Abstract
Both natively folded and intrinsically disordered proteins (IDPs) destined for the nucleus need to transport through the nuclear pore complexes (NPCs) in eukaryotic cells. NPCs allow for passive diffusion of small folded proteins while barricading large ones, unless they are facilitated by nuclear transport receptors. However, whether nucleocytoplasmic transport of IDPs would follow these rules remains unknown. By using a high‐speed super‐resolution fluorescence microscopy, we have measured transport kinetics and 3D spatial locations of transport routes through native NPCs for various IDPs. Our data revealed that the rules executed for folded proteins are not well followed by the IDPs. Instead, both large and small IDPs can passively diffuse through the NPCs. Furthermore, their diffusion efficiencies and routes are differentiated by their content ratio of charged (Ch) and hydrophobic (Hy) amino acids. A Ch/Hy‐ratio mechanism was finally suggested for nucleocytoplasmic transport of IDPs.
Keywords: intrinsically disordered proteins (IDPs), nuclear pore complex (NPC), nucleocytoplasmic transport, super‐resolution
1. INTRODUCTION
Bioinformatics studies indicate that more than half of all eukaryotic proteins contain regions of disorder,1, 2 in which approximately one third of them largely lack secondary structure.3 Many of these intrinsically disordered proteins (IDPs) are known to localize in the sub‐nuclear compartments, such as cleavage body, nuclear body, chromatin, Cajal body (CB), and nuclear pore complexes (NPCs).4 Studies have shown that IDPs play important roles in signaling, genome fidelity, and regulatory functions in these compartments.5, 6, 7 For instance, Coilin, which is a highly disordered and essential protein component of CB, functions in adhering the CB to the nucleolus and conducting post‐transcriptional modification of RNA within mammalian cells.8 Similarly, calcium‐responsive transactivator (CREST) localizes to the nuclear bodies and plays a role in dendritic development and other cellular processes in mammalian cells by interacting with histones and DNA remodeling proteins.9 Another important nuclear IDP, SLD2, associates with chromatins and participates in DNA replication initiation in Saccharomyces cerevisiae.10 Following the central dogma of transcription and translation, nuclear bound IDPs must translocate from the cytoplasm into the nucleus by crossing the nuclear envelope (NE).11, 12
Acting as nuclear gatekeepers, thousands of NPCs are embedded in the NE and regulate the trafficking of folded and unfolded proteins between the nucleoplasm and cytoplasm. The NPC is a large assembly of approximately 30 different proteins, known as nucleoporins (Nups), with each present in multiples of eight copies per pore.13, 14, 15 One third of these Nups are IDPs with domains rich in phenylalanine‐glycine (FG) repeats.16 The FG Nups form the critical permeability and selectivity barrier (FG‐Nup barrier) in the NPC to mediate transport of macromolecules approaching the NPC from either cytoplasm or nucleus.17, 18
In past years, intensive studies on the permeability and selectivity of NPC revealed a size‐dependent criterion for transportation of folded proteins through the NPC. If folded protein cargos are larger than ~40 kDa, they can be barricaded by the FG‐Nup barrier13, 14, 15 unless they carry either nuclear localization sequence (NLS) for nuclear import or nuclear export signal (NES) for nuclear export.19 Nuclear transport receptors, also known as karyopherins, recognize the NLS or NES and facilitate folded protein cargos to transport through the NPCs by directly interacting with the FG‐Nup barrier. The directionality of facilitated transport through the NPC is regulated by a gradient of Ran‐GDP/Ran‐GTP across the NE.19, 20, 21 In contrast, if folded proteins lack the above signals and are smaller than ~40 kDa, they are capable of passively diffusing through the NPC independent of energy consumption and transport receptors.22 However, it is still unknown if a size‐dependent rule of exclusion, observed in folded protein transport, is applicable for IDP transport through the NPC. By employing a high‐speed super‐resolution fluorescence microscopy, termed single‐point edge‐excitation sub‐diffraction (SPEED) microscopy,23, 24 we measured transport kinetics and 3D spatial locations of transport routes for various IDP candidates through the NPCs with a spatiotemporal resolution of 400 μs and <10 nm. We found that the size‐exclusion rule for folded proteins is not followed by the IDPs as they move through the NPC. Instead, both large and small IDPs can diffuse through the NPC neither consuming energy nor utilizing transport receptors. Specifically, the IDP's transport efficiencies and spatial diffusion routes through the NPCs strongly depend on their content ratio between charged and hydrophobic amino acids (AAs).
