Abstract
Traditionally, structures of cytoskeletal components have been studied ex situ, that is, with biochemically purified materials. There are compelling reasons to develop approaches to study them in situ in their native functional context. In recent years, cryo‐electron tomography emerged as a powerful method for visualizing the molecular organization of unperturbed cellular landscapes with the potential to attain near‐atomic resolution. Here, we review recent works on the cytoskeleton using cryo‐electron tomography, demonstrating the power of in situ studies. We also highlight the potential of this method in addressing important questions pertinent to the field of cytoskeletal biomechanics.
Keywords: actin filaments, cryo‐electron tomography, in situ architecture, intermediate filaments, microtubules
Abbreviations
- 3D
three‐dimensional
- ATP
adenosine triphosphate
- CEMOVIS
cryo‐electron microscopy of vitreous sections
- cryo‐EM
cryo‐electron microscopy
- cryo‐ET
cryo‐electron tomography
- EM
electron microscopy
- ET
electron tomography
- FH2
formin homology 2
- FIB
focused ion beam
- IFs
intermediate filaments
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- MAPs
microtubule‐associated proteins
- MTs
microtubules
- PFs
protofilaments
- TAT
tubulin acetyltransferase
- TEM
transmission electron microscopy
- VASP
vasodilator‐stimulated phosphoprotein
- VPP
Volta phase plate
- WASP
Wiskott–Aldrich syndrome protein
- WAVE
WASP‐family verprolin‐homologous protein
1. INTRODUCTION
In their seminal work on cellular mechanics, Francis Crick and Arthur Hughes proposed a model for the cytoskeleton: “If we were compelled to suggest a model (of cell mechanics) we would propose Mother's Work Basket—a jumble of beads and buttons of all shapes and sizes, with pins and threads for good measure, all jostling about and held together by colloidal forces.” 1 In fact, around the middle of the 20th century, the idea gained momentum that the cell is not just passive material but a hierarchically ordered system of interacting molecular species (beads and buttons) and supramolecular entities (threads). Life depends on their interaction networks. Owing to advances in biochemistry and biophysics, we have now identified many of those beads and buttons, pins, and threads. These are the numerous cytoskeletal proteins, motor proteins as well as their regulatory molecules.
The cytoskeleton has three major constituents: actin filaments, microtubules (MTs), and intermediate filaments (IFs), which are organized into higher‐order assemblies. Each filament is structurally and functionally unique, interacts with its own set of proteins involved in filament crosslinking, bundling, capping, or severing, and performs specialized tasks within the cell. The regulation of the on–off kinetics of these interactors, the polymerization–depolymerization dynamics, polarity and chirality of the filaments, and the action of molecular motors is what makes the cytoskeleton a multifunctional supramolecular assembly. 2 The traditional reductionist approach—studying one molecule at a time in isolation 3 —cannot lead to a comprehensive understanding of how cells use the cytoskeleton to perform exquisitely complex mechanical tasks such as division, locomotion, or shape remodeling. 4 For example, weak or transient interactions between filaments or their binding partners are lost when the effects of macromolecular crowding are reduced. 5 This might lead to nonphysiological conformational or oligomeric states, and their spatiotemporal modulation remains elusive when taken out of context.
The history of cytoskeletal research revolves around the pursuit of tools and techniques that promise an ever closer view of these elaborate structures. Studies in the 17th century using early microscopes revealed a network of fibrils in muscles 6 and other tissue types. Since then, researchers strived to understand the three‐dimensional (3D) organization of cytoskeletal elements that forms the underlying robust, yet dynamic, scaffold of all cells. Along with it, the importance of establishing and maintaining order over different length‐ and time‐scales became apparent. With the emergence of electron microscopy (EM),7, 8, 9 unprecedented views of the cytoskeleton were provided (Figure 1). As EM techniques improved, they revealed an increasingly complicated cytoskeletal network in cells, composed of intermingled actin filaments, IFs and MTs, and numerous crosslinkers and associated proteins.18, 19, 22, 23, 24, 25, 26, 27 The major cytoskeletal elements, actin filaments, 14 and MTs,15, 16 were first discovered in nonmuscle cells after the introduction of chemical fixation and heavy metal staining 13 (Figure 1). However, this came with a price. Structural studies in this field have relied principally on two approaches: immunohistochemical techniques53, 54 and transmission electron microscopy (TEM) of plastic embedded or deep‐etched 11 and metal‐shadowed specimens. 12 First, some of the fixatives used in EM, although invaluable for the visualization of the cytoskeleton, are harsh compounds leading to structural alterations. Second, proteins associated with the cytoskeleton cannot be identified easily by these techniques. Although immuno‐EM provides a partial solution to the latter problem, the methodology is challenging and idiosyncratic. Fluorescence microscopy, when combined with cytoskeleton‐binding probes, offers a number of ways to highlight the cytoskeleton. 55 However, the cytoskeleton is tightly packed, and it is not easy to discern individual structures owing to the diffraction limit of optical microscopy (approximately 300 nm). This issue has been resolved with the emergence of optical super‐resolution imaging techniques, 56 although these techniques are still limited in resolution (~20–50 nm). 57
FIGURE 1.
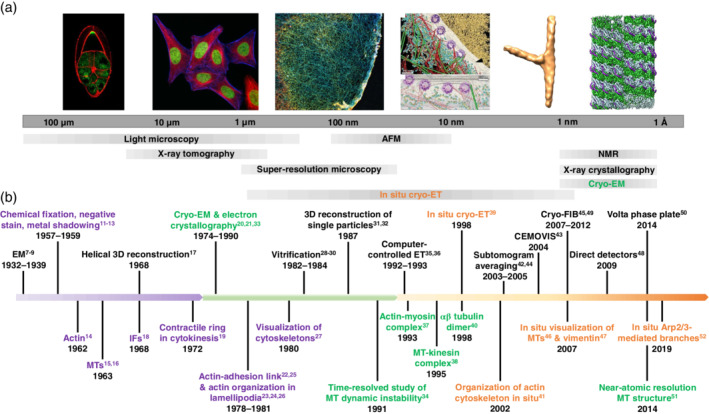
Evolution of the electron microscopy (EM) field with special emphasis on milestones in cytoskeleton research. (a) Biological length‐scales accessible through different imaging techniques. In situ cryo‐electron tomography (cryo‐ET; orange) spans length‐scales from several microns to the subnanometer range making it suitable for the exploration of cellular architecture and biomolecules. From left to right: A Drosophila oocyte stained for actin (red; courtesy of Dr. Daniel St. Johnston, University of Cambridge, UK), HeLa cells labeled for actin (blue), and microtubules (MTs; red), a Cos7 cell stained for actin (courtesy of Prof. Ralf Jungmann, LMU and MPIB, Germany), the nuclear periphery of a HeLa cell (reproduced with permission from reference [10], copyright (2016) AAAS), the in situ subtomogram average of Arp2/3 complex‐mediated branch junctions from Dictyostelium discoideum (EMD 4790) and the cryo‐EM reconstruction of the MT‐tau complex (EMD 7523). (b) The EM field is divided into three periods, starting from the invention of EM and the development of conventional sample preparation techniques (purple) to the emergence of single‐particle cryo‐EM (green) followed by in situ cryo‐ET (orange). The development of the EM field goes hand‐in‐hand with milestone discoveries of cytoskeletal elements and architectures highlighted with the same color (see references [7, 8, 9] and [11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52])
Therefore, there was a need for a method that provides faithful representations of functional modules and their interplay in a cellular context. This can be achieved through structural studies performed in situ, that is, in unperturbed environments. Cryo‐electron tomography (cryo‐ET) fulfills these criteria: it provides molecular resolution information of cells and organelles unadulterated by specimen preparation.58, 59 Rapid freezing ensures the best structural preservation that is physically possible to achieve.28, 29, 30 Although the idea to use ET for native samples was there for decades, 60 the realization of the vision followed only much later39, 61 (Figure 1). Technological advances such as computer‐controlled transmission electron microscopes made it possible to develop automated data acquisition procedures minimizing exposure to the electron beam.35, 36 The advent of focused ion beam (FIB) milling adapted to cryogenic conditions45, 49 permitted the reproducible preparation of thin vitrified cellular samples without the notorious artefacts of cryo‐sectioning such as sample compression. 62 With the development of direct electron detection 48 and advances in image processing,17, 20, 21, 31, 32, 33, 42, 44 we are now entering the realm of subnanometer resolution for structural studies of the cellular interior. 58 Cryo‐ET technologies have already started to provide new insights into the 3D architecture of the cytoskeleton in situ (Figure 1). In this review, we discuss recent progress toward a structural understanding of the cytoskeleton; in particular, we show how the application of in situ approaches has led to new insights into the organization and function of cytoskeletal filaments that had remained elusive so far.
2. THE ARCHITECTURE OF THE ACTIN CYTOSKELETON
The actin cytoskeleton is essential for motile cells to modulate their shape and move within complex environments. It adopts a variety of architectures that contribute to protrusion, adhesion, contraction, and retraction of the cell. 63 At the leading edge, branched and crosslinked networks form a lamellipodium that, by pushing the plasma membrane forward, promotes cell movement. 64 Thin actin‐rich, finger‐like membrane protrusions called filopodia assemble from peripheral regions of the cell in response to chemical stimuli, providing initial cell‐substrate contact sites.65, 66 At the basal cell membrane, self‐organized actin waves propagate, 67 and, in invasive cells, large filopodia‐like protrusions called invadopodia can penetrate through the extracellular matrix. 68 Podosomes extend a core of branched and crosslinked actin filaments into the cytoplasm for mechanosensing. 69 Cell contractility arises from the association of actin with myosin II 37 as exemplified in stress fibers, thick antiparallel bundles anchored at focal adhesion sites where they sense, generate, and transmit tension to the extracellular matrix. 70 The actomyosin cortex lying beneath the plasma membrane contributes to changes and maintenance of cell shape. 71 When membranes occasionally detach from the cortex and inflate, spherical protrusions, called blebs, are transiently generated; upon the reassembly of an actin cortex the blebs can be retracted. 72
The assembly of the diverse cellular actin architectures is finely tuned by a large array of actin‐associated proteins. Actin nucleation and elongation factors comprise the Arp2/3 complex, formins, and Ena/vasodilator‐stimulated phosphoprotein (VASP), which generate branched or linear filaments, respectively. 63 Several bundling and crosslinking proteins, including fascin, fimbrins, alpha‐actinins, and filamins, can connect filaments over a wide range of distances, contributing to the macroscale organization of the networks. 63 In vitro studies are key to decipher the architectural properties of actin arrays arising from a defined pool of accessory proteins. Visualizing actin network architectures within the complex cellular environment is essential to understand physiological higher‐order actin assemblies in relation to actin function. Since the first in situ observation of the actin cytoskeleton by Medalia et al. almost two decades ago, 41 the cryo‐ET field has evolved considerably through a series of technological developments that include direct electron detectors, 48 cryo‐FIB milling,45, 49, 73 and Volta phase plates (VPPs). 50 In the following sections, we summarize what we have learnt so far about the nanoscale organization of actin bundles and branched networks inside cells using cryo‐ET or ET. Additionally, we discuss the ultrastructure of two composite actin networks, Listeria comet tails, and self‐organized actin waves.
