Abstract
Our understanding of the plasma membrane structure has undergone a major change since the proposal of the fluid mosaic model of Singer and Nicholson in the 1970s. In this model, the membrane, composed of over thousand lipid and protein species, is organized as a well‐equilibrated two‐dimensional fluid. Here, the distribution of lipids is largely expected to reflect a multicomponent system, and proteins are expected to be surrounded by an annulus of specialized lipid species. With the recognition that a multicomponent lipid membrane is capable of phase segregation, the membrane is expected to appear as patchwork quilt pattern of membrane domains. However, the constituents of a living membrane are far from being well equilibrated. The living cell membrane actively maintains a trans‐bilayer asymmetry of composition, and its constituents are subject to a number of dynamic processes due to synthesis, lipid transfer as well as membrane traffic and turnover. Moreover, membrane constituents engage with the dynamic cytoskeleton of a living cell, and are both passively as well as actively manipulated by this engagement. The extracellular matrix and associated elements also interact with membrane proteins contributing to another layer of interaction. At the nano‐ and mesoscale, the organization of lipids and proteins emerge from these encounters, as well as from protein–protein, protein–lipid, and lipid–lipid interactions in the membrane. New methods to study the organization of membrane components at these scales have also been developed, and provide an opportunity to synthesize a new picture of the living cell surface as an active membrane composite.
Keywords: actin binding proteins, actin cytoskeleton, fluid mosaic model, GPI‐anchored proteins, mesoscale organization, nanoclustering, picket fence model, plasma membrane, plasma membrane organization, transmembrane proteins
1. INTRODUCTION
Understanding the structure of the animal cell membrane is of paramount importance to decipher how a cell interacts with its external milieu, and has, consequently, a very long history.1, 2 Beginning with the development of the Pockels cell by Agnes Pockels in the 1880s, to the measurement of the surface area of a phospholipid by the use of a modified version of the same by Langmuir (1935), the proposal that the cell is covered by a bi‐layered film of membrane lipids 3 was a major breakthrough. To account for the presence of membrane proteins, a tri‐lamellar structure was also proposed. 4 A true synthesis was achieved in an influential model proposed by Singer and Nicholson, 5 wherein the membrane is depicted as a sea of lipids forming a fluid lipid bilayer in which proteins are dissolved (Figure 1). This history reveals an intertwined relationship between concepts borrowed from the physics of artificial membranes and observations made on cell membranes.
FIGURE 1.
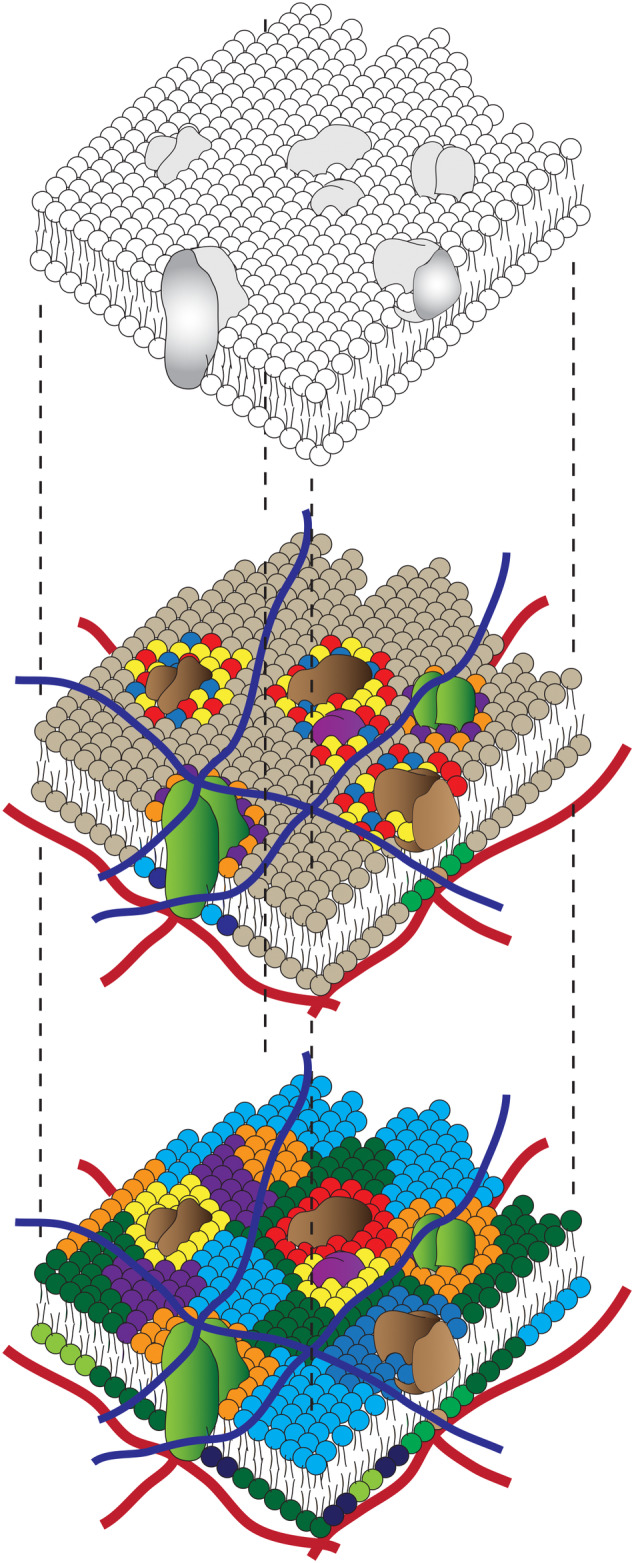
The fluid mosaic and patchwork quilt models of the cell membrane: The Singer Nicholson model of the membrane where proteins are dissolved in a two‐dimensional fluid sea of lipids (top), has given rise over the years to a more variegated view of the membrane where the membrane is today considered as a patchwork quilt of multiple domains of proteins and lipids of a variety of compositions (middle and bottom). These models are predominantly based on thermodynamic equilibrium principles, and embody an equilibrium steady state expectation of this complex mixture of proteins and lipids. The light brown and other multicolored balls represent different lipid head group species that populate the membrane. Transmembrane proteins are shown as green blobs that traverse the membrane bilayer and dark brown proteins denote various kinds of extrinsic surface proteins. Note how various proteins are associated with subsets of multicolored lipid species (middle and bottom panel) while membrane lipids are also thought to form a patchwork quilt (bottom panel). The extracellular matrix is shown as a cross‐linked network of blue filaments, and the cytoskeletal elements are detailed as red filaments
The picture has been naturally extended to encompass the physical consequences of a multicomponent system capable of phase segregation, giving rise to a patchwork quilt model of a plasma membrane 6 (Figure 1), where these lateral heterogeneities or membrane domains are imbued with functional consequences. 7 While the mechanism for the generation of these lateral heterogeneities has been hotly debated, the constituents of a living membrane are not in chemical equilibrium. This membrane maintains a trans‐bilayer asymmetry of composition by the use of active pumps, and its constituents are subject to a number of dynamic processes due to synthesis, lipid transfer protein interactions, especially at membrane contact sites, as well as membrane traffic and turnover. Therefore, purely equilibrium principles of phase segregation are unlikely to serve as the sole mechanism for creating the organization that is observable in the membrane of a living cell. Indeed, Edidin had stated “we are awaiting a new model that integrates the numerous features of a eukaryotic cell membrane which have emerged since they were first characterized.” 6 Any new model must also take into account the cause and consequence of the multilayered structuring of the material that is in continuous contact with the components of the membrane in the living cell, and this is the main focus of this review.
