Abstract
The HECT‐type ubiquitin ligase E6AP (UBE3A) is critically involved in several neurodevelopmental disorders and human papilloma virus‐induced cervical tumorigenesis; the structural mechanisms underlying the activity of this crucial ligase, however, are incompletely understood. Here, we report a crystal structure of the C‐terminal lobe (“C‐lobe”) of the catalytic domain of E6AP that reveals two molecules in a domain‐swapped, dimeric arrangement. Interestingly, the molecular hinge that enables this structural reorganization with respect to the monomeric fold coincides with the active‐site region. While such dimerization is unlikely to occur in the context of full‐length E6AP, we noticed a similar domain swap in a crystal structure of the isolated C‐lobe of another HECT‐type ubiquitin ligase, HERC6. This may point to conformational strain in the active‐site region of HECT‐type ligases with possible implications for catalysis.
Significance Statement
The HECT‐type ubiquitin ligase E6AP has key roles in human papilloma virus‐induced cervical tumorigenesis and certain neurodevelopmental disorders. Here, we present a crystal structure of the C‐terminal, catalytic lobe of E6AP, providing basic insight into the conformational properties of this functionally critical region of HECT‐type ligases.
Keywords: dimerization, domain swapping, E3 enzyme, UBE3A, X‐ray crystallography
1. INTRODUCTION
The ubiquitin system regulates protein functions at all levels, thereby orchestrating countless physiological and disease‐associated cellular pathways. Among the components of the ubiquitination machinery, ubiquitin ligases (E3 enzymes, E3s) are pivotal in determining the specificity of substrate selection and modification. E3s of the HECT (homologous to E6AP C‐terminus)‐subfamily share a C‐terminal catalytic domain that is comprised of two lobes—a ~30 kDa N‐lobe and a ~14 kDa C‐lobe—tethered by a flexible linker.1 Both lobes cooperate with each other in mediating sequential macromolecular interactions during ubiquitin transfer; in addition, the C‐lobe contains a catalytic cysteine that forms a thioester with the C‐terminus of ubiquitin in an intermediate step of catalysis. The specific recognition of substrates and regulatory factors by HECT‐type E3s is mediated by the diverse regions N‐terminal to the catalytic domain that are poorly characterized at a structural level.2
The founding member of the HECT E3 family, E6AP (UBE3A), has key roles in human diseases: its activity is hijacked by the E6 protein from high‐risk human papilloma viruses to promote the proteasomal degradation of the tumor suppressor p53, thereby driving cervical tumorigenesis; genetic up‐regulation of E6AP has been linked to autism spectrum disorders; and the deletion or down‐regulation of this ligase in the brain causes Angelman's syndrome.3 While the interactions of E6AP with ubiquitin have been analyzed and structural knowledge of its interplay with certain substrates is emerging,4, 5, 6, 7, 8 no specific inhibitors targeting this crucial ligase are available.9
Here, we report a crystal structure of the isolated C‐lobe of E6AP, revealing a three‐dimensionally domain‐swapped dimer. While distinct in structural detail, an overall similar crystallographic dimer is observed for the C‐lobe of HERC6 (PDB ID: http://firstglance.jmol.org/fg.htm?mol=5W87), suggesting that the conformational rearrangements with respect to the native, monomeric fold reflect dynamic properties of HECT‐type E3s beyond E6AP.
2. RESULTS AND DISCUSSION
We determined a crystal structure of the C‐lobe of E6AP at 1.3 Å resolution (Table 1). The structure reveals a dimeric arrangement, in which the C‐terminal α‐helix (H14) and adjacent β‐strand (S10) of two symmetry‐related molecules have undergone three‐dimensional domain swapping (Fig. 1a, top panel). This term describes a phenomenon where identical proteins exchange part of their structure to give rise to an oligomer, of which individual subunits have a similar fold as an isolated monomer.10 Each composite subunit of the crystallographic C‐lobe dimer thus recapitulates the canonical, globular α/β‐fold seen in the context of the HECT domain of E6AP (RMSD of 0.48 Å in 104 equivalent Cα‐positions with respect to the C‐lobe structure extracted from PDB ID: http://firstglance.jmol.org/fg.htm?mol=1C4Z, chain A)1 (Fig. 1b). Interestingly, the hinge region that mediates the domain swap coincides with the active‐site region (Thr819, Cys820, Phe821, and Asn822) and adopts two conformations (Fig. 1a, bottom panel). As a consequence, the catalytic cysteine (Cys820) residues of the two subunits are either in immediate proximity or at a sulfur–sulfur distance of 4.7 Å, which we interpret as a mixture of disulfide‐bonded and reduced states. We speculate that these alternative oxidation states originate from radiation‐induced, partial cleavage of an intermolecular disulfide bond formed during the crystallization process.