2.1. Nanoscale organization of cellular actin bundles
Before the emergence of cryo‐FIB milling sample preparation, cryo‐ET was limited to thin peripheral parts of the cell. Cryo‐ET could be employed to show the organization of filopodia in Dictyostelium cells. 74 The resolution (4–6 nm) at the time was sufficient to resolve individual actin filaments (diameter of 7–10 nm) within the tomograms. Using a correlative approach, Patla et al. examined chemically fixed actin bundles associated with integrin‐mediated focal adhesion sites at the periphery of REF52 cells. 75 They observed flat pseudopodial protrusions filled with unidirectional actin bundles and adhesion complexes.
This initial work lead to the development of an automated segmentation method allowing for robust description of filament networks. 76 The method is based on template matching of a generic filament, providing cross‐correlation maps for the extraction of the filament centerlines, which can either be used for visualization purposes or for a quantitative description of the networks. Taking advantage of this automated segmentation procedure, methodologies were developed to quantitatively analyze the nanoscale architecture of filopodia, stress fibers, and Listeria comet tails imaged at the periphery of intact Ptk2 cells using cryo‐ET. 77 The study revealed the existence of nanoscopic bundles within these networks, with interfilament distances of 12–13 nm, suggesting that nanoscopic bundles are a generic feature of actin assemblies involved in motility (Figure 2a,b). In filopodia and Listeria protrusions, actin bundles are hexagonally packed (Figure 2c), whereas in stress fibers and cytoplasmic comets tails, they rather form sheets. The data further indicate that bundling proteins, fascin, and fimbrin are commensurate with a narrow range of interfilament distances and allow for hexagonal packing. In Dictyostelium cells, short bundles of actin filaments persist during membrane pearling induced by actin depolymerization using latrunculin A. 79
FIGURE 2.
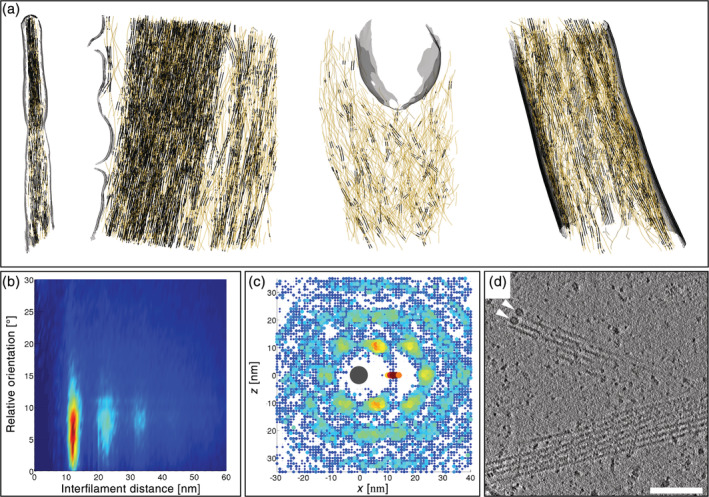
Nanoscale architecture of cellular actin bundles explored by cryo‐electron tomography. (a) Three‐dimensional (3D) architecture of peripheral actin networks in Ptk2 cells. From left to right: actin filaments in a filopodium, a stress fiber, a cytoplasmic Listeria comet tail, and a Listeria protrusion. Actin filaments are shown in yellow with bundled portions displayed in black. The plasma membrane and the cell wall of Listeria are shown in grey. (b) 2D histogram showing distances between filopodial actin filaments as a function of their relative orientation, and revealing a population of parallel filaments with center‐to‐center distances of ~12 nm. Second‐ and third‐order peaks are visible indicating long‐range order. Reproduced from reference [77]. (c) Packing analysis for actin filaments belonging to a filopodium revealing a hexagonal arrangement of neighboring filaments. Reproduced from reference [77]. (d) Slice through a tomogram of a neuronal process acquired with the Volta phase plate. Two annular structures (white arrowheads) resembling formin homology 2 domains are visible at the tip of actin filaments. The periodicity of the helical repeat of actin filaments is evident in the lower bundle. Scale bar: 100 nm. Reproduced with permission from reference [78], copyright (2015) Elsevier
Combining direct detection with VPP imaging, Fukuda et al. pushed the frontiers of quality for tomographic images of actin filaments in primary neurons. 78 The resolution (better than 3 nm) permitted to resolve the periodicity of actin filaments. They visualized two annular structures (~11 nm in size) at the end of actin filaments forming a hexagonal bundle (Figure 2d). These densities are likely associated with the formin homology 2 (FH2) domain responsible for the binding of formin to the barbed end of actin filaments.80, 81, 82 Future in situ cryo‐ET studies may help uncover the structure of the FH2 domain bound to an actin filament, and to explore how formins contribute to the generation of diverse cellular actin organizations.
Recently, photo‐micropatterning of EM grids enabled controlled positioning of cells with predictable actin organization.83, 84 By applying cryo‐FIB milling to a RP1E cell grown on a crossbow micropattern, actin transverse arcs and internal stress fibers could be targeted by cryo‐ET. 83 This approach will be instrumental in future studies to unravel the relationship between cytoskeleton organization and biomechanics at the nanoscale.
2.2. Classical branched organization mediated by Arp2/3 complex
To date, most of the structural knowledge on cellular actin organization derives from studies on the lamellipodia of keratocytes and fibroblasts. First, conventional EM work on negatively stained fibroblasts suggested that the lamellipodium is made of long diagonally oriented actin filaments.23, 24, 26 Two decades later, platinum‐replica EM work showed that this flat (0.1–0.2 μm) cellular protrusion is made of branched filament arrays, 85 contributing to the emergence of the dendritic nucleation model for the mechanism of actin assembly by Arp2/3 complex at the leading edge of the cell. 86 The model proposes that upon recruitment and activation at the protruding cell membrane, the Arp2/3 complex initiates actin polymerization by branching off new filaments from the side of mother filaments at an angle of 70°.87, 88 Both mother and daughter filaments grow with their barbed ends facing the cell membrane with an incidence angle of 55° until they get capped.86, 89 Filament growth toward the membrane results in network expansion whereas disassembly deeper in the cell provides a pool of subunits for the growth of dendritic arrays. 86 Years later, Small and coworkers argued that the visualization of branched networks by platinum‐replica EM is an artefact of critical‐point drying. 90 They challenged the dendritic nucleation model for lamellipodia propulsion using ET data of vitreous and negatively stained lamellipodial networks, in which they found that filaments were mostly unbranched. 90 However, using their primary ET data, Yang and Svitkina detected a significant number of branch junctions within one tomogram, 91 later confirmed by Small et al. 92
Recently, Mueller et al. used ET to quantify how the lamellipodium of fish keratocytes responds to varying forces associated with changes in membrane tension. 93 They showed that the incident angle of the branched filaments with respect to the membrane varies with the load, from the typical 55° angle to 90° in case of reduced membrane tension. In contrast, an increased load on the lamellipodium produces a dense dendritic network with a broadened range of angles relative to the membrane. The load adaptation of the lamellipodium is a direct consequence of the nucleation geometry of branched networks at the membrane. 93 A recent study showed that force generation in lamellipodia depends additionally on formin‐like protein (FMNL) formin activity. 94 Future in situ cryo‐ET studies could help reveal the fine 3D architectural changes associated with the loss of specific formins and how this may result in changes in force production. More generally, they would provide valuable insights into the native 3D architecture of the lamellipodium, its transition to a lamella 95 and how these two types of actin networks overlap during the protrusion of the cell. 96
Apart from the lamellipodium, ET permitted to reveal the architecture of baculovirus actin comet tails assembled by Arp2/3 complex. 97 Mueller et al. showed that the small virus (~50 nm in diameter) assembles a fishbone‐like array of actin filaments generated by the formation of branch junctions (Figure 3a). They proposed a model for baculovirus propulsion based on branch tethering at the virus surface.
FIGURE 3.