2. RESULTS
2.1. IDPs display different characteristics from folded proteins in nucleocytoplasmic transport
To test whether the size‐dependent rule of nucleocytoplasmic transport for folded proteins applies to IDPs, we tracked single IDPs with different sizes as they transport through the native NPCs of HeLa cells. In our tests, both large (>40 kDa) and small (<40 kDa) IDPs with and without FG domains contained in their disordered regions were employed as candidates, including Coilin (63 kDa), Nsp1 (1–603, 62 kDa), SLD2 (52 kDa), Nup159 (441–881, 45 kDa), Nsp1 (173–603, 45 kDa), CREST (43 kDa), Nup42 (1–372, 37 kDa), Nsp1 (1–172, 17 kDa), and Nup116 (348–458, 11 kDa) (Table 1). These IDP candidates possess a high‐level of disorder and were isolated with a high purity (Figure S1, Section 4). Prior to imaging, these IDPs were chemically labeled by Alexa Fluor 647 succinimidyl ester or maleimide dyes. Native NPCs embedded into the NE were labeled through genetically tagging a green fluorescent protein (GFP) to a scaffold Nup POM121 (Figure 1a, Section 4). Next, SPEED microscopy was utilized to illuminate a single GFP‐labeled NPC on the NE and track individual Alexa‐Fluor‐647 labeled IDPs as they transport through the illuminated NPC in digitonin‐permeabilized HeLa cells (Figures 1b and S2, Supplementary Notes).
Table 1.
Characteristic list of IDPs sorted by Ch/Hy ratio
| Candidate | MW (kDa) | Net charge | Charged AA (%) | Hydrophobic AA (%) | Spatial transport route | Ch/Hy ratio | |
|---|---|---|---|---|---|---|---|
| Periphery (%) | Central channel (%) | ||||||
| SLD2 | 52 | 4.2 ± 1.5 | 35 | 32 | 28 ± 2 | 72 ± 2 | 1.08 |
| Nsp1 (173‐603) | 45 | ‐8.5 ± 4.5 | 30 | 32 | 20 ± 3 | 80 ± 3 | 0.94 |
| Coilin | 63 | 12.2 ± 1.5 | 28 | 34 | 30 ± 2 | 70 ± 2 | 0.80 |
| Nup116 (348 – 458) charged mutation | 11 | ‐5.5 ± 4.5 | 31 | 41 | 17 ± 3 | 83 ± 3 | 0.74 |
| Nsp1 (1‐603) | 62 | ‐5.5 + 4.5 | 22 | 31 | 28 ± 2 | 72 ± 2 | 0.73 |
| Nup159 (441‐881) | 45 | ‐28.3 ± 4.5 | 17 | 34 | 17 ± 3 | 83 ± 3 | 0.51 |
| CREST | 43 | ‐9.6 ± 1.5 | 8 | 27 | 91 ± 3 | 9 ± 3 | 0.30 |
| Nup42 (1‐372) | 37 | ‐3.5 ± 4.5 | 4 | 31 | 82 ± 3 | 18 ± 3 | 0.11 |
| WT Nup116 (348 – 458) | 11 | ‐10.5 ± 4.5 | 3 | 28 | 92 ± 3 | 8 ± 3 | 0.09 |
| Nsp1 (1–172) | 17 | ‐10.5 ± 4.5 | 26 | 72± 2 | 28 ± 2 | 0.07 | |
Note: Disorder level score was calculated using IUPRED.58 Approximately 500‐1,200 spatial locations were collected in order to obtain reliable 3D spatial transport route for each candidate in our measurements based on computation simulation (Section 4).
Abbreviations: Ch/Hy, charged/hydrophobic; CREST, calcium‐responsive transactivator; IDP, intrinsically disordered protein; IUPRED, intrinsically unstructured proteins prediction.
Figure 1.
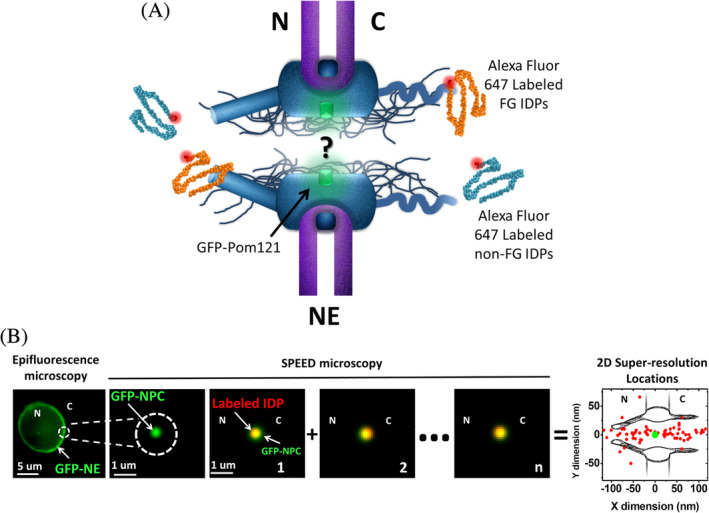
Nuclear transport of IDPs through the NPC illuminated by SPEED microscopy. (a) The fundamental mechanism for nuclear transport of IDPs through the NPCs remains unknown. Native NPCs embedded into the NE were labeled through genetically tagging green fluorescent protein (GFP) to a scaffold Nup POM121, in which the fluorescence of GFP‐POM121was utilized to localize the NPC. Alexa Fluor 647 dye molecules were used to label FG‐ and non‐FG‐IDPs. N, nucleus; C, cytoplasm. (b) Wide‐field epifluorescence image shown a green fluorescent ring of NE in a permeabilized HeLa cell expressing GFP‐POM121. SPEED microscopy illuminated only a single NPC on the NE. Labeled IDPs were tracked as they diffused through the NPC and their dwelling positions (red dots) were localized and then superimposed to obtain a 2D super‐resolution distribution around the NPC centroid (green dot). N, nucleus; C, cytoplasm. IDP, intrinsically disordered protein; NE, nuclear envelope; NPC, nuclear pore complex; SPEED, single‐point edge‐excitation sub‐diffraction