Table 1.
X‐ray crystallographic data collection and refinement statistics
| Data collection | |
| Wavelength | 0.9184 |
| Space group | I 4 2 2 |
| Unit cell parameters | |
| a, b, c (Å) | 72.28, 72.28, 98.97 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 30.03–1.30 (1.35–1.30) |
| Total reflections | 65,057 (6,414) |
| Unique reflections | 32,542 (3,211) |
| R pim | 0.01279 (0.5023) |
| CC1/2 | 1 (0.676) |
| Mean I/σ(I) | 26.56 (1.49) |
| Completeness (%) | 99.92 (99.63) |
| Multiplicity | 2.0 (2.0) |
| Wilson B‐factor | 17.13 |
| Refinement | |
| R work | 17.8 (28.8) |
| R free | 20.7 (29.8) |
| No. of non‐hydrogen atoms | 1,051 |
| Protein | 954 |
| Ligand | 9 |
| Water | 88 |
| Average B‐factors | 23.57 |
| Protein | 22.79 |
| Ligands | 30.43 |
| Water | 31.35 |
| RMS deviations from ideality | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.9 |
| Ramachandran statistics | |
| Favored (%) | 97.09 |
| Disallowed (%) | 0.0 |
| MolProbity clash score | 3.09 |
| MolProbity overall score | 1.18 |
Note: Values in parentheses correspond to the highest‐resolution shell. Data were collected from a single crystal.
Figure 1.
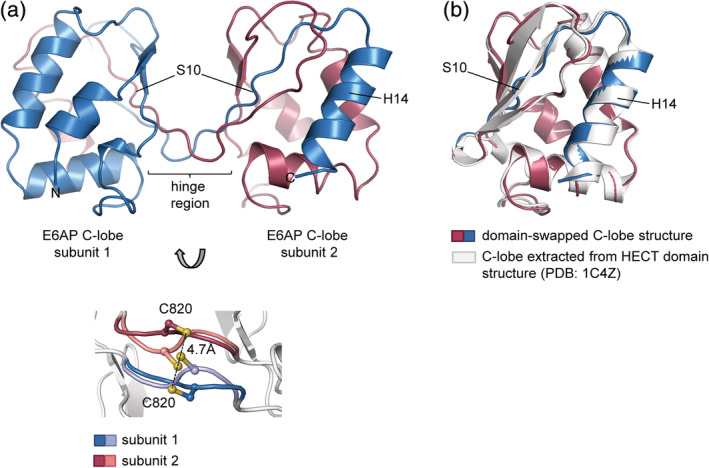
Crystal structure of a domain‐swapped E6AP C‐lobe dimer. (a) Crystal structure of the E6AP C‐lobe dimer determined in this study; the swapped secondary structure elements are labeled; only one conformation of the active‐site region is displayed (top). Detail of the active‐site region showing the two alternate conformations of residues 819–822 (colored) and the side chain of the catalytic Cys820 (bottom); the sulfur–sulfur distance of the non‐disulfide bonded state is indicated. (b) Superposition of one domain‐swapped unit extracted from (a) with the crystal structure of the (monomeric) C‐lobe, extracted from a structure of the E6AP HECT domain (PDB ID: http://firstglance.jmol.org/fg.htm?mol=1C4Z 1; Cα‐RMSD: 0.48 Å)
We and others have previously reported that the C‐lobe and the HECT domain of E6AP are monomeric in solution,1, 4 consistent with cell‐based studies on the full‐length ligase.11, 12 Yet, evidence for a trimeric state in vitro and oligomerization of E6AP in the cell has also been presented.6, 13, 14, 15, 16 Either way, we posit that the observed domain swapping specifically occurs in the context of the isolated C‐lobe and is unlikely to happen in the presence of the N‐lobe. In line with this notion, the C‐lobe dimer is incompatible with the position of the N‐lobe in several alternative conformations in which HECT domains have been crystallized, including an “L”‐shaped state (PDB ID: http://firstglance.jmol.org/fg.htm?mol=1C4Z 1) and a “T”‐shaped complex with an E2 (UBCH7), as required for the trans‐thioesterification reaction (analogous to PDB ID: http://firstglance.jmol.org/fg.htm?mol=3JVZ 17).
Interestingly, a similar domain‐swapped, crystallographic dimer is formed by the isolated C‐lobe of another HECT‐type E3 HERC6 (Fig. 2b), which may present a bona fide ubiquitin‐directed ligase in the human system (while its murine orthologue is specific for the ubiquitin‐like modifier ISG1519, 20). As for E6AP, the hinge region in HERC6 comprises the active site, yet the swapped region is slightly offset and the subunits are tilted by ~40° with respect to the E6AP C‐lobe dimer, thus positioning the catalytic cysteine residues of the two subunits at a distance of 11 Å (Fig. 2a,b). This comparison implies that domain swapping can occur in the isolated C‐lobes of HECT‐type E3s beyond E6AP and independently of intermolecular disulfide formation.