2. EARLY EVIDENCE OF MULTISCALE ORGANIZATION OF PLASMA MEMBRANE MOLECULES
The discussion regarding the existence of heterogeneous distribution of membrane molecules at multiple spatial scales: nano and the meso, goes back decades. 2 This has relevance, since compelling evidence for functional domains, often termed “rafts,” are implicated in creating platforms for sorting and signaling.8, 9, 10 Equilibrium phase segregation occurs in multicomponent artificial membranes of appropriate composition and concentration, at both the nano‐ and mesoscale. 11 However, there appears to be little evidence that domains consisting of specific proteins and lipids form by equilibrium phase segregation driven processes in living cell membranes. 10
A number of methodologies have been deployed to ascertain the existence of these membrane domains, relying on measuring consequences of sequestering membrane components in such domains. These range from visual inspection of enrichment in domains, to exploring enhanced proximity between molecules that visit these domains, to studying the diffusion of molecules proposed to be part of these domains. 12
Multiple modes of diffusion are exhibited by membrane molecules, simple Brownian, confined diffusion as well as ballistic diffusion (Figure 2). Using high‐speed single particle tracking (SPT) studies, Kusumi et al. observed multiple modes of diffusion.14, 15 Findings from a large number of studies using different membrane molecules indicated that transmembrane proteins or glycosylphosphatidylinositol‐anchored proteins and phospholipids, exhibited a dependence of diffusion time scales on observation area that did not scale according to expectations from simple Brownian diffusion. Large diffusion coefficients were detected in compartment sizes of less than hundred nanometers and an order of magnitude smaller diffusion coefficients over micrometers.14, 16 This led to the development of the hop diffusion model where it was proposed that molecules would exhibit fast diffusion within a compartment, but would be limited by barriers to hop/transition to neighboring compartments. The anchored picket fence model of the plasma membrane was evoked to explain such anomalous diffusion behavior of membrane lipids and proteins. 15 In this model, transmembrane actin‐binding proteins are anchored to the underlying cortical actin (CA) filaments, creating actin corrals, which in turn were also corroborated in electron microscopy (EM)‐based studies. 17 The actin mesh is relatively static and undergoes thermally driven fluctuations and polymerization and depolymerization driven turnover regulated by activity of cytoskeletal motors, actin‐nucleating, and severing proteins.15, 18
FIGURE 2.
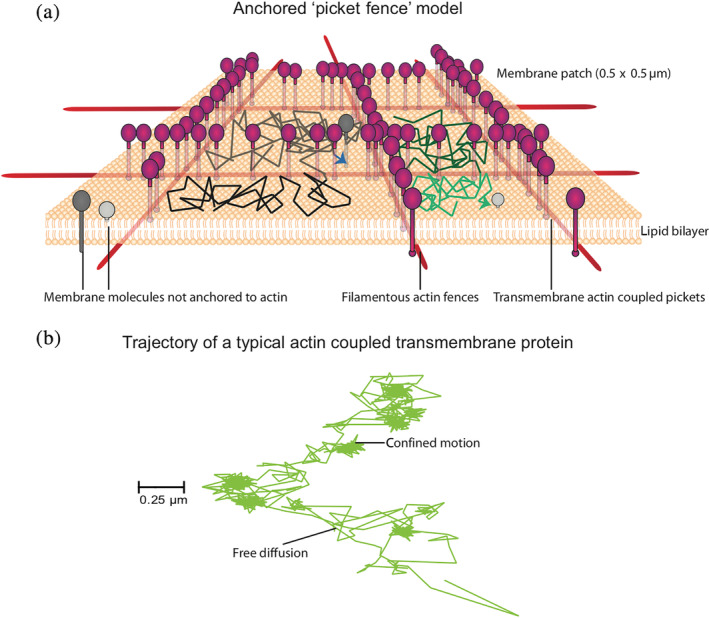
The “anchored picket‐fence” model of Kusumi and a number of membrane biophysicists have proposed that the membrane is draped on a filamentous actin meshwork. This meshwork creates fences which are a consequence of the attachment of membrane components to the underlying meshwork, for example, by transmembrane actin coupled “picket” molecules (shown in magenta). Molecules not coupled to the cytoskeleton can collide with the fences and the picketed molecules and can also cross over to adjoining compartments (shown in gray), resulting in hop‐diffusion. This architecture has profound influence on the typical trajectory of a protein that has the capacity to serve as such an anchored picket molecule. Note the transient confinement zones and regions of free diffusion (marked by arrowheads) of one such protein, CD44 (b), which provides an appreciation of the influence of the multiple scales and modes of diffusion. The trajectory has been adapted from Reference 13
Studies with multiple techniques, including fluorescence correlation spectroscopy (FCS) with varying observation volumes, and with different membrane probes have, further corroborated the heterogenous diffusion behavior of lipid‐anchored proteins and fluorescently labeled lipids.19, 20, 21, 22, 23, 24 The steady‐state organization of several lipid‐linked proteins and glycolipids including the outer leaflet GPI‐anchored proteins and the inner leaflet Ras isoforms indicates that there is a more local scale of organization.25, 26, 27 These membrane components form nanoclusters whose statistical distribution between monomers and clusters does not conform to equilibrium chemical oligomerization. 28 Furthermore, in many instances this nanoscale organization is dependent on the activity of the underlying cortical actomyosin cytoskeleton.29, 30 These observations suggest a more intimate link between the membrane bilayer and the activity of the actin cortex juxtaposed to it.31, 32 The spatial distribution of these nanoclusters are nonequilibrium; they exhibit concentration‐independent clustering behavior, and considerable spatial heterogeneity forming “hotspots” of extremely high enrichment, surrounded by regions that are depleted of these nanoclusters.30, 33 The clustering in turn appears to be regulated by inputs from the external environment.33, 34, 35
3. THE ACTIVE ACTIN‐MEMBRANE COMPOSITE MODEL