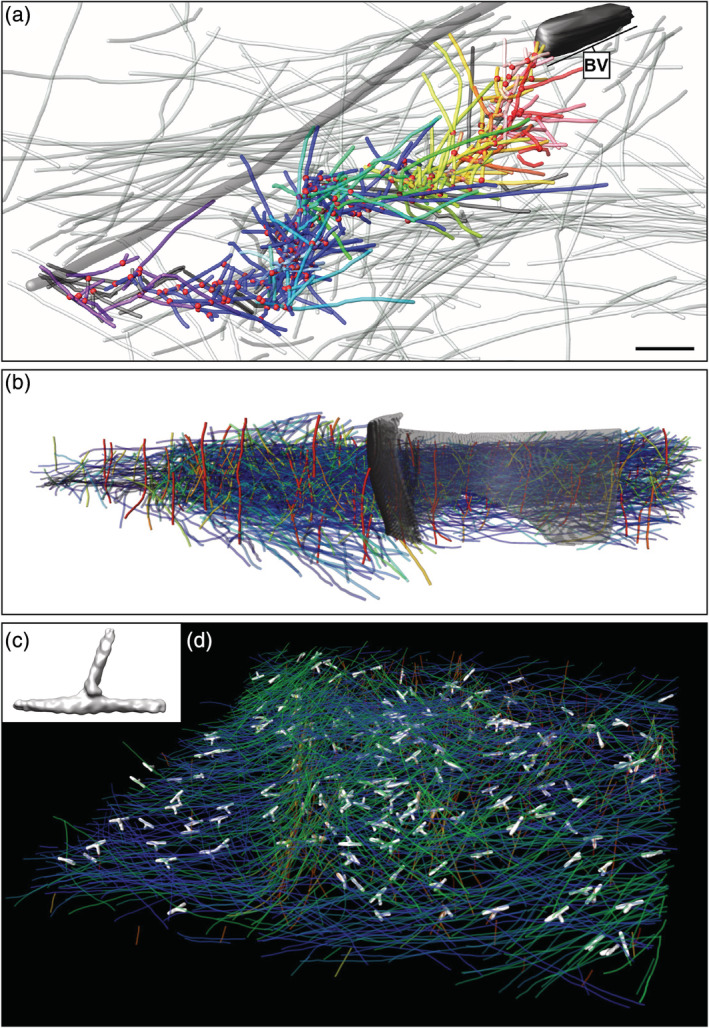
Branched actin structures revealed using cryo‐electron tomography. (a) Three‐dimensional (3D) architecture of a baculovirus (“BV”) actin tail assembled in a B16 melanoma cell. Branch points (red dots), actin filaments of the host cytoskeleton (translucent lines) and the comet tail (colored lines), and one microtubule (grey tube) are represented. Reproduced from reference [97]. (b) 3D organization of a Listeria comet tail assembled in Xenopus egg extracts. Actin filaments are represented in a blue‐to‐red color map as a function of their elevation angle relative to the support film. The cell wall of Listeria is shown in grey. Reproduced with permission from reference [98], copyright (2016) Elsevier. (c) Density map of branch junctions mediated by the Arp2/3 complex from Dictyostelium cells (EMD 4790) in solid‐surface representation. (d) Branch junctions (white) in an actin wave traveling at the basement membrane of Dictyostelium cells. Actin filaments are rendered in the same color as in (b)
In addition to providing architectural insights into branched networks, ET was used to obtain the structure of reconstituted and negatively stained branch junctions of Acanthamoeba castellanii with a resolution of 2.6 nm showing a uniform branching angle of 78°. 99 A similar structure was obtained from chemically fixed and negatively stained lamellipodia of NIH3T3 cells. 100 Using cryo‐FIB milling, cryo‐CLEM and cryo‐ET, Jasnin and colleagues targeted actin waves traveling at the basal membrane of Dictyostelium cells. 52 Template matching and subtomogram averaging42, 44 served to identify branch junctions in different orientations. The in situ structure of branch junctions obtained at a resolution of 3.1 nm (EMD 4790, Figure 3c) showed a conserved branch angle of 70° in agreement with previous in vitro measurements using electron and light microscopy86, 101 and ET of lamellipodia.90, 91 The angle differs from the uniform 78° angle obtained for the reconstituted amoeba samples, 99 which may derive from the limited range of orientations contributing to the reconstruction. In higher eukaryotes, Arp2/3 complex is a family of complexes with a pleiotropic impact on actin filament dynamics. 102 This raises the possibility that there exist Arp2/3 complexes inducing different branching conformations, a question that could be addressed through cryo‐ET. The work on actin waves paves the way for further in situ structural investigations of branched networks, and holds promise for obtaining a native high‐resolution structure of Arp2/3 complex‐mediated branch junctions.
2.3. Listeria comet tails and actin waves: Two examples of collaborative actin assembly
2.3.1. Cytoplasmic tails and protrusions assembled by Listeria monocytogenes
Listeria movement and propagation in host cells is facilitated by the ability of the bacteria to hijack the Arp2/3 complex activity. Listeria expresses at its surface the transmembrane protein ActA, which recruits Arp2/3 complex, VASP, and G‐actin, and mimics the Wiskott‐Aldrich Syndrome protein (WASP)/WASP‐family verprolin‐homologous protein (WAVE) family of nucleation promoting factors. 103 Cryo‐ET studies demonstrated that the ultrastructure of Listeria tails assembled inside cells 77 and in Xenopus egg extracts 98 (Figure 3b) is different from a classical dendritic organization as well as from the fishbone structure reported for the baculovirus tails 97 (Figure 3a). Unlike lamellipodia and baculovirus tails, in Listeria comet tails, filament intersections showing a branch geometry are randomly oriented with respect to the polymerizing surface and the direction of movement. 98 In addition, nanoscopic actin bundles were observed within the comets (Figure 2a,b), some of them hexagonally packed (Figure 2c), indicating that Arp2/3 complex alone does not determine the tail architecture. 98 In a reconstituted motility system, increasing the ratio of VASP to Arp2/3 at the polymerizing surface induces filament alignment parallel to the direction of movement. 104 The architecture of Listeria tails observed during intracellular motility and cell‐to‐cell spread may therefore reflect the combined action of Arp2/3 complex and VASP at the bacterial surface. 98 Higher‐order organization of the aligned filaments into hexagonal bundles may arise from the diverse bundlers and cross‐linkers present in Listeria tails. 98
Other viral and bacterial pathogens, including Shigella flexneri, Rickettsia rickettsii, and the vaccinia virus, have developed similar strategies to promote their dissemination into host cells. 105 The structural diversity of actin tails assembled by intracellular pathogens illustrates the malleability of the actin machinery in generating a broad spectrum of architectures through fine differences in actin‐associated factors and nucleation geometry. Exploring pathogen subversion of host cell cytoskeletal machinery using in situ cryo‐ET can provide valuable insights into the molecular and structural basis of actin‐based motility.
2.3.2. Traveling actin waves
Another phenomenon linked to composite actin networks is provided by the propagation of self‐organized actin waves at the cell cortex. Since their initial observation in Dictyostelium cells, 106 actin waves were reported in a variety of motile cells and correlated with cell protrusion, polarization, migration, and adhesion.107, 108 Our understanding of these excitable dynamic actin patterns comes mostly from live‐imaging experiments and mathematical modeling. Recently, cryo‐ET has been used to explore the architecture of actin waves assembled at the basement membrane of Dictyostelium cells 52 and how they respond to geometrical cues. 109 These waves consist of oblique tent‐like arrays within a dense horizontal meshwork. Subtomogram averaging was used to identify Arp2/3 complex‐mediated branch junctions within the waves (Figure 3d). Spatial pattern analysis revealed that branch points cluster strongly in the waves. Most branches are nucleated from filaments oriented parallel to the membrane, and most daughter filaments grow either toward or parallel to the membrane. This differs from the classical dendritic organization observed in lamellipodia and associated with pure Arp2/3 complex nucleation, in which both mother and daughter filaments point toward the membrane at an angle of ~55°.86, 89 Live‐imaging data showed that VASP forms patches followed by actin polymerization at the wave front and associated with Arp2/3 complex clustering. 52 This suggests that VASP generates at least part of the filaments from which Arp2/3 complex nucleates branches. In addition, formin B is localized to the actin waves 110 and may complement VASP in supplying the Arp2/3 complex with mother filaments.
The wave architecture exemplifies a complex actin network organization derived from the combined action of actin nucleation and elongation factors. The variety of actin waves associated with diverse cell systems provides an opportunity to explore the versatile actin machinery at work and reveal the interplay of actin assembly factors, architecture, and function. Additionally, other composite cellular actin structures such as podosomes and phagocytic cups could help expand our understanding of actin assembly and force generation in situ.
3. NANOSCALE ARCHITECTURE OF THE MT CYTOSKELETON
The MT cytoskeleton constitutes the underlying skeletal framework that is crucial for many cellular functions including cell division,111, 112 motility, 113 and cell architecture. 114 MTs are polymerized filaments assembled of α‐ and β‐tubulin monomers in a head‐to‐tail arrangement which organize into a hollow cylinder with a diameter of 25 nm.40, 51, 115 Conventional EM with stained MTs has been instrumental in studying filaments formed by tubulins in cellular sections. 15 Once cryo‐EM was developed as a near‐physiological sample preparation method, cryo‐EM images of reconstituted MTs yielded key insights into their nucleotide‐dependent dynamics and their interactions with their binding partners including molecular motors.38, 116
In order to relate MT architecture to its dynamics and function, it is necessary to elucidate MT structure along with its interacting proteins in their functional context. Early tomographic studies of MTs were performed on intact neuronal cells, 117 fibroblasts, 118 and vitreous sections of animal cells. 46 They revealed several novel structural features: (a) MT lumen is not empty but often contains electron dense particles; (b) MT plus ends are mostly frayed structures and differ from reconstituted plus ends; and (c) cellular MTs are not exactly cylindrically symmetric. In the following paragraphs, we will report some of these findings, with a special emphasis on cryo‐ET studies.
3.1. Architecture of the MT cytoskeleton
The architecture of the MT cytoskeleton depends on the cell type and physiological state. An early ET study provided insights into the organization of interphase MTs in plastic‐embedded Schizosaccharomyces pombe cells. 119 Höög et al. obtained a 3D reconstruction of a whole interphase cell revealing a bundled MT organization. 119 MT polarity was determined based on their plus end appearances, with most cytoplasmic MTs open at one end and capped at the other. Several connections between MTs and between MTs and the nuclear envelope were identified, the latter suggesting a role in organelle positioning.
The most elaborate MT architecture is found in mitotic cells in the form of a spindle. Mitotic spindle organization is vital for chromosome segregation. 120 Yet, the structure–function relationship of a spindle has remained largely unexplored. Ward et al. performed ET of fission yeast spindles, revealing that structural integrity comes from MT organization into a rigid hexagonally packed transverse array. 121 Such arrangement maximizes the interactions between a MT and its neighbors, making the spindle stronger and preventing it from buckling under compressive forces. However, structural integrity of MTs was found to be compromised in the spindle of the smallest eukaryote, Ostreococcus tauri. 122 Using cryo‐ET of high‐pressure frozen cells, Gan et al. found that spindle MTs in this organism are short and mostly incomplete (C‐shaped cross‐sections), comprising fewer than 13 protofilaments (PFs). 122
Recently, cryo‐FIB milling and cryo‐ET were harnessed to explore the nanoscale architecture of MTs inside different mammalian cells and at different cell‐cycle stages using the VPP. 123 In interphase HeLa cells, MTs are spatially entangled in a composite cytoskeletal matrix of actin filaments and IFs (Figure 4a). Similar organization was found for astral MTs at the cortex of mitotic cells. Further away from the cortex, mitotic cells have a cytoskeletal organization dominated by the high abundance of parallel arrays of MTs forming a spindle (Figure 4b), with an interfilament distance of 65 nm. Smaller distances (~50 nm) were observed in MT bundles assembled in thin cellular parts of neuronal cells. In the presence of Taxol, the distance between parallel MTs in both mitotic and interphase HeLa cells is reduced.
FIGURE 4.
