] [Google Scholar]
- 29.Mao LH, Chen SY, Li XQ, Xu F, Lei J, Wang QL, Luo LY, Cao HY, Ge X, Ran T, et al. LncRNA-LALR1 upregulates small nucleolar RNA SNORD72 to promote growth and invasion of hepatocellular carcinoma. Aging (Albany NY) 2020;12:4527–4546. doi: 10.18632/aging.102907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan D, Jin F, Lin Y. lncRNA HAND2-AS1 inhibits liver cancer cell proliferation and migration by upregulating SOCS5 to inactivate the JAK-STAT pathway. Cancer Biother Radiopharm. 2020;35:143–152. doi: 10.1089/cbr.2019.2958. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang S, Zhou X, Chen Z, Wang M, Zheng X, Xie M. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72. doi: 10.1186/s12935-019-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godinho MF, Wulfkuhle JD, Look MP, Sieuwerts AM, Sleijfer S, Foekens JA, Petricoin EF, III, Dorssers LC, van Agthoven T. BCAR4 induces antioestrogen resistance but sensitises breast cancer to lapatinib. Br J Cancer. 2012;107:947–955. doi: 10.1038/bjc.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Yan L, Sun K, Sun X, Zhang X, Cai K, Song T. lncRNA BCAR4 increases viability, invasion, and migration of non-small cell lung cancer cells by targeting glioma-associated oncogene 2 (GLI2) Oncol Res. 2019;27:359–369. doi: 10.3727/096504018X15220594629967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y, Wang Z, Zong Y, Deng D, Chen P, Lu J. LncRNA MFI2-AS1 promotes HCC progression and metastasis by acting as a competing endogenous RNA of miR-134 to upregulate FOXM1 expression. Biomed Pharmacother. 2020;125:109890. doi: 10.1016/j.biopha.2020.109890. [DOI] [PubMed] [Google Scholar]
- 35.Moyret-Lalle C, Drouet Y, Puisieux A. The catalytic subunit of the anaphase-promoting complex, APC11, is involved in CIN+ CRC progression. Med Sci (Paris) 2019;35:118–122. doi: 10.1051/medsci/2019020. In French. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or used during the present study are available from the corresponding author on reasonable request.