To explain the nonequilibrium nanoclustering of GPI‐anchored proteins, an active actin‐membrane composite model was proposed.31, 32, 36 It offers a new paradigm for how we understand membrane organization, encompassing the picket fence model proposed by Kusumi et al.14, 37 According to this, the cell surface should be viewed as a composite of the multicomponent plasma membrane that rests on a multicomponent CA cytoskeleton beneath it. Thus, in addition to the long bundled actin filaments that constitute a well‐defined stable mesh, 17 this model invokes the existence of a pool of smaller, highly dynamic actin filaments. Energy consuming processes, such as treadmilling (of polar actin filaments) and myosin dependent contractility and alignment, drive these filaments into spontaneous patterns that include inward pointing “asters”.36, 38 The noise arising from stochasticity in motor activity and binding and unbinding rates of the motors with actin filaments as well as length distributions of the actin filaments, results in aster remodeling. These shorter dynamic actin filaments transiently attach to the inner leaflet of the cell membrane via linker proteins. Cell surface molecules that can interact with this dynamic CA, either directly (such as transmembrane proteins [TM] with actin binding domains [ABD]) or indirectly (such as GPI‐anchored proteins via trans bilayer interactions 39 or transmembrane proteins that recruit cytoskeletal adaptors such as Ezrin‐Radixin‐Moesin [ERM] proteins), are advected into the core of such asters and thereby result in their ability to form nanoclusters.36, 40 In vitro reconstitution of the essential ingredients of such an actin‐membrane composite, albeit using components that would allow a direct visualization of the actin patterns, indeed recapitulate many of the features observed in cell membranes. 40 The coupling to this dynamic CA network to the membrane therefore generates active stresses and currents on the membrane that can in turn affect local membrane composition and shape. 36
The active composite model recognizes the role of energy consuming CA localized acto‐myosin driven processes in maintaining the distribution of cell membrane components out of equilibrium. It adds an additional layer to the regulation of chemical signaling processes 41 ; along with intermolecular interactions, the interaction with actin governs the localization, movement, and the environment of cell surface molecules. Thus, along with providing a consistent explanation for the nonequilibrium organization of membrane components, this model also provides a framework to classify cell surface molecules on the basis of their ability to couple to this dynamic CA network, that is, inert, passive, and active (Figure 3).
FIGURE 3.
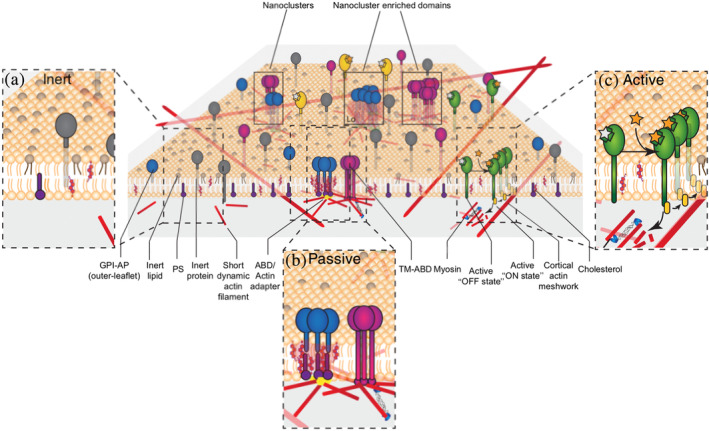
The active actin‐membrane composite model proposes that cortical acto‐myosin activity templates the organization of membrane components that interact with it, for example, TM‐ABD (magenta) and the outer leaflet GPI‐APs (blue) that couple with actin (red) via a transbilayer interaction involving cholesterol and inner leaflet PS lipids (purple). The latter likely results in the construction of a local lo like nanodomain. This model classifies molecules at the cell surface (orange lipids represent generic membrane lipid molecules) into (i) inert (inset a) molecules (brown lipids and gray transmembrane proteins) that randomly diffuse on the cell membrane; (ii) passive (inset b) molecules like GPI‐APs and TM‐ABD; and (iii) active molecules (inset c) green which when activated can activate signaling machinery that regulate the creation of the acto‐myosin machinery that templates the nanoclusters. Glycosylphosphatidylinositol‐anchored protein‐APs, GPI‐anchored proteins; PS, phosphatidylserine; TM‐ABD, transmembrane proteins with actin binding domains
The active composite model provides a consistent explanation for the peculiar properties of GPI‐anchored protein organization. 36 It also makes some key predictions that have been experimentally validated. For instance, a key assumption of this model is the existence of short dynamic actin filaments, in addition to a more stable CA meshwork, juxtaposed to the membrane. FCS‐and SPT‐based measurements using fluorescent‐actin‐binding probes have provided evidence for existence of short (~100–300 nm) actin filaments 36 consistent with earlier biochemical and EM‐based work.42, 43 Also, consistent with the prediction of this model, the introduction of an actin‐binding domain (ABD) into the cytosolic region of a model transmembrane protein was sufficient to induce its nanoclustering. 36 Reconstitution of the essential ingredients of this model, namely short filaments driven by myosin activity, juxtaposed to membrane lipids and bilayer anchored ABD in vitro, has provided us valuable insights into membrane organization templated by actin reorganization44, 45 and activity. 40 The role for actin remodeling in dynamic clustering of membrane tethered proteins on supported lipid bilayers has also been reconstituted. 40 Temperature‐independent diffusion of GPI‐anchored proteins at the length scales of the dynamic actin 46 and the statistically anomalous spatial distribution of GPI‐anchored protein nanoclusters 36 provide further validation of the model.