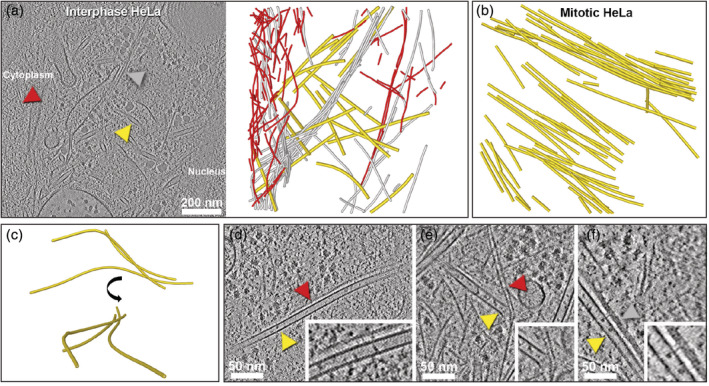
Architecture of the microtubule (MT) cytoskeleton revealed by cryo‐electron tomography. (a) Slice through a tomogram acquired in a focused ion beam‐milled interphase HeLa cell with the Volta phase plate. Arrowheads indicate a MT (yellow), an intermediate filament (IF; grey), and an actin filament (red). Cytoskeletal filaments were segmented and displayed using the same color code on the right panel. (b) Bundled organization of MTs in a mitotic HeLa cell. (c) Curvilinear trajectories of MTs in an interphase HeLa cell resembling short wavelength buckling and shown in two different orientations. (d) MT‐actin wall‐to‐wall interaction in an interphase HeLa cell. Inset: Close up showing putative crosslinkers connecting the filaments. (e) Actin‐MT plus end interaction in an interphase P19 cell. Inset: Zoomed image of the interaction zone. (f) MT‐IF wall‐to‐wall interaction in an interphase HeLa cell. Inset: Close up showing putative thin stalk‐like densities connecting the filaments. Panels (a) and (d‐f) reproduced from reference [123]
3.2. MT curvature in situ
MT curvature plays a crucial role for biological processes which require the MT cytoskeleton to function as a scaffold. MTs need to be sufficiently stiff to create and maintain cell shape, especially for extended morphologies such as neuronal processes. MTs also need to remain fairly straight to function as long‐range tracks for efficient molecule and organelle transport by cargo‐carrying motor proteins. Consistent with this, isolated MTs have persistence length on the order of a 1–6 mm, 124 whereas typical mammalian cells measure only tens of microns.
However, MTs were occasionally found in highly bent configurations inside cells.125, 126 In a recent cryo‐ET study, MT curvature was quantitatively analyzed inside interphase and mitotic cells. 123 The apparent persistence length of MTs in interphase HeLa cells is ~2–3 orders of magnitude lower than their in vitro persistence length, consistent with previous reports based on fluorescence microscopy investigations.127, 128 In a subset of MTs residing at the nucleocytoplasmic boundary of HeLa cells, highly localized bends with sinusoidal wave‐like trajectories (named “short wavelength bending”) were found (Figure 4c). This contradicts the long‐arc‐like bending predicted by classical mechanics. 129 Astral MTs entangled with actin filaments and IFs at the cortex of mitotic cells have similar curvatures as MTs in interphase cells. In contrast, spindle and neuronal MTs show long‐arc‐like bending. Apparently, the matrix surrounding the MTs can substantially modulate the curvature. 123
3.3. MT‐actin crosstalk
Biophysical and biological properties of the actin and MT cytoskeletons have been studied extensively over the past decades. It is accepted that their functional dynamical properties are intimately intertwined. Actin‐MTs crosstalk is known to promote symmetry‐breaking polarization for cell shape changes, division, and motility. 130 Early observations with conventional TEM revealed that MTs and actin filaments interact in the presence of MT‐associated proteins (MAPs) ex situ 131 and in situ. 132 Yet, how these two cytoskeletal systems work together during cellular processes remains to be elucidated.
In situ cryo‐ET of eukaryotic cells provided insights into MT and actin coordination inside cells. 123 MTs and actin filaments were found to interact primarily through passive entanglements (Figure 4a). Such entanglements are known to create steric interactions between the two types of filaments inside the cytosol, and thereby influence cell shape and mechanics by restraining filament buckling and coordinated reinforcement of the cytoskeleton. 133 Indeed, the ability of MTs to withstand strong compressive forces relies on the reinforcement provided by the surrounding elastic actin network that constrains MT bending. 126 MTs were also found to interact with actin filaments via crosslinkers, either through wall‐to‐wall interactions with a spacing of about 11 nm (Figure 4d) or via their plus ends (Figure 4e). 123 The latter interaction is very common at the actomyosin cortex, providing a physical barrier that prevents growing MTs from targeting the plasma membrane 130 and can induce MT bending and catastrophe. 134 It is also involved in actin‐mediated MT guidance and growth along actin bundles. 135 The physical connections could be mediated by proteins with binding sites for both the actin filaments and MTs or MT‐end binding proteins such as CLIP‐170. 136 Visualization of both types of interactions using in situ cryo‐ET supports the tensegrity model, in which tension propagation requires physical crosslinks between actin filaments and MTs. 137
3.4. Insights into the native MT structure
In vitro‐assembled tubulins are known to yield polymorphic MT structures with PFs varying between 11 and 18.138, 139 Early conventional EM studies in 1970s using glutaraldehyde fixation and negative staining have shown that MTs in animal cells are mostly made of 13 PFs. 140 Using cryo‐electron microscopy of vitreous sections (CEMOVIS) 43 and cryo‐ET, Bouchet‐Marquis et al. explored MT ultrastructure in vitreous sections of high‐pressure frozen CHO cells. 46 MT chirality was visible in the raw data and enhanced by 13‐fold rotational averaging, revealing a “clockwise slew” (structure seen from the minus end) or an “anticlockwise slew” (structure seen from the plus end; Figure 5a). The authors showed that tomography of vitreous sections can achieve a similar resolution as with plunge‐frozen samples, despite the compression artifacts associated with the cutting process. The same MT structure was later observed in thin cellular peripheries of primary fibroblasts and neurons using cryo‐ET. 118 When McIntosh et al. visualized Eg5‐decorated interphase MTs in 3T3 cells using cryo‐ET, visual inspection of the tomograms and power spectra of the single‐projection images confirmed the presence of B‐lattices, in which the α‐tubulins of one PF lie next to α‐tubulins in the neighboring PFs 141 (Figure 5b). As a result, 13‐PFs MTs with B‐lattices must include a “seam,” that is, a pair of PFs where α‐tubulin lies beside β‐tubulin, and therefore breach cylindrical symmetry (Figure 5b).
FIGURE 5.
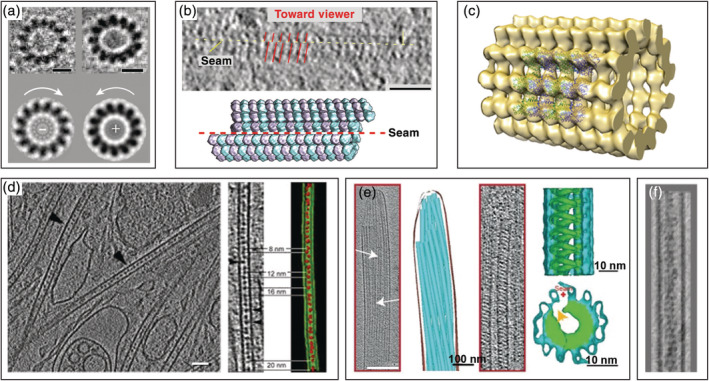
Native microtubule (MT) structure revealed by cryo‐electron tomography. (a) Cross‐sections through MTs in CHO cells (top) and “clockwise slew” (bottom left) or “anticlockwise slew” (bottom right) structure obtained by 13‐fold rotational averaging revealing MT polarity. Scale bars: 10 nm. Reproduced with permission from reference [46], copyright (2012) John Wiley and Sons. (b) Seam observed in cytoplasmic MTs decorated with Eg5 motor domains in an interphase 3T3 cell. Scale bar: 50 nm. Reproduced with permission from reference [141], copyright (2009) Elsevier. (c) MT structure of U87MG neuronal cells obtained by subtomogram averaging, with the fit of a MT segment (EMD 6O2R) in the density map. The structure confirmed the presence of 13 protofilaments in situ. (d) Heterogeneous lumenal densities observed in MTs of neuronal cells. Right: Close up in the MT lumen showing segmented material and interparticle distances. Scale bar: 100 nm. Reproduced with permission from reference [117], copyright (2006) Rockefeller University Press. (e) Singlet MTs observed at the tip of a sperm cell containing diagonal lumenal densities with 8 nm periodicity. Density map of the tail axoneme intra‐lumenal spirals complex obtained by subtomogram averaging (right). Reproduced from reference [142]. (f) Actin filament segments in the MT lumen of MT‐based cellular projections in HAP1 cells. Reproduced from reference [143]
Cryo‐ET also offers an unbiased way to determine 3D structures using subtomogram averaging.42, 44 MT structure was elucidated in situ at 18–24 Å from a single tomogram 144 (Figure 5c). The resulting MT structure is consistent with the current understanding of MT structure in vivo. Given the recent technical advancements, cryo‐ET will be the method of choice to generate in situ structures of MTs with their binding partners routinely, at resolutions facilitating a mechanistic understanding of MT‐related functions.
3.5. Lumenal densities
MAPs contribute to the stability of MTs and regulate their dynamics whereas MT‐based motors mediate the transport of cargoes along MT tracks. Although all MAPs and motors studied so far bind to the outside of the MT wall, small molecules such as Taxol can associate with the lumenal side of MTs. 145 The MT lumen is physically separated from the cytoplasm, except from the lateral pores and two 200 nm 2 entrances at the MT ends. The presence of electron‐dense material within the MT lumen was first observed in plastic‐embedded and stained preparations of insect epithelia, 146 spermatids, 147 blood platelets, 148 and neuronal cells. 149 However, the existence of the lumenal densities was largely thought to be an artifact of the staining procedures. Moreover, there was no evidence of any electron‐dense material in the lumen of in vitro‐nucleated MTs. 117
Cryo‐ET of frozen sections of cells confirmed the existence of these particles in the MT lumen. In neuronal cells, discrete, closely spaced globular particles of 6–7 nm in diameter were observed 117 (Figure 5d). The typical distances between particles were apparent multiples of 4 nm, ranging from 8 to 20 nm (Figure 5d). Since 4 nm corresponds to the size of a tubulin monomer, it suggested that lumenal particles decorate the tubulins along the MT wall. Subtomogram averaging revealed points of contact between lumenal particles and the MT wall. 117 The existence and frequency of lumenal particles was found to vary between cell types, with neuronal cells having the highest abundance. 117 Depolymerizing MTs contain more lumenal particles as compared to growing MTs, with a minimal interparticle distance of ~8 nm, and have the same “beaded fiber” appearance as observed in conventionally prepared EM specimens. 150
Identification of intralumenal components is challenging. Recent reports provided evidence for the presence of tubulin acetyltransferase (TAT). 151 TAT is known to transfer an acetyl group from acetyl‐CoA to the Lys40 of α‐tubulin at the lumenal side 152 and to regulate MT stiffness, 153 which is thought to be useful in mechanosensory and cilia functions.