Figure 2.
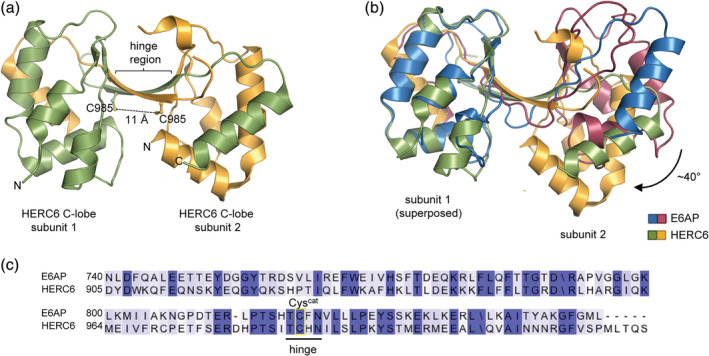
Comparison of the crystallographic, domain‐swapped dimers of the C‐lobe of E6AP and HERC6. (a) Crystal structure of a domain‐swapped dimer of the C‐lobe of HERC6 (PDB ID: http://firstglance.jmol.org/fg.htm?mol=5W87). The side chain of the catalytic Cys985 is shown for each molecule, along with the sulfur–sulfur distance (b) Superposition of the domain‐swapped, dimeric structures of the C‐lobe of E6AP and HERC6. (c) Sequence alignment for the C‐lobes of E6AP and HERC6 (rendered with Jalview18 and colored according to conservation), highlighting the hinge region and the catalytic cysteine (Cyscat)
While its biological relevance needs to be evaluated on a case‐by‐case basis, domain swapping typically reports on conformational tension and can thus unveil energetic properties of a monomeric structure that are hard to prove otherwise.21 We assume that the domain‐swapped dimers observed for the C‐lobes of HECT‐type E3s represent an artifact of extracting this region from the native protein context. At the same time, the structural reorganization may indicate inherent conformational strain in the hinge region within the monomeric fold. In line with this notion, the hinge region is the shortest among the loops connecting individual β‐strands of the C‐lobe and may thus resemble a loaded spring that releases conformational tension upon swapping.21 In the context of the full‐length protein—where we expect domain swapping is inhibited—the conformational tension in the active‐site region may affect the energetics and dynamic behavior of the entire C‐lobe. Further studies will be required to explore this hypothesis and interrogate whether these “hidden” conformational properties affect catalysis.
3. MATERIALS AND METHODS
3.1. Protein preparation and crystallization
The nucleotide sequence encoding the C‐lobe (residues 741‐852; numbering according to isoform 1) of E6AP was cloned into a pET‐28a vector (Merck), modified to encode an N‐terminal 3C protease‐cleavable His6‐tag. The protein was recombinantly expressed in E. coli BL21 (DE3) and purified as previously described.4 After removal of the His6‐tag, the protein was subjected to size‐exclusion chromatography (Superdex HiLoad 26/600 75 pg column, GE Healthcare, Uppsala, Sweden) in 75 mM Tris, pH 7.5, 200 mM NaCl. The purified protein crystallized at concentrations ≥30 mg/ml and 20°C in sitting drops containing 0.1 M calcium chloride, 0.1 M sodium acetate pH 4.6, and 30% PEG400; the particular crystal used for structure determination was obtained at 60 mg/ml protein concentration; no additional cryo‐protection was required.
3.2. X‐ray crystallographic data collection and structure determination
Diffraction data were collected at 100 K, beamline BL14.1 at the BESSY II, Helmholtz‐Zentrum Berlin and processed with XDS.22 Molecular replacement was performed with Phaser,23 as implemented in the collaborative computational project no. 4 (ccp4) suite (RRID: SCR_007255),24 using a structure of the E6AP C‐lobe as a search model (extracted from PDB ID: http://firstglance.jmol.org/fg.htm?mol=1C4Z 1); refinement with Phenix (RRID: SCR_014224)25 and REFMAC5 (RRID: SCR_014225)26 with individual B‐factors and TLS (translation/libration/screw); and model building with Coot (RRID: SCR_014222).27
4. ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the PDB under accession code http://firstglance.jmol.org/fg.htm?mol=6TGK.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
ACKNOWLEDGMENTS
L.K.R. held a Kékulé doctoral fellowship (694152) from the Fonds der Chemischen Industrie and was a member of the GSLS, Würzburg. A.K.L.L. is affiliated with the GRK2243 (“Understanding ubiquitylation: from molecular mechanisms to disease”; DFG) and the GSLS. D.E.S. is supported by the National Institutes of Health (R15GM126432) and start‐up funds from Clark University. S.L. is supported by the Emmy Noether Program (DFG; LO 2003/1‐1) and the EMBO Young Investigator Program.