In the context of GPI‐anchored proteins, Raghupathy et al. have shown that when either the GPI‐anchor or phosphatidylserine (PS) is immobilized, there is a significant co‐segregation and trans‐bilayer interaction of GPI‐anchor lipids and PS, in the presence of adequate cholesterol. This was accompanied by an increase in local chain stiffness and a nonlinear increase in the length scale over which the lipids maintain the liquid order (lo)‐like order (as ascertained in simulations of asymmetric bilayers). 39 All these point to the construction of a local lo‐domain when long saturated acyl chain containing GPIs or PS are immobilized by actin. Thus, in addition to regulating the nanoscale organization of membrane components, the active composite model can account for the generation of local lo‐like domains.
The cortical acto‐myosin network thus directs the organization of membrane components and at the same time membrane composition can regulate cytoskeletal dynamics. 47 It is this dynamic interplay between membrane organization and the dynamic actin cytoskeleton that provides the cell with a stable, yet flexible cell surface that can continuously adapt to its environment. 31
4. REGULATION OF MEMBRANE DOMAINS BY “ACTIVE” MEMBRANE MOLECULES
Another important prediction of the active composite model is that molecules that can regulate the dynamic acto‐myosin machinery could potentially direct the patterning of passive components at the plasma membrane. 36 Many signaling receptors through their ability to recruit and regulate cytoskeletal modifiers48, 49 are predicted to behave as such regulators. 31 The integrin family of heterodimeric type 1 transmembrane proteins that facilitate the adhesion of cells to the extra‐cellular matrix (ECM) and to other cells is a prime example of one such regulator.50, 51 Experiments from various groups have demonstrated that the sites of integrin activation such as focal adhesions are associated with membranes with an lo‐like characteristic.52, 53, 54 Cell detachment resulted in the decrease in membrane order suggesting that integrin ECM attachment regulates this process. 55
Near‐field scanning optical microscopy (NSOM) revealed that when the ligand ICAM‐1 binds to its cognate integrin receptor LFA‐1 (αLβ2) in immune cells, there was an increase in the fraction of GPI‐anchored proteins that formed oligomers, from 30% in the resting state to 80% in the integrin activated state. These GPI‐anchored protein nanoclusters occurred within spatial proximity of the activated LFA‐1 nanoclusters that were spatially correlated over a larger scale, forming “hotspots.” 33 Using homo‐forsters resonance energy transfer imaging 56 to monitor the extent of nanocluster formation in living cells, it was found that activation of the β1 class of integrins also lead to enhanced nanoclustering of GPI‐anchored proteins and the model transmembrane protein with ABD, in the immediate vicinity of the activated receptor. 35 This occurs via triggering of the typical integrin signaling process that includes several kinases (src family kinases and the focal adhesion kinase), the RhoGTPase RhoA, and a specific actin nucleator formin, as well as rho‐kinase ROCK activation. While necessary, this arm of integrin signaling is not sufficient to trigger nanoclustering of GPI‐anchored proteins. Additionally, this requires the activation of the mechanosensitive proteins, talin and vinculin. 35 Mutant cells that were unable to efficiently nanoclusters their GPI‐anchored proteins exhibited defects in integrin mediated function such as a cell spreading response on ligand coated surfaces and delayed migration in a scratch‐wound assay, potentially due to the failure to generate lipid ordered domains. 35 These actively regulated GPI‐anchored proteins nanoclusters are therefore functional, indicating the role played by such active membrane receptors in regulating their membrane environment as thereby modulating their own function.
5. NANO‐DOMAINS ARE UBIQUITOUS AND NONUNIFORMLY DISTRIBUTED
The existence of signaling nano‐domains of molecules such as epidermal growth factor receptor, Ras, or immune cell receptors,10, 27 observed using a variety of super‐resolution based techniques, illustrate the importance of cluster formation at the nanoscale (~5–100 nm). It is yet unclear, whether there is a higher level of organization of these nano‐clusters, namely the mesoscale (100 nm to microns length scale). Assuming a homogeneous distribution of actin filaments and active components that are necessary for nanocluster formation of membrane proteins, it is logical to expect a uniform distribution of nanoclusters at the cell membrane. However, the plasma membrane is tightly juxtaposed to the complex nonuniform mesh‐like CA. 57 Furthermore, this heterogeneity also evolves over time generating new nodes of high filamentous actin, motor protein and cytoskeletal adaptor density or depleting the same at different patches, of even a resting cell membrane. Coupled with the triggered local activation of the actomyosin machinery, it is likely that there will be fluctuations in the local densities of these nanoscale clusters, further creating another level in the hierarchical organization of membrane components. This will be dictated by the local concentrations of key CA and associated cytoskeletal adaptor components that could have an intrinsic role in turning over and redistributing membrane components that directly or indirectly interact with the actin meshwork.