Another occurrence of lumenal material was observed in the singlet region of human spermatozoon tails by cryo‐ET. 142 In these cells, axonemal MTs display at their distal end a singular interior structure made of diagonal densities with 8 nm periodicity 142 (Figure 5e). Subtomogram averaging revealed that this lumenal structure, named Tail Axoneme Intra‐Lumenal Spirals, consists of an interrupted left‐handed helix that follows the pattern of the MT lattice 142 (Figure 5e).
A recent cryo‐ET study of chemically induced, MT‐based cellular projections in HAP1 cells exposed the presence of actin filament segments in the MT lumen 143 (Figure 5f). Although the physiological relevance of this cell‐based extrusion system remains to be established, it exemplifies the power of cryo‐ET to discover unexpected structures inside cells.
3.6. Structure of the MT plus end
MTs exhibit dynamic instability that forms the basis of most MT functions including force generation 154 (Figure 6a). MTs have the ability to grow or shrink at their plus ends by the net addition or loss of tubulin dimers. Dynamic instability results from the intrinsic GTPase activity of the tubulin dimer inside the polymer. 158 Conventional EM and later cryo‐EM studies on reconstituted MTs have shown that growing MT ends are structurally heterogeneous 34 (Figure 6b). Most of them have either a sheet‐like extension that curves away from the long axis of the MT or blunt ends.159, 160 In contrast, shrinking ends are frayed. 161 Based on their sheet‐like appearance, the “closing sheet model” for MT polymerization has been proposed where long sheet‐like PFs curl up and form a tube. 159 Several ET and cryo‐ET studies have challenged this model.162, 163 It was suggested that the in vitro relationship between MT plus end structure and dynamics is not necessarily identical to the situation in vivo where MT dynamics is regulated by a variety of MAPs. 164 Elucidating MT end structure in cells can therefore help develop mechanistic models for MT dynamics.
FIGURE 6.
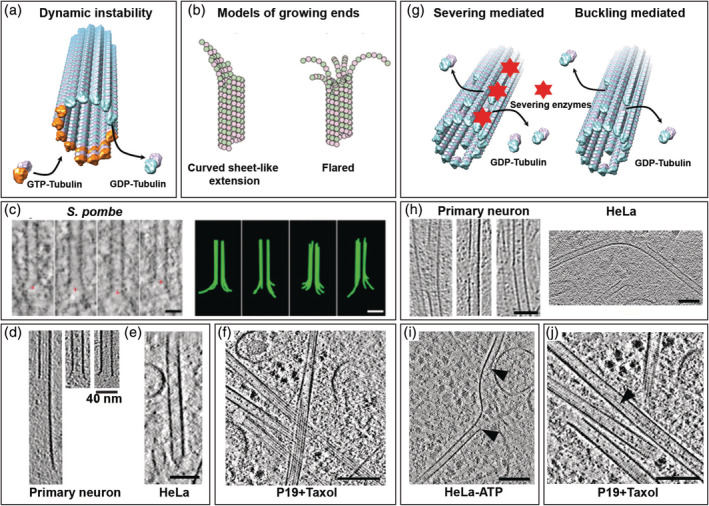
Lattice plasticity of microtubules (MTs) revealed by cryo‐electron tomography. (a) Schematic representation of MT dynamic instability. (b) Proposed models of MT growing ends. Reproduced with permission from reference [155], copyright (2018) Rockefeller University Press. (c) Flared morphology of growing MT plus ends observed in Schizosaccharomyces pombe cells. Scale bars: 50 nm. Reproduced with permission from reference [156], copyright (2018) Rockefeller University Press. (d) Curved extension (left), flared (middle), or blunt (right) MT plus ends observed in hippocampal neurons. Reproduced from reference [157]. (e) Capped MT plus end in a mitotic HeLa cell. Scale bars: 40 nm. (f) Sheet‐like extensions of MT plus ends observed in a Taxol‐treated HeLa cell. Scale bars: 100 nm. (g) Proposed mechanisms of lattice defects in MTs. (h) Lattice defects observed in primary neurons (reproduced from reference [157]) and a HeLa cell (reproduced from reference [123]). Scale bar: 40 nm. (i) Protofilament (PF) segments lost in an adenosine triphosphate (ATP)‐depleted HeLa cell indicated by black arrowheads. Scale bar: 100 nm. Reproduced from reference [123]. (j) 13‐to‐14 PF transition (black arrowhead) observed in a Taxol‐treated cell. Scale bars: 100 nm. GTP, guanosine triphosphate; GDP, guanosine diphosphate
ET studies of MT plus ends in sections of freeze‐substituted cells provided new insights and changed the way we understand MT plus end dynamics. In S. pombe cells, when MTs are allowed to repolymerize growing MT plus ends display largely a flared morphology. 165 McIntosh et al. observed short curved extensions at the ends of growing interzone anaphase MTs in cells from five species 156 (Figure 6c). They have shown that the extensions are curled PFs growing independently of one another and away from the MT axis. Because the curvatures of PFs were similar between growing and shrinking MTs (being the most curved at the PF tip), they suggested that guanosine triphosphate (GTP)‐bound tubulin is bent in solution and must uncurl to form the MT polymer.
Recent cryo‐ET studies showed curved extensions at the growing MT end in a variety of cells, including plasmodium sporozoites, 166 fibroblasts, 118 and hippocampal neurons 157 (Figure 6d). The observed structures were mostly flared and, sometimes, capped (Figure 6e), sheet‐like, or blunt. The sheet‐like extension was also observed in Taxol‐treated cells (unpublished data; Figure 6f).
Altogether, these studies emphasize the importance of exploring MT structure in its native environment. Cryo‐ET holds promise for the structural determination of cellular MTs including their plus end and end‐binding partners.
3.7. Lattice defects
Apart from dynamic instability that creates structural heterogeneity at the plus end, in vitro EM studies have revealed the presence of a wide range of irregularities in MT core lattice structures, commonly known as lattice defects 167 (Figure 6g). These include variations in PF number within a MT 168 as well as between MTs. 139 Recent in vitro studies have shown that the MT lattice could also lose a tubulin dimer or several of them, thereby creating a nanoscale hole in the lattice. These defects are hypothesized to arise from cross‐sectional flattening due to mechanical bending, 169 packing defects, 167 or the activity of severing proteins 170 (Figure 6g). In vitro lattice vacancies and dislocations weaken the ability of MTs to withstand compressive forces 171 and has been shown to affect the efficiency of kinesin‐based transport. 172 However, in vivo relevance of lattice defects remained obscure.
Recent cryo‐ET studies revealed MT lattice defects of various sizes in neurons 157 and HeLa cells 123 (Figure 6h). Because these defects occurred in strongly buckled growing MTs, a causal relationship between MT bending and lattice defects was proposed, although an action of MT severing enzymes could not be ruled out. 123 In ATP depleted cells, the loss of multiple PF segments was observed (Figure 6i) and may be a long‐lived intermediate resulting from the incomplete action of severing enzymes. 123 Irrespective of their origin, all these defects could contribute to the apparent strong curvature of MTs in situ. Growing evidence suggests that lattice defects are exploited as lattice control points where fresh GTP‐tubulins can be introduced and thereby rescue/regrow MTs. 173
PF transition within one MT has only been reported in vitro. 168 Recent in situ cryo‐ET work showed that it occurs in Taxol‐treated cells, with a transition from 13 to 14 PFs (unpublished data; Figure 6j).
4. NANOSCALE ARCHITECTURE OF THE IF CYTOSKELETON
IFs represent a major and structurally diverse group of nonpolar cellular filaments. 174 They were originally referred to as IFs because their diameter is intermediate between the diameters of actin and myosin filaments forming myofibrils in muscle cells. 18 IFs are made of smooth PFs composed of antiparallel arranged α‐helical coiled‐coil bundles flanked by small globular domains at either end. 175 These features make it challenging to study IFs using EM‐based averaging procedures.17, 42, 44
The 3D architecture of IFs within an intact cellular environment remains poorly studied, with only a handful ET and cryo‐ET studies. Cytoplasmic bundles of IFs were observed in ultrathin sections of high‐pressure frozen CHO and 3T3 A31 cells. 176 Vitrified IFs have a diameter of 12–13 nm on average, 176 larger than the values reported with conventional EM methods (~7–11 nm). 177
Cryo‐EM and ET of vitrified sections of human epidermis permitted to visualize bundles of keratin filaments.178, 179 Keratin filaments have a diameter of 8–10 nm and a center‐to‐center distance between neighboring filaments of about 15 nm. 178 They present a central core surrounded by an annular ring composed of PFs in a quasi‐hexagonal arrangement. 179 Keratin structure is different from the 4‐PFs structure found for vimentin. 47
Cryo‐ET exploration of FIB‐milled HeLa cells showed a dense bundled network of IFs at the perinuclear region 123 (Figure 7a). Diameter and cross‐section analysis suggested that they are vimentin filaments, although proper identification of the IF type requires correlative approaches. Physical connections between IFs and MTs were observed123, 181 (Figure 4f) and are found to be more abundant in cells depleted of actin filaments. 123 These contacts are hypothesized to be responsible for transducing compressive forces onto the MT network, inducing MT bending inside cells.123, 182
FIGURE 7.