Ries LK, Liess AKL, Feiler CG, Spratt DE, Lowe ED, Lorenz S. Crystal structure of the catalytic C‐lobe of the HECT‐type ubiquitin ligase E6AP. Protein Science. 2020;29:1550–1554. 10.1002/pro.3832
Christian G. Feiler, present address: Macromolecular Crystallography, BESSY, Helmholtz‐Zentrum Berlin für Materialien und Energie, 12489 Berlin, Germany.
Funding information Deutsche Forschungsgemeinschaft, Grant/Award Number: LO 2003/1‐1; Fond der chemischen Industrie, Grant/Award Number: 694152; National Institutes of Health, Grant/Award Number: R15GM126432; EMBO Young Investigator Program
REFERENCES
- 1. Huang L, Kinnucan E, Wang G, et al. Structure of an E6AP‐UbcH7 complex: Insights into ubiquitination by the E2‐E3 enzyme cascade. Science. 1999;286:1321–1326. [DOI] [PubMed] [Google Scholar]
- 2. Lorenz S. Structural mechanisms of HECT‐type ubiquitin ligases. Biol Chem. 2017;399:127–145. [DOI] [PubMed] [Google Scholar]
- 3. Scheffner M, Kumar S. Mammalian HECT ubiquitin‐protein ligases: Biological and pathophysiological aspects. Biochim Biophys Acta. 2014;1843:61–74. [DOI] [PubMed] [Google Scholar]
- 4. Ries LK, Sander B, Deol KK, Letzelter M‐A, Strieter ER, Lorenz S. Analysis of ubiquitin recognition by the HECT ligase E6AP provides insight into its linkage specificity. J Biol Chem. 2019;294:6113–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mortensen F, Schneider D, Barbic T, et al. Role of ubiquitin and the HPV E6 oncoprotein in E6AP‐mediated ubiquitination. Proc Natl Acad Sci U S A. 2015;112:9872–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sailer C, Offensperger F, Julier A, et al. Structural dynamics of the E6AP/UBE3A‐E6‐p53 enzyme‐substrate complex. Nat Commun. 2018;9:4441–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez‐Zapien D, Ruiz FX, Poirson J, et al. Structure of the E6/E6AP/p53 complex required for HPV‐mediated degradation of p53. Nature. 2016;529:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen D, Gehringer M, Lorenz S. Developing small‐molecule inhibitors of HECT‐type ubiquitin ligases for therapeutic applications: Challenges and opportunities. ChemBioChem. 2018;19:2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennett MJ, Choe S, Eisenberg D. Domain swapping: Entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994;91:3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of P53 with the E6 oncoprotein of human papillomavirus type‐16 or type‐18. EMBO J. 1991;10:4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinez‐Noel G, Galligan JT, Sowa ME, et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32:3095–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nuber U, Schwarz SE, Scheffner M. The ubiquitin‐protein ligase E6‐associated protein (E6‐AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. [DOI] [PubMed] [Google Scholar]
- 14. Chan A‐L, Grossman T, Zuckerman V, et al. c‐Abl phosphorylates E6AP and regulates its E3 ubiquitin ligase activity. Biochemistry. 2013;52:3119–3129. [DOI] [PubMed] [Google Scholar]
- 15. Yi JJ, Berrios J, Newbern JM, et al. An autism‐linked mutation disables phosphorylation control of UBE3A. Cell. 2015;162:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ronchi VP, Klein JM, Edwards DJ, Haas AL. The active form of E6‐associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J Biol Chem. 2014;289:1033–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamadurai HB, Souphron J, Scott DC, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin‐HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2‐a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oudshoorn D, van Boheemen S, Sánchez‐Aparicio MT, Rajsbaum R, García‐Sastre A, Versteeg GA. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One. 2011;7:e29870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ketscher L, Basters A, Prinz M, Knobeloch K‐P. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem Biophys Res Commun. 2012;417:135–140. [DOI] [PubMed] [Google Scholar]
- 21. Rousseau F, Schymkowitz J, Itzhaki LS. Implications of 3D domain swapping for protein folding, misfolding and function. Adv Exp Med Biol. 2012;747:137–152. [DOI] [PubMed] [Google Scholar]
- 22. Kabsch W. XDS. Acta Crystallogr. 2010;D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: A comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov GN, Skubák P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 2011;D67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]