An implication of the picket fence model of the plasma membrane is that if the membrane is detached from the underlying cortical fences, one may observe large‐scale coalescence of lipid ordered domains on the plasma membrane. However, the active composite model proposes a mechanism for spatiotemporal regulation of the formation of potentially lo‐like nano‐domains by engagement with the dynamic cytoskeleton. The connection between these two models of the plasma membrane‐actin linkage can be established by reconciling observations made regarding the nano‐ and mesoscale diffusion behavior of the GPI‐anchored proteins and their nanoscale clustering. It is observed by stimulated emission depletion‐fluorescence correlation spectroscopy‐based studies that lipids and GPI‐anchored proteins exhibit anomalous hop diffusion behavior on the membrane.19, 20, 21, 58 GPI‐anchored protein nanoclusters formed in proximity of activated LFA‐1 integrins exhibit a cluster size of <100 nm 33 as detected using NSOM. Moreover, based on homo‐FRET imaging, GPI‐anchored proteins not only form nanoclusters but such nanoclusters are further enriched within domains of~450 nm in size. 30 Furthermore, induced clusters of CD59 and other GPI‐anchored proteins exhibit STALL (stimulation induced temporary arrest of lateral diffusion) where the clusters are transiently immobilized. Such STALL behavior has been linked to the triggering of signaling by the recruitment of the inner leaflet Lyn‐kinase in case of CD59.59, 60 Interestingly, immobilization is a feature that has also been observed for the naturally existing nanoclusters on live cell membranes. 30 Lastly, the generative mechanism for nanocluster formation also results in major cytoskeletal rearrangement, opening up questions about how would the mesoscale distribution of clusters of lipid anchored GPI‐anchored proteins and transmembrane proteins with ABD respond to such cytoskeletal changes. Synthesizing these observations, it is reasonable to envision that GPI‐anchored proteins exhibit actomyosin based nanoclustering and immobilization by a pool of dynamic actin filaments, in compartments demarcated by the more static mesh decorated with transmembrane actin binding pickets. This way the active actin‐composite system may be envisioned to work in concert with the more static picket‐fence architecture underlying the plasma membrane.
6. HIERARCHICAL NANO‐ AND MESOSCALE ORGANIZATION OF A CELL ADHESION PROTEIN
Recent studies on the cell adhesion protein, CD44 have revealed such a hierarchical structuring of the membrane.13, 61 The extra‐cellular domain of CD44 binds the ECM component hyaluronic acid, 62 while its cytoplasmic domain binds to the actin cytoskeleton. Several studies have demonstrated the existence of CD44 nanoclusters at the plasma membrane using a multitude of localization based super‐resolution microscopy 63 as well as FRET‐based studies.13, 64 CD44 forms nanoclusters in a Galectin‐3 dependent manner on mouse embryonic fibroblast cell membrane, and secreted Galectins are known to be associated with the ECM. The cluster size distribution of ~50 nm, (limited by the sensitivity of the STORM super resolution technique), suggests hotspots of proteins at the plasma membrane. 63 FRET‐based measurements identified the existence of much higher density clustered distribution of the protein, indicative of proteins at <5 nm distances from each other. These studies found that the cytoplasmic domain‐based interactions, especially with the cytoskeletal adaptor, Ezrin, is key for clustering of CD44 in an actin‐dependent manner.13, 64 Ezrin is a member of the ERM family of actin binding proteins. It binds to CD44 by its ERM binding domain, and with a high affinity to the filamentous actin via its actin binding domain. 65 SPT studies showed that CD44 exhibits non‐Brownian diffusion at the plasma membrane with frequent temporal and spatial confinements subject to its ability to interact with the actin cytoskeleton via Ezrin, thereby acting as membrane pickets of the picket fence model of the plasma membrane proposed by Kusumi and colleagues.13, 61 This function of CD44 enabled the restriction of FcγR1 mobility and thereby promoting phagocytic function in macrophages. Such picketing activity was enhanced upon activation of Rho GTPase, suggesting an involvement of Formin activity in the picketing function of the protein. 61 Nanocluster formation of CD44 also required formin nucleated actin polymerization. 13 Thus, the involvement of shared molecular determinants for diffusion as well as the clustering of CD44 is suggestive of the interdependence of the two parameters in determining the ultimate distribution of this protein at the plasma membrane with additional functional consequences.
Combining analysis of the spatial distribution of single molecules of CD44 using a high‐density single molecule localization technique 66 with its clustering potential at nanoscale, we generated dynamic exploration cartography of CD44 at the plasma membrane. 13 This analysis showed that regions of high frequency (density) of localizations of CD44 correlated with regions of enrichment of nano‐clusters of the protein. Although intuitive, the combination of distinct techniques of localization and nanoscale cluster detection provides a powerful tool to visualize the distribution of a molecule at multiple spatial scales. 13
Studying CD44 distribution by building cartography maps in two distinct cell types and cellular contexts demonstrated that CD44 exhibits a mesh‐like distribution at the plasma membrane. This was dependent on cytoplasmic domain dependent interactions. Formin‐driven actin polymerization appears to be necessary to form the mesh‐like distribution and regulates the turnover of the same. Formin activity is also instrumental in the turnover of the underlying actin cortex. 18 These observations taken together suggest an involvement of CA meshwork dynamics in regulating the distribution of CD44 at the mesoscale. These findings open up possibilities that several other cytoskeleton‐coupled transmembrane proteins that function as pickets may reflect the footprint of the underlying cytoskeleton, awaiting further validation by direct visualization (Figure 4). These experiments are, however, fraught with challenges of being able to label the actin cytoskeleton suitably in living cells with improvised fluorescent probes that are stable and bright enough to enable high contrast imaging. Moreover, correlating the actin cytoskeletal meshwork size and F‐actin turnover with the membrane protein mesh size and turnover will be important determinants to establish the connection.
FIGURE 4.