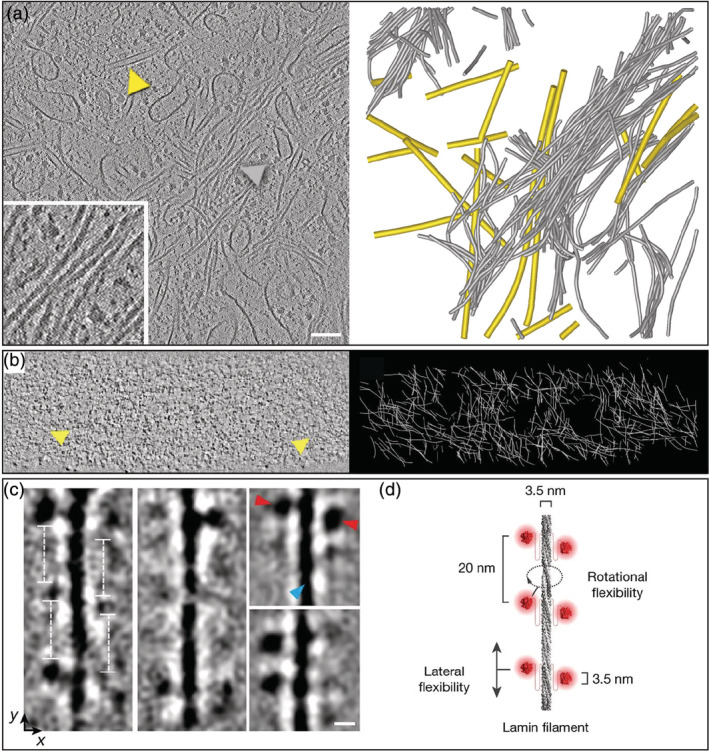
Organization of intermediate filaments (IFs) explored by cryo‐electron tomography. (a) Slice through a tomogram acquired in a focused ion beam‐milled interphase HeLa cell with the Volta phase plate. Inset: Close up on a bundle of smooth cytoplasmic IFs. Arrowheads indicate a microtubule (yellow) and an IF (grey). Cytoskeletal filaments were segmented and displayed using the same color code on the right panel. (b) Slice through the lamina of an interphase HeLa cell. Yellow arrowheads indicate lamin filaments. Three‐dimensional network of lamins are represented in white on the right panel. Reproduced with permission from reference [10], copyright (2016) AAAS. (c) Classes of lamin filaments obtained in vimentin‐null MEFs showing repetitive globular domains spaced 20 nm apart along the filament. Scale bar: 5 nm. Reproduced with permission from reference [180], copyright (2017) Springer Nature. (d) Structural model of lamin filaments. Reproduced with permission from reference [180], copyright (2017) Springer Nature
Another type of IFs, known as lamins, is a fundamental constituent of metazoan nuclear lamina. In situ cryo‐ET of HeLa cells revealed that these 4‐nm thick filaments are organized in a mesh‐like structure just underneath the nuclear membrane 10 (Figure 7b). Cryo‐ET of vimentin‐null mouse embryonic fibroblasts showed that individual lamin filaments have a globular‐decorated fiber appearance (Figure 7c,d), which differs from cytoplasmic IFs. 180
Future in situ cryo‐ET work could help reveal the native structure and organization of the different classes of IFs across cell types and tissues.
5. OUTLOOK
Advancements in cryo‐ET technologies have enabled in situ imaging of cytoskeletal networks with molecular resolution. We can now retrieve both structural and spatial information about cytoskeletal filaments within the cell, linking filament structure to supramolecular assemblies. Future studies may bring insights into “forgotten” cytoskeletal elements such as septins and spectrins. They will also open new avenues to understand the architectural and mechanistic links between cytoskeleton and organelles. Cryo‐ET can be expected to become indispensable for exploring the nanoscale architecture of composite cytoskeletal assemblies in diverse biological systems, and help generate quantitative models for cellular biomechanics. It holds promise for understanding the guiding principles that enable the cytoskeleton to orchestrate physiological processes as diverse as nerve conduction and infection.
AUTHOR CONTRIBUTIONS
Saikat Chakraborty: Conceptualization; writing‐original draft; writing‐review and editing. Marion Jasnin: Conceptualization; supervision; writing‐original draft; writing‐review and editing. Wolfgang Baumeister: Conceptualization; funding acquisition; supervision; writing‐original draft.
ACKNOWLEDGMENTS
We thank Julia Mahamid for her collaboration in studying MTs in situ, and Renaud Poincloux for his critical comments on the manuscript. This work was funded by the Human Frontier Science Program Grant RGP0035/2016 and the Max Planck Society.
Chakraborty S, Jasnin M, Baumeister W. Three‐dimensional organization of the cytoskeleton: A cryo‐electron tomography perspective. Protein Science. 2020;29:1302–1320. 10.1002/pro.3858
Saikat Chakraborty and Marion Jasnin contributed equally to this study.
Funding information HFSP, Grant/Award Number: RGP0035/2016
Contributor Information
Marion Jasnin, Email: jasnin@biochem.mpg.de.
Wolfgang Baumeister, Email: baumeist@biochem.mpg.de.
REFERENCES
- 1. Crick FHC, Hughes AFW. The physical properties of cytoplasm. Exp Cell Res. 1950;1:37–80. [Google Scholar]
- 2. Steven AC, Baumeister W, Johnson LN, Perham RN. Molecular biology of assemblies and machines. Garland Science: New York, NY, 2016. [Google Scholar]
- 3. Bausch AR, Kroy K. A bottom‐up approach to cell mechanics. Nat Phys. 2006;2:231–238. [Google Scholar]
- 4. Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andrault R. Mathématiser l’anatomie: La myologie de Stensen (1667). Early Sci Med. 2010;15:505–536. [DOI] [PubMed] [Google Scholar]
- 7. Knoll M, Ruska E. Das Elektronenmikroskop. Z Phys. 1932;78:318–339. [Google Scholar]
- 8. Borries B, Ruska E, Ruska H. Bakterien und Virus in Übermikroskopischer Aufnahme. Klin Wochenschr. 1938;17:921–925. [Google Scholar]
- 9. Kausche GA, Pfankuch E, Ruska H. Die Sichtbarmachung von pflanzlichem Virus im Übermikroskop. Naturwissenschaften. 1939;27:292–299. [Google Scholar]
- 10. Mahamid J, Pfeffer S, Schaffer M, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. [DOI] [PubMed] [Google Scholar]
- 11. Steere RL. Electron microscopy of structural detail in frozen biological specimens. J Biophys Biochem Cytol. 1957;3:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bradley DE. Simultaneous evaporation of platinum and carbon for possible use in high‐resolution shadow‐casting for the electron microscope. Nature. 1958;181:875–877. [DOI] [PubMed] [Google Scholar]
- 13. Brenner S, Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959;34:103–110. [DOI] [PubMed] [Google Scholar]
- 14. Wohlfarth‐Bottermann KE. Weitreichende, fibrilläre Protoplasmadifferenzierungen und ihre Bedeutung für die Protoplasmaströmung ‐ I. Elektronenmikroskopischer Nachweis und Feinstruktur. Protoplasma. 1962;54:514–539. [Google Scholar]
- 15. Ledbetter MC, Porter KR. A “microtubule” in plant cell fine structure. J Cell Biol. 1963;19:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabatini DD, Bensch K, Barrnett RJ. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963;17:19–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Rosier DJ, Klug A. Reconstruction of three dimensional structures from electron micrographs. Nature. 1968;217:130–134. [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa H, Bischoff R, Holtzer H. Mitosis and intermediate‐sized filaments in developing skeletal muscle. J Cell Biol. 1968;38:538–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schroeder TE. The contractile ring: II. determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J Cell Biol. 1972;53:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1037. [DOI] [PubMed] [Google Scholar]
- 21. Glaeser RM, Taylor KA. Radiation damage relative to transmission electron microscopy of biological specimens at low temperature: A review. J Microsc. 1978;112:127–138. [DOI] [PubMed] [Google Scholar]
- 22. Heath JP, Dunn GA. Cell to substratum contacts of chick fibroblasts and their relation to the microfilament system. A correlated interference‐reflexion and high‐voltage electron‐microscope study. J Cell Sci. 1978;29:197–212. [DOI] [PubMed] [Google Scholar]
- 23. Small JV, Isenberg G, Celis JE. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. [DOI] [PubMed] [Google Scholar]
- 24. Small JV, Celis JE. Filament arrangements in negatively stained cultured cells: The organization of actin. Cytobiologie. 1978;16:308–325. [PubMed] [Google Scholar]
- 25. Singer II. The fibronexus: A transmembrane association of fibronectin‐containing fibers and bundles of 5 nm microfilaments in hamster and human fibroblasts. Cell. 1979;16:675–685. [DOI] [PubMed] [Google Scholar]
- 26. Small JV. Organization of actin in the leading edge of cultured cells: Influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J Cell Biol. 1981;91:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heuser JE, Kirschner MW. Filament organization revealed in platinum replicas of freeze‐dried cytoskeletons. J Cell Biol. 1980;86:212–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dubochet J, Lepault J, Freeman R, Berriman JA, Homo JC. Electron microscopy of frozen water and aqueous solutions. J Microsc. 1982;128:219–237. [Google Scholar]
- 29. Dubochet J, Chang JJ, Freeman R, Lepault J, McDowall AW. Frozen aqueous suspensions. Ultramicroscopy. 1982;10:55–61. [Google Scholar]
- 30. Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo‐electron microscopy of viruses. Nature. 1984;308:32–36. [DOI] [PubMed] [Google Scholar]
- 31. Van Heel M. Angular reconstitution: A posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. [DOI] [PubMed] [Google Scholar]
- 32. Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three‐dimensional reconstruction from a single‐exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. [DOI] [PubMed] [Google Scholar]
- 33. Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high‐resolution electron cryo‐microscopy. J Mol Biol. 1990;213:899–929. [DOI] [PubMed] [Google Scholar]
- 34. Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: A time‐resolved cryo‐electron microscopy study. J Cell Biol. 1991;114:977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dierksen K, Typke D, Hegerl R, Koster AJ, Baumeister W. Towards automatic electron tomography. Ultramicroscopy. 1992;40:71–87. [Google Scholar]
- 36. Dierksen K, Typke D, Hegerl R, Baumeister W. Towards automatic electron tomography II. Implementation of autofocus and low‐dose procedures. Ultramicroscopy. 1993;49:109–120. [Google Scholar]
- 37. Rayment I, Holden HM, Whittaker M, et al. Structure of the actin‐myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. [DOI] [PubMed] [Google Scholar]
- 38. Kikkawa M, Ishikawa T, Wakabayashi T, Hirokawa N. Three‐dimensional structure of the kinesin head‐microtubule complex. Nature. 1995;376:274–277. [DOI] [PubMed] [Google Scholar]
- 39. Grimm R, Singh H, Rachel R, Typke D, Zillig W, Baumeister W. Electron tomography of ice‐embedded prokaryotic cells. Biophys J. 1998;74:1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. [DOI] [PubMed] [Google Scholar]
- 41. Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. [DOI] [PubMed] [Google Scholar]
- 42. Grünewald K, Desai P, Winkler DC, et al. Three‐dimensional structure of herpes simplex virus from cryo‐electron tomography. Science. 2003;302:1396–1398. [DOI] [PubMed] [Google Scholar]
- 43. Al‐Amoudi A, Chang JJ, Leforestier A, et al. Cryo‐electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo‐electron tomography. Proc Natl Acad Sci U S A. 2005;102:4729–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused‐ion‐beam thinning of frozen‐hydrated biological specimens for cryo‐electron microscopy. Nat Methods. 2007;4:215–217. [DOI] [PubMed] [Google Scholar]
- 46. Bouchet‐Marquis C, Zuber B, Glynn AM, et al. Visualization of cell microtubules in their native state. Biol Cell. 2007;99:45–53. [DOI] [PubMed] [Google Scholar]
- 47. Goldie KN, Wedig T, Mitra AK, Aebi U, Herrmann H, Hoenger A. Dissecting the 3‐D structure of vimentin intermediate filaments by cryo‐electron tomography. J Struct Biol. 2007;158:378–385. [DOI] [PubMed] [Google Scholar]
- 48. McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rigort A, Bauerlein FJB, Villa E, et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A. 2012;109:4449–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in‐focus phase contrast transmission electron microscopy. Proc Natl Acad Sci U S A. 2014;111:15635–15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High‐resolution microtubule structures reveal the structural transitions in αβ‐tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jasnin M, Beck F, Ecke M, et al. The architecture of traveling actin waves revealed by cryo‐electron tomography. Structure. 2019;27:1211–1223.e5. [DOI] [PubMed] [Google Scholar]
- 53. Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proc Soc Exp Biol Med. 1941;47:200–202. [Google Scholar]
- 54. Ramos‐Vara JA. Principles and methods of immunohistochemistry. Methods Mol Biol. 1641;2011:115–128. [DOI] [PubMed] [Google Scholar]
- 55. Leterrier C, Dubey P, Roy S. The nano‐architecture of the axonal cytoskeleton. Nat Rev Neurosci. 2017;18:713–726. [DOI] [PubMed] [Google Scholar]
- 56. Huang B, Bates M, Zhuang X. Super‐resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shtengel G, Galbraith JA, Galbraith CG, et al. Interferometric fluorescent super‐resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A 2009;106:3125–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beck M, Baumeister W. Cryo‐electron tomography: Can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. [DOI] [PubMed] [Google Scholar]
- 59. Lučić V, Rigort A, Baumeister W. Cryo‐electron tomography: The challenge of doing structural biology in situ. J Cell Biol. 2013;202:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hart RG. Electron microscopy of unstained biological material: The polytropic montage. Science. 1968;159:1464–1467. [DOI] [PubMed] [Google Scholar]
- 61. Mahamid J, Baumeister W. Cryo‐electron tomography: The realization of a vision. Microsc Anal. 2012;26:45–48. [Google Scholar]
- 62. Al‐Amoudi A, Studer D, Dubochet J. Cutting artefacts and cutting process in vitreous sections for cryo‐electron microscopy. J Struct Biol. 2005;150:109–121. [DOI] [PubMed] [Google Scholar]
- 63. Blanchoin L, Boujemaa‐Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. [DOI] [PubMed] [Google Scholar]
- 64. Mogilner A, Barnhart EL, Keren K. Experiment, theory, and the keratocyte: An ode to a simple model for cell motility. Semin Cell Dev Biol. 2020;100:143–151. [DOI] [PubMed] [Google Scholar]
- 65. Jacquemet G, Stubb A, Saup R, et al. Filopodome mapping identifies p130Cas as a mechanosensitive regulator of filopodia stability. Curr Biol. 2019;29:202–216.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gallop JL. Filopodia and their links with membrane traffic and cell adhesion. Semin Cell Dev Biol. 2020;102:81–89. [DOI] [PubMed] [Google Scholar]
- 67. Yang Y, Wu M. Rhythmicity and waves in the cortex of single cells. Philos Trans R Soc B Biol Sci. 2018;373:20170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Poincloux R, Lizárraga F, Chavrier P. Matrix invasion by tumour cells: A focus on MT1‐MMP trafficking to invadopodia. J Cell Sci. 2009;122:3015–3024. [DOI] [PubMed] [Google Scholar]
- 69. van den Dries K, Linder S, Maridonneau‐Parini I, Poincloux R. Probing the mechanical landscape ‐ New insights into podosome architecture and mechanics. J Cell Sci. 2019;132:jcs236828. [DOI] [PubMed] [Google Scholar]
- 70. Tojkander S, Gateva G, Lappalainen P. Actin stress fibers ‐ Assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. [DOI] [PubMed] [Google Scholar]
- 71. Salbreux G, Charras G, Paluch E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 2012;22:536–545. [DOI] [PubMed] [Google Scholar]
- 72. Charras G, Paluch E. Blebs lead the way: How to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. [DOI] [PubMed] [Google Scholar]
- 73. Rigort A, Bäuerlein FJB, Leis A, et al. Micromachining tools and correlative approaches for cellular cryo‐electron tomography. J Struct Biol. 2010;172:169–179. [DOI] [PubMed] [Google Scholar]
- 74. Medalia O, Beck M, Ecke M, et al. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. [DOI] [PubMed] [Google Scholar]
- 75. Patla I, Volberg T, Elad N, et al. Dissecting the molecular architecture of integrin adhesion sites by cryo‐electron tomography. Nat Cell Biol. 2010;12:909–915. [DOI] [PubMed] [Google Scholar]
- 76. Rigort A, Günther D, Hegerl R, et al. Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J Struct Biol. 2012;177:135–144. [DOI] [PubMed] [Google Scholar]
- 77. Jasnin M, Asano S, Gouin E, et al. Three‐dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci U S A. 2013;110:20521–20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fukuda Y, Laugks U, Lučić V, Baumeister W, Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol. 2015;190:143–154. [DOI] [PubMed] [Google Scholar]
- 79. Heinrich D, Ecke M, Jasnin M, Engel U, Gerisch G. Reversible membrane pearling in live cells upon destruction of the actin cortex. Biophys J. 2014;106:1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu Y, Moseley JB, Sagot I, et al. Crystal structures of a formin homology‐2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. [DOI] [PubMed] [Google Scholar]
- 81. Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. [DOI] [PubMed] [Google Scholar]
- 82. Thompson ME, Heimsath EG, Gauvin TJ, Higgs HN, Kull FJ. FMNL3 FH2‐actin structure gives insight into formin‐mediated actin nucleation and elongation. Nat Struct Mol Biol. 2013;20:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Toro‐Nahuelpan M, Zagoriy I, Senger F, Blanchoin L, Théry M, Mahamid J. Tailoring cryo‐electron microscopy grids by photo‐micropatterning for in‐cell structural studies. Nat Methods. 2020;17:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Engel L, Gaietta G, Dow LP, et al. Extracellular matrix micropatterning technology for whole cell cryogenic electron microscopy studies. J Micromech Microengin. 2019;29:115018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin‐myosin II system in fish epidermal keratocytes: Mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin‐binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Amann KJ, Pollard TD. Direct real‐time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001;98:15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maly IV, Borisy GG. Self‐organization of a propulsive actin network as an evolutionary process. Proc Natl Acad Sci U S A. 2001;98:11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. [DOI] [PubMed] [Google Scholar]
- 91. Yang C, Svitkina T. Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol. 2011;13:1012–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Small JV, Winkler C, Vinzenz M, Schmeiser C. Reply: Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol. 2011;13:1013–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mueller J, Szep G, Nemethova M, et al. Load adaptation of lamellipodial actin networks. Cell. 2017;171:188–200.e16. [DOI] [PubMed] [Google Scholar]
- 94. Kage F, Winterhoff M, Dimchev V, et al. FMNL formins boost lamellipodial force generation. Nat Commun. 2017;8:14832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ponti A, Machacek M, Gupton SL, Waterman‐Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. [DOI] [PubMed] [Google Scholar]
- 96. Giannone G, Dubin‐Thaler BJ, Rossier O, et al. Lamellipodial actin mechanically links myosin activity with adhesion‐site formation. Cell. 2007;128:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mueller J, Pfanzelter J, Winkler C, et al. Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 2014;12:e1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jasnin M, Crevenna AH. Quantitative analysis of filament branch orientation in Listeria actin comet tails. Biophys J. 2016;110:817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rouiller I, Xu XP, Amann KJ, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vinzenz M, Nemethova M, Schur F, et al. Actin branching in the initiation and maintenance of lamellipodia. J Cell Sci. 2012;125:2775–2785. [DOI] [PubMed] [Google Scholar]
- 101. Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. [DOI] [PubMed] [Google Scholar]
- 102. Abella JV, Galloni C, Pernier J, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. [DOI] [PubMed] [Google Scholar]
- 103. Radoshevich L, Cossart P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018;16:32–46. [DOI] [PubMed] [Google Scholar]
- 104. Plastino J, Olivier S, Sykes C. Actin Filaments align into hollow comets for rapid VASP‐mediated propulsion. Curr Biol. 2004;14:1766–1771. [DOI] [PubMed] [Google Scholar]
- 105. Welch MD, Way M. Arp2/3‐mediated actin‐based motility: A tail of pathogen abuse. Cell Host Microbe. 2013;14:242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vicker MG. F‐actin assembly in Dictyostelium cell locomotion and shape oscillations propagates as a self‐organized reaction‐diffusion wave. FEBS Lett. 2002;510:5–9. [DOI] [PubMed] [Google Scholar]
- 107. Inagaki N, Katsuno H. Actin waves: Origin of cell polarization and migration? Trends Cell Biol. 2017;27:515–526. [DOI] [PubMed] [Google Scholar]
- 108. Allard J, Mogilner A. Traveling waves in actin dynamics and cell motility. Curr Opin Cell Biol. 2013;25:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jasnin M, Ecke M, Baumeister W, Gerisch G. Actin organization in cells responding to a perforated surface, revealed by live imaging and cryo‐electron tomography. Structure. 2016;24:1031–1043. [DOI] [PubMed] [Google Scholar]
- 110. Ecke M, Prassler J, Tanribil P, et al. Formins specify membrane patterns generated by propagating actin waves. Mol Biol Cell. 2020;31:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Forth S, Kapoor TM. The mechanics of microtubule networks in cell division. J Cell Biol. 2017;216:1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wittmann T, Hyman AA, Desai A. The spindle: A dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–E34. [DOI] [PubMed] [Google Scholar]
- 113. Etienne‐Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. [DOI] [PubMed] [Google Scholar]
- 114. Kapitein LC, Hoogenraad CC. Building the neuronal microtubule cytoskeleton. Neuron. 2015;87:492–506. [DOI] [PubMed] [Google Scholar]
- 115. Nogales E, Whittaker M, Milligan RA, Downing KH. High‐resolution model of the microtubule. Cell. 1999;96:79–88. [DOI] [PubMed] [Google Scholar]
- 116. Moores C. Studying microtubules by electron microscopy. Methods Cell Biol. 2008;88:299–317. [DOI] [PubMed] [Google Scholar]
- 117. Garvalov BK, Zuber B, Bouchet‐Marquis C, et al. Luminal particles within cellular microtubules. J Cell Biol. 2006;174:759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Koning RI, Zovko S, Bárcena M, et al. Cryo electron tomography of vitrified fibroblasts: Microtubule plus ends in situ. J Struct Biol. 2008;161:459–468. [DOI] [PubMed] [Google Scholar]