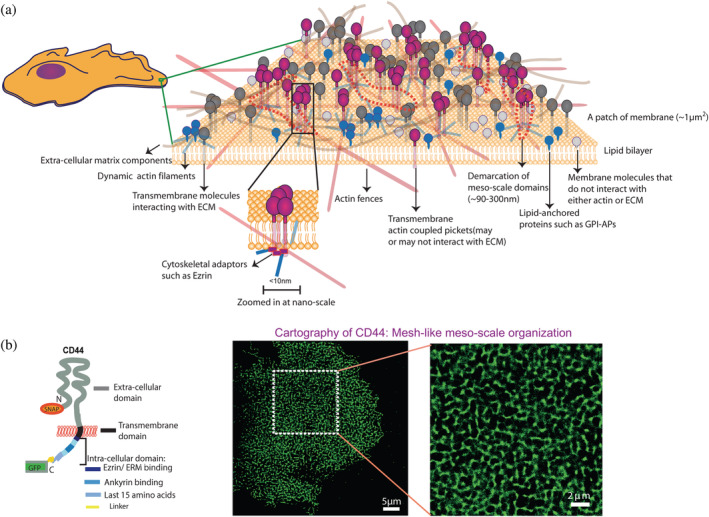
(a) Schematic depicting the meso‐ and nanoscale organization of different membrane molecules: (i) Transmembrane proteins that can couple to the actin cytoskeleton and may or may not interact with the overlying extra‐cellular mesh (magenta), (ii) transmembrane proteins that do not interact with the actin cytoskeleton but bind extra‐cellular matrix components (gray), (iii) membrane molecules that are lipid anchored and are indirectly influenced by actin dynamics (blue), such as GPI anchored proteins, (iv) membrane molecules that are not coupled to either the extracellular matrix or the actin cytoskeleton (light gray). The mesoscale domains (demarcated in dotted regions) show association with nano‐clusters of the cytoskeleton interacting proteins that in turn may be generated by the nanoscale clustering mechanism described in Figure 3. Schematic adapted from Reference 13. (b) A schematic of the transmembrane cell adhesion protein CD44 (on the left), tagged with a GFP and SNAP‐tag. The SNAP‐tag was exogenously labeled to obtain cartography of the protein on the cell surface, shown in two different magnifications (on the right). This figure has been derived from figure 1 of Reference 13
7. ACTIN‐TEMPLATED HIERARCHY IN SPATIAL ORGANIZATION AT THE CELL MEMBRANE
CXCR‐4, a chemokine receptor on T cells exists as monomers, dimers, or nanoclusters of a few molecules at the plasma membrane of resting T cells. Upon activation by the chemokine ligand CXCL‐12, the receptor enriches in larger nano‐clusters with the aid of the actin cytoskeleton and modulated by the CD4 coreceptor. Mutations that prevented the large‐scale cluster formation of CXCR‐4 prevented T cells from performing adhesion and migration function. This study reveals how regulation of higher order organization is crucial to mediate signaling function. However, it remains unknown how changes in the underlying CA is correlated to changes in the degree of clustering of the receptor. CXCR‐4 is often enriched in the leading edge of the migrating T cell and hence offers an excellent system to probe the correlation of changes in cortex and consequent organization of signaling relevant receptors. 67
On the other hand, a lot of work has been carried out to understand cytoskeletal reorganization during T cell activation. A recent study shows how large‐scale actin cytoskeletal network ramification follows the activation and synapse formation process in T cells, characterized by changes in mesh size and turnover times. 68 This system is hence excellently poised to study the adaptation of the organization of myriad coreceptors, receptor tyrosine kinases, and phosphatases relevant to T cell signaling that may or may not directly couple to the changing cytoskeleton underneath.
8. EXTRA‐CELLULAR INTERACTIONS TEMPLATE MESOSCALE ORGANIZATION OF CELL SURFACE RECEPTORS
Given that lectins such as multivalent Galectins can serve to organize glycosylated proteins,63, 69 and that the ECM harbors binding sites for cell surface adhesion receptors, it is likely that the ECM and the glycocalyx of the cell will pattern molecules from the outside. For example, interactions of the N‐glycosylated CD44 extra‐cellular domain with Galectin 3 drive the formation of clusters of CD44, and when these interactions are inhibited either by treatment with lactose (that competes for the Galactose binding sites of Galectins) or glycosylation inhibitors and mutants, CD44 clusters are depleted. 63 However, complete removal of the extra‐cellular domain of CD44 enhanced the mobility of the protein on the cell surface, but neither altered the size of the localization hotpots in live cells nor the defined meshwork like appearance of the distribution. 13
On the contrary, in the context of dendritic cells, it was observed that the glycan based mesoscale compartments defined the distribution of the tetrameric dendritic cell specific intercellular adhesion molecule‐3‐grabbing nonintegrin (DC‐SIGN) without influencing its nanocluster formation. 66 In this study, the authors identified that removal of extra‐cellular interactions mediated by Galectin‐9 disrupts the meshwork‐like mesoscale pattern of DC‐SIGN receptors, preventing their enrichment in clathrin‐coated patches on the plasma membrane, consequently affecting its endocytosis. The DC‐SIGN receptor is a classic example of tetramers 70 organizing in nano‐clusters (2–3 tetramers) and further incorporated into micron‐scale localization hotspots, driven at least in two different length scales by extra‐cellular domain interactions. 66 Artificially engineering different ABDs to transmembrane proteins with such defined extra‐cellular interactions, in the appropriate cell system, will help understand the interplay or which interactions (extra‐cellular or indirect cytoskeletal barriers) will emerge as a stronger determinant of the nanoscale and emerging mesoscale distribution of the protein. It is also speculated that organization at different length scales serves different functions: while the nano‐domains serve to capture pathogens, the mesoscale domains serve as hotspots for endocytosis.71, 72, 73, 74
9. CONCLUSION
It is clear today, that the membrane of a living cell is highly organized, and “there's (always) more to this than meets the eye.” From the nanoscale to the mesoscale, to even higher order organization, the cell membrane and its constituents appear to be influenced by interactions with cytoplasmic and extracellular components on either side of the membrane bilayer. 1 Assembly and disassembly of the active actin machinery is likely to be a major driving force for the dynamic remodeling of the cell surface. However, there are many other modes of patterning the cell surface with passive templates. The cortical meshwork at the cytoplasmic surface and the ECM (along with glycan polymers with their myriad binding sites for multiple cell surface proteins and lipids) are but two types of extra‐membranous material that serve to play such a patterning role. Both membrane receptors mediated outside‐in as well as inside‐out signaling pose a huge potential for tuning local membrane organization. This promotes a new picture of the cell membrane, of one that resembles a composite of all these interactions in space and time.
AUTHOR CONTRIBUTIONS
Joseph Mathew Kalappurakkal: Conceptualization; writing‐review and editing. Parijat Sil: Conceptualization; writing‐review and editing. Satyajit Mayor: Conceptualization; writing‐review and editing.
ACKNOWLEDGMENTS
S. M. acknowledges a Margdarshi fellowship from the DBT‐Wellcome Trust India Alliance (IA/M/15/1/502018), and insightful discussions with Thomas van Zanten, Madan Rao (NCBS), and Maria Garcia Parajo (ICFO, Barcelona). J. M. K. and P. S. acknowledge predoctoral funding from NCBS, TIFR.
Kalappurakkal JM, Sil P, Mayor S. Toward a new picture of the living plasma membrane. Protein Science. 2020;29:1355–1365. 10.1002/pro.3874
Joseph Mathew Kalappurakkal and Parijat Sil contributed equally to this study.