- 119. Höög JL, Schwartz C, Noon AT, et al. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell. 2007;12:349–361. [DOI] [PubMed] [Google Scholar]
- 120. Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat Rev Mol Cell Biol. 2017;18:187–201. [DOI] [PubMed] [Google Scholar]
- 121. Ward JJ, Roque H, Antony C, Nédélec F. Mechanical design principles of a mitotic spindle. Elife. 2014;3:e03398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gan L, Ladinsky MS, Jensen GJ. Organization of the smallest eukaryotic spindle. Curr Biol. 2011;21:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chakraborty S, Mahamid J, Baumeister W. Cryo‐electron tomography reveals nanoscale organization of the cytoskeleton and its relation to microtubule curvature inside cells. Structure. 2020;accepted. [DOI] [PubMed] [Google Scholar]
- 124. Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Odde DJ, Ma L, Briggs AH, DeMarco A, Kirschner MW. Microtubule bending and breaking in living fibroblast cells. J Cell Sci. 1999;112:3283–3288. [DOI] [PubMed] [Google Scholar]
- 126. Brangwynne CP, MacKintosh FC, Kumar S, et al. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol. 2006;173:733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brangwynne CP, Mackintosh FC, Weitz DA. Force fluctuations and polymerization dynamics of intracellular microtubules. Proc Natl Acad Sci U S A. 2007;104:16128–16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pallavicini C, Levi V, Wetzler DE, et al. Lateral motion and bending of microtubules studied with a new single‐filament tracking routine in living cells. Biophys J. 2014;106:2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Landau LD, Lifshitz EM, Sykes JB, Reid WH, Dill EH. Theory of elasticity: Vol. 7 of course of theoretical physics. Phys Today. 1960;13:44–46. [Google Scholar]
- 130. Dogterom M, Koenderink GH. Actin–microtubule crosstalk in cell biology. Nat Rev Mol Cell Biol. 2019;20:38–54. [DOI] [PubMed] [Google Scholar]
- 131. Griffith LM, Pollard TD. Evidence for actin filament‐microtubule interaction mediated by microtubule‐associated proteins. J Cell Biol. 1978;78:958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Pollard TD, Selden SC, Maupin P. Interaction of actin filaments with microtubules. J Cell Biol. 1984;99:33s–37s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Huber F, Boire A, López MP, Koenderink GH. Cytoskeletal crosstalk: When three different personalities team up. Curr Opin Cell Biol. 2015;32:39–47. [DOI] [PubMed] [Google Scholar]
- 134. Janson ME, de Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J Cell Biol. 2003;161:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. López MP, Huber F, Grigoriev I, et al. Actin–microtubule coordination at growing microtubule ends. Nat Commun. 2014;5:4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Henty‐Ridilla JL, Rankova A, Eskin JA, Kenny K, Goode BL. Accelerated actin filament polymerization from microtubule plus ends. Science. 2016;352:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ingber DE. Tensegrity: The architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. [DOI] [PubMed] [Google Scholar]
- 138. Wade RH, Chrétien D, Job D. Characterization of microtubule protofilament numbers. J Mol Biol. 1990;212:775–786. [DOI] [PubMed] [Google Scholar]
- 139. Pierson GB, Burton PR, Himes RH. Alterations in number of protofilaments in microtubules assembled in vitro. J Cell Biol. 1978;76:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tilney LG, Bryan J, Bush DJ, et al. Microtubules: Evidence for 13 protofilaments. J Cell Biol. 1973;59:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. McIntosh JR, Morphew MK, Grissom PM, Gilbert SP, Hoenger A. Lattice structure of cytoplasmic microtubules in a cultured mammalian cell. J Mol Biol. 2009;394:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zabeo D, Heumann JM, Schwartz CL, et al. A lumenal interrupted helix in human sperm tail microtubules. Sci Rep. 2018;8:2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Paul DM, Mantell J, Borucu U, Coombs J, Katherine J, Surridge KJ, Squire JM, Verkade P, Dodding MP. In situ cryo‐electron tomography reveals filamentous actin within the microtubule lumen. bioRxiv. 2019. 10.1101/844043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Grange M, Vasishtan D, Grünewald K. Cellular electron cryo tomography and in situ sub‐volume averaging reveal the context of microtubule‐based processes. J Struct Biol. 2017;197:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Amos LA, Löwe J. How Taxol stabilises microtubule structure. Chem Biol. 1999;6:R65–R69. [DOI] [PubMed] [Google Scholar]
- 146. Afzelius BA. Microtubules in the spermatids of stick insects. J Ultrastruct Res Mol Struct Res. 1988;98:94–102. [DOI] [PubMed] [Google Scholar]
- 147. Bassot JM, Martoja R. Données histologiques et ultrastructurales sur les microtubules cytoplasmiques du canal éjaculateur des insectes orthoptères. Z Zellforsch Mikrosk Anat. 1966;74:145–181. [PubMed] [Google Scholar]
- 148. Behnke O. Incomplete microtubules observed in mammalian blood platelets during microtubule polymerization. J Cell Biol. 1967;34:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Burton PR. Luminal material in microtubules of frog olfactory axons: Structure and distribution. J Cell Biol. 1984;99:520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Peters A, Proskauer CC, Kaiserman‐Abramof IR. The small pyramidal neuron of the rat cerebral cortex: The axon hillock and initial segment. J Cell Biol. 1968;39:604–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Akella JS, Wloga D, Kim J, et al. MEC‐17 is an α‐tubulin acetyltransferase. Nature. 2010;467:218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. L’Hernault SW, Rosenbaum JL. Chlamydomonas α‐tubulin is posttranslationally modified by acetylation on the ε‐amino group of a lysine. Biochemistry. 1985;24:473–478. [DOI] [PubMed] [Google Scholar]
- 153. Portran D, Schaedel L, Xu Z, Théry M, Nachury MV. Tubulin acetylation protects long‐lived microtubules against mechanical ageing. Nat Cell Biol. 2017;19:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. [DOI] [PubMed] [Google Scholar]
- 155. Rice LM. A new look for the growing microtubule end? J Cell Biol. 2018;217:2609–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. McIntosh JR, O’Toole E, Morgan G, et al. Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments. J Cell Biol. 2018;217:2691–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Atherton J, Stouffer M, Francis F, Moores CA. Microtubule architecture in vitro and in cells revealed by cryo‐electron tomography. Acta Crystallogr Sect D Struct Biol. 2018;74:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Burbank KS, Mitchison TJ. Microtubule dynamic instability. Curr Biol. 2006;16:R516–R517. [DOI] [PubMed] [Google Scholar]
- 159. Chrétien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: Two‐dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Müller‐Reichert T, Chrétien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: Information from a slowly hydrolyzable analogue of GTP, guanylyl (α,β)methylenediphosphonate. Proc Natl Acad Sci U S A. 1998;95:3661–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol. 2006;16:1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. McIntosh JR, Grishchuk EL, Morphew MK, et al. Fibrils connect microtubule tips with kinetochores: A mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JAG. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: Two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. [DOI] [PubMed] [Google Scholar]
- 165. Höög JL, Huisman SM, Sebö‐Lemke Z, et al. Electron tomography reveals a flared morphology on growing microtubule ends. J Cell Sci. 2011;124:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Cyrklaff M, Kudryashev M, Leis A, et al. Cryoelectron tomography reveals periodic material at the inner side of subpellicular microtubules in apicomplexan parasites. J Exp Med. 2007;204:1281–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Cross RA. Microtubule lattice plasticity. Curr Opin Cell Biol. 2019;56:88–93. [DOI] [PubMed] [Google Scholar]
- 168. Chrétien D, Metoz F, Verde F, Karsenti E, Wade RH. Lattice defects in microtubules: Protofilament numbers vary within individual microtubules. J Cell Biol. 1992;117:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Memet E, Hilitski F, Morris MA, Schwenger WJ, Dogic Z, Mahadevan L. Microtubules soften due to cross‐sectional flattening. Elife. 2018;7:e34695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Vemu A, Szczesna E, Zehr EA, et al. Severing enzymes amplify microtubule arrays through lattice GTP‐tubulin incorporation. Science. 2018;361:eaau1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Schaedel L, John K, Gaillard J, Nachury MV, Blanchoin L, Théry M. Microtubules self‐repair in response to mechanical stress. Nat Mater. 2015;14:1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Liang WH, Li Q, Rifat Faysal KM, King SJ, Gopinathan A, Xu J. Microtubule defects influence kinesin‐based transport in vitro. Biophys J. 2016;110:2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Aumeier C, Schaedel L, Gaillard J, John K, Blanchoin L, Théry M. Self‐repair promotes microtubule rescue. Nat Cell Biol. 2016;18:1054–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Herrmann H, Bär H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: From cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. [DOI] [PubMed] [Google Scholar]
- 175. Parry DA, Steinert PM. Intermediate filaments: Molecular architecture, assembly, dynamics and polymorphism. Q Rev Biophys. 1999;32:99–187. [DOI] [PubMed] [Google Scholar]
- 176. Kirmse R, Bouchet‐Marquis C, Page C, Hoenger A. Three‐dimensional cryo‐electron microscopy on intermediate filaments. Methods Cell Biol. 2010;96:565–589. [DOI] [PubMed] [Google Scholar]
- 177. Herrmann H, Aebi U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem. 2004;73:749–789. [DOI] [PubMed] [Google Scholar]
- 178. Masich S, Östberg T, Norlén L, Shupliakov O, Daneholt B. A procedure to deposit fiducial markers on vitreous cryo‐sections for cellular tomography. J Struct Biol. 2006;156:461–468. [DOI] [PubMed] [Google Scholar]
- 179. Norlén L, Al‐Amoudi A. Stratum corneum keratin structure, function, and formation: The cubic rod‐packing and membrane templating model. J Invest Dermatol. 2004;123:715–732. [DOI] [PubMed] [Google Scholar]
- 180. Turgay Y, Eibauer M, Goldman AE, et al. The molecular architecture of lamins in somatic cells. Nature. 2017;543:261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–613. [DOI] [PubMed] [Google Scholar]
- 182. Brodland GW, Gordon R. Intermediate filaments may prevent buckling of compressively loaded microtubules. J Biomech Eng. 1990;112:319–321. [DOI] [PubMed] [Google Scholar]