Funding information National Centre for Biological Sciences, Tata Institute of Fundamental Research (NCBS, TIFR); India Alliance of the DBT‐Wellcome Trust, Grant/Award Number: IA/M/15/1/502018
REFERENCES
- 1. Jacobson K, Liu P, Lagerholm BC. The lateral organization and mobility of plasma membrane components. Cell. 2019;177:806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edidin M. Lipids on the frontier: A century of cell‐membrane bilayers. Nat Rev Mol Cell Biol. 2003;4:414–418. [DOI] [PubMed] [Google Scholar]
- 3. Gorter E, Grendel F. On biomolecular layers of lipoids on the chromocytes of the blood. J Exp Med. 1925;41:439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danielli JF, Davson H. A contribution to the theory of permeability of thin films. J Cell Comp Physiol. 1935;5:495–508. [Google Scholar]
- 5. Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. [DOI] [PubMed] [Google Scholar]
- 6. Edidin M. The state of lipid rafts: From model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. [DOI] [PubMed] [Google Scholar]
- 7. Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. [DOI] [PubMed] [Google Scholar]
- 8. Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. [DOI] [PubMed] [Google Scholar]
- 9. Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. [DOI] [PubMed] [Google Scholar]
- 10. Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heberle FA, Feigenson GW. Phase separation in lipid membranes. Cold Spring Harb Perspect Biol. 2011;3:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Zanten TS, Mayor S. Current approaches to studying membrane organization. F1000Res. 2015;4:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sil P, Mateos N, Nath S, et al. Dynamic actin‐mediated nano‐scale clustering of CD44 regulates its meso‐scale organization at the plasma membrane. Mol Biol Cell. 2020;31:561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kusumi A, Suzuki KGN, Kasai RS, Ritchie K, Fujiwara TK. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem Sci. 2011;36:604–615. [DOI] [PubMed] [Google Scholar]
- 15. Kusumi A, Fujiwara TK, Chadda R, et al. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of Singer and Nicolson's fluid‐mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. [DOI] [PubMed] [Google Scholar]
- 16. Eggeling C. Super‐resolution optical microscopy of lipid plasma membrane dynamics. Essays Biochem. 2015;57:69–80. [DOI] [PubMed] [Google Scholar]
- 17. Morone N, Fujiwara T, Murase K, et al. Three‐dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol. 2006;174:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fritzsche M, Lewalle A, Duke T, Kruse K, Charras G. Analysis of turnover dynamics of the submembranous actin cortex. Mol Biol Cell. 2013;24:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eggeling C, Ringemann C, Medda R, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. [DOI] [PubMed] [Google Scholar]
- 20. Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C. Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc Natl Acad Sci U S A. 2010;107:6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andrade DM, Clausen MP, Keller J, et al. Cortical actin networks induce spatio‐temporal confinement of phospholipids in the plasma membrane—A minimally invasive investigation by STED‐FCS. Sci Rep. 2015;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lagerholm BC, Andrade DM, Clausen MP, Eggeling C. Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED‐FCS. J Phys D Appl Phys. 2017;50:063001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenne PF, Wawrezinieck L, Conchonaud F, et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wenger J, Conchonaud F, Dintinger J, et al. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys J. 2007;92:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Varma R, Mayor S. GPI‐anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. [DOI] [PubMed] [Google Scholar]
- 26. Parton RG, Hancock JF. Lipid rafts and plasma membrane microorganization: Insights from Ras. Trends Cell Biol. 2004;14:141–147. [DOI] [PubMed] [Google Scholar]
- 27. Garcia‐Parajo MF, Cambi A, Torreno‐Pina JA, Thompson N, Jacobson K. Nanoclustering as a dominant feature of plasma membrane organization. J Cell Sci. 2014;127:4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma P, Varma R, Sarasij RC, et al. Nanoscale organization of multiple GPI‐anchored proteins in living cell membranes. Cell. 2004;116:577–589. [DOI] [PubMed] [Google Scholar]
- 29. Plowman SJ, Muncke C, Parton RG, Hancock JF. H‐ras, K‐ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci U S A. 2005;102:15500–15505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goswami D, Gowrishankar K, Bilgrami S, et al. Nanoclusters of GPI‐anchored proteins are formed by cortical actin‐driven activity. Cell. 2008;135:1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rao M, Mayor S. Active organization of membrane constituents in living cells. Curr Opin Cell Biol. 2014;29:126–132. [DOI] [PubMed] [Google Scholar]
- 32. Köster DV, Mayor S. Cortical actin and the plasma membrane: Inextricably intertwined. Curr Opin Cell Biol. 2016;38:81–89. [DOI] [PubMed] [Google Scholar]
- 33. Van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia‐Parajo MF. Hotspots of GPI‐anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci U S A. 2009;106:18557–18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high‐fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. [DOI] [PubMed] [Google Scholar]
- 35. Kalappurakkal JM, Anilkumar AA, Patra C, van Zanten TS, Sheetz MP, Mayor S. Integrin mechano‐chemical signaling generates plasma membrane nanodomains that promote cell spreading. Cell. 2019;177:1738–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gowrishankar K, Ghosh S, Saha SCR, Mayor S, Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. [DOI] [PubMed] [Google Scholar]
- 37. Kusumi A, Nakada C, Ritchie K, et al. Paradigm shift of the plasma membrane concept from the two‐dimensional continuum fluid to the partitioned fluid: High‐speed single‐molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. [DOI] [PubMed] [Google Scholar]
- 38. Husain K, Rao M. Emergent structures in an active polar fluid: Dynamics of shape, scattering, and merger. Phys Rev Lett. 2017;118:078104. [DOI] [PubMed] [Google Scholar]
- 39. Raghupathy R, Anilkumar AA, Polley A, et al. Transbilayer lipid interactions mediate nanoclustering of lipid‐anchored proteins. Cell. 2015;161:581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Köster DV, Husain K, Iljazi E, et al. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc Natl Acad Sci U S A. 2016;113:E1645–E1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chaudhuri A, Bhattacharya B, Gowrishankar K, Mayor S, Rao M. Spatiotemporal regulation of chemical reactions by active cytoskeletal remodeling. Proc Natl Acad Sci U S A. 2011;108:14825–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cano ML, Lauffenburger DA, Zigmond SH. Kinetic analysis of F‐actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulation increases actin filament number without altering the filament length distribution. J Cell Biol. 1991;115:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Podolski JL, Steck TL. Length distribution of F‐actin in Dictyostelium discoideum . J Biol Chem. 1990;265:1312–1318. [PubMed] [Google Scholar]
- 44. Murrell MP, Gardel ML. F‐actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc Natl Acad Sci U S A. 2012;109:20820–20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vogel SK, Greiss F, Khmelinskaia A, Schwille P. Control of lipid domain organization by a biomimetic contractile actomyosin cortex. Elife. 2017;6:e24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saha S, Lee I‐H, Polley A, Groves JT, Rao M, Mayor S. Diffusion of GPI‐anchored proteins is influenced by the activity of dynamic cortical actin. Mol Biol Cell. 2015;26:4033–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bezanilla M, Gladfelter AS, Kovar DR, Lee WL. Cytoskeletal dynamics: A view from the membrane. J Cell Biol. 2015;209:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vicente‐Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration—The actin connection. J Cell Sci. 2009;122:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hynes RO. Integrins: Bidirectional, allosteric signaling machines in their roles as major adhesion receptors. Cell. 2002;110:673–687. [DOI] [PubMed] [Google Scholar]
- 51. Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275:21785–21788. [DOI] [PubMed] [Google Scholar]
- 52. Gaus K, Le Lay S, Balasubramanian N, Schwartz MA. Integrin‐mediated adhesion regulates membrane order. J Cell Biol. 2006;174:725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leitinger B, Hogg N. The involvement of lipid rafts in the regulation of integrin function. J Cell Sci. 2002;115:963–972. [DOI] [PubMed] [Google Scholar]
- 54. Ge Y, Gao J, Jordan R, Naumann CA. Changes in cholesterol level alter integrin sequestration in raft‐mimicking lipid mixtures. Biophys J. 2018;114:158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. del Pozo MA, Balasubramanian N, Alderson NB, et al. Phospho‐caveolin‐1 mediates integrin‐regulated membrane domain internalization. Nat Cell Biol. 2005;7:901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ghosh S, Saha S, Goswami D, Bilgrami S, Mayor S. Dynamic imaging of homo‐FRET in live cells by fluorescence anisotropy microscopy Methods in enzymology. P. Michael Conn (eds), volume 505 1st ed. Elsevier, Academic Press, United States, 2012; p. 291–327. [DOI] [PubMed] [Google Scholar]
- 57. Fritzsche M, Erlenkämper C, Moeendarbary E, Charras G, Kruse K. Actin kinetics shapes cortical network structure and mechanics. Sci Adv. 2016;2:e1501337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de la Serna JB, Schütz GJ, Eggeling C, Cebecauer M. There is no simple model of the plasma membrane organization. Front Cell Dev Biol. 2016;4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki KGN, Fujiwara TK, Sanematsu F, Iino R, Edidin M, Kusumi A. GPI‐anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: Single‐molecule tracking study 1. J Cell Biol. 2007;177:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suzuki KGN, Fujiwara TK, Edidin M, Kusumi A. Dynamic recruitment of phospholipase Cγ at transiently immobilized GPI‐anchored receptor clusters induces IP3‐Ca2+ signaling: Single‐molecule tracking study 2. J Cell Biol. 2007;177:731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Freeman SA, Vega A, Riedl M, et al. Transmembrane pickets connect cyto‐ and pericellular skeletons forming barriers to receptor engagement. Cell. 2018;172:305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ponta H, Sherman L, Herrlich PA. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. [DOI] [PubMed] [Google Scholar]
- 63. Lakshminarayan R, Wunder C, Becken U, et al. Galectin‐3 drives glycosphingolipid‐dependent biogenesis of clathrin‐independent carriers. Nat Cell Biol. 2014;16:595–606. [DOI] [PubMed] [Google Scholar]
- 64. Wang Y, Yago T, Zhang N, et al. Cytoskeletal regulation of CD44 membrane organization and interactions with E‐selectin. J Biol Chem. 2014;289:35159–35171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mori T, Kitano K, Terawaki S, Maesaki R, Fukami Y, Hakoshima T. Structural basis for CD44 recognition by ERM proteins. J Biol Chem. 2008;283:29602–29612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Manzo C, van Zanten TS, Saha S, Torreno‐Pina JA, Mayor S, Garcia‐Parajo MF. PSF decomposition of nanoscopy images via Bayesian analysis unravels distinct molecular organization of the cell membrane. Sci Rep. 2015;4:4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martínez‐Muñoz L, Rodríguez‐Frade JM, Barroso R, et al. Separating actin‐dependent chemokine receptor nanoclustering from dimerization indicates a role for clustering in CXCR4 signaling and function. Mol Cell. 2018;70:106–119. [DOI] [PubMed] [Google Scholar]
- 68. Fritzsche M, Fernandes RA, Chang VT, et al. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci Adv. 2017;3:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Garner OB, Baum LG. Galectin–glycan lattices regulate cell‐surface glycoprotein organization and signalling. Biochem Soc Trans. 2008;36:1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Itano MS, Graus MS, Pehlke C, et al. Super‐resolution imaging of C‐type lectin spatial rearrangement within the dendritic cell plasma membrane at fungal microbe contact sites. Front Phys. 2014;2:612–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Manzo C, Torreno‐Pina JA, Joosten B, et al. The neck region of the C‐type lectin DC‐SIGN regulates its surface spatiotemporal organization and virus‐binding capacity on antigen‐presenting cells. J Biol Chem. 2012;287:38946–38955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cambi A, De Lange F, Van Maarseveen NM, et al. Microdomains of the C‐type lectin DC‐SIGN are portals for virus entry into dendritic cells. J Cell Biol. 2004;164:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu P, Wang X, Itano MS, Neumann AK, Jacobson K, Thompson NL. The formation and stability of DC‐SIGN microdomains require its extracellular moiety. Traffic. 2012;13:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Neumann AK, Thompson NL, Jacobson K. Distribution and lateral mobility of DC‐SIGN on immature dendritic cells ‐ Implications for pathogen uptake. J Cell Sci. 2008;121:634–643. [DOI] [PubMed] [Google Scholar]