Digitonin‐permeabilized cells containing intact nuclei and porous cytoplasm were chosen for re‐building a controllable and functional nucleocytoplasmic transport system for folded proteins.23, 25, 26 Before applying the permeabilized cell system for our candidates, we measured the NPC's permeability and selectivity for various dextrans and folded proteins ranging in size from 10 to 70 kDa, and tested if the NPC's FG‐Nup barrier remains functional under our permeabilization conditions. As shown in Figures S2 and S3, we found that the NPCs in permeabilized cells are permeable to a 10‐kDa dextran and a 27‐kDa GFP, while less permeable to a 40‐kDa dextran and almost impermeable for 54‐kDa 2xGFP and 70‐kDa dextran. Meanwhile, the nuclear import efficiencies significantly decreased from ~50% down to ~4% as the molecular weights increased from 10 to 70 kDa, which further confirmed the NPC's functional permeability in permeabilized cells (Table 2). In this case, nuclear import efficiency is defined as the number of successful tracking events (the particle originates in the cytoplasm, interacts with the NPC, and then successfully crosses to reach the nucleoplasm) divided by the total number of tracking events (also including those abortive events where the particle failed to reach the nucleoplasm). In addition to testing passively diffusing particles, the NPC's FG‐Nup barrier was also determined to be functional for facilitated translocation after permeablization. In contrast to the failure of 2xGFP to penetrate the NPCs by itself (Figure S3C), 2xGFP equipped with a nuclear localization signal (NLS) was successfully carried through the pore by Importin α/Importin β1 when these nuclear transport receptors were introduced to the permeabilized cell (Figure S2C,D).23, 24 Overall, the above tests reaffirmed that the NPCs maintain their functional permeable selectivity barriers under our permeabilization conditions. Moreover, to the best of our knowledge, no specific transport receptors are suggested for these IDP candidates we tested.
Table 2.
Nuclear transport kinetics of protein candidates
| Candidate | IDPs | Dextrans and folded proteins | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SLD2 | CREST | Charged Nup116 (348–458) | WT Nup116 (348–458) | Dextran | Dextran | Dextran | GFP | 2xGFP | |
| MW (kDa) | 52 | 43 | 11 | 11 | 10 | 40 | 70 | 27 | 54 |
| Ch/Hy ratio | 1.08 | 0.30 | 0.74 | 0.09 | ‐ | ‐ | 0.12 | 0.12 | |
| Major spatial transport route | Central | Peripheral | Central | Peripheral | Central | Central | ‐ | Central | ‐ |
| Dwell time (ms) | 1.4 ± 0.5 | 3.0 ± 1.0 | 1.4 ± 0.8 | 2.7 ± 1.2 | 1.2 ± 0.2 | 2.0 ± 0.4 | ‐ | 2.7 ± 1.2 | ‐ |
| Import efficiency (%) | 36 ± 4 | 56 ± 4 | 39 ± 4 | 51 ± 5 | 51 ±4 | 26 ± 4 | 4 ± 2 | 47 ± 5 | 6 ± 6 |
| Export efficiency (%) | 50 ± 4 | 49 ±4 | 39 ± 4 | 50 ± 5 | 51 ±4 | 26 ± 4 | ‐ | 47 ± 5 | ‐ |
Note: In our measurements, more than 200 transport events were collected from 10 different NPCs of 10 cells to determine the transport kinetics for each candidate.
Abbreviations: Ch/Hy, charged/hydrophobic; CREST, calcium‐responsive transactivator; GFP, green fluorescent protein; IDP, intrinsically disordered protein; NPC, nuclear pore complex.
With the above control experiments, we started to determine the transport kinetics and localize the dwell positions of individual IDPs traversing the NPC by adding in vitro purified IDPs (~0.1 nM) into the permeabilized cell system. The localized positions were then superimposed to form a 2D super‐resolution spatial distribution of the IDP in the NPC (Figures 1b and 2). Following an established transform (or back‐projection) algorithm, this 2D distribution can be further converted into their 3D transport routes (computed probability density maps) in the NPC (Figures S4–S6, Supplementary Notes, README).23, 27 First, because of large disordered regions and behaving as highly dynamic IDPs,28 isolated FG segments of FG Nups can be used as IDP candidates to explore the size‐exclusion rule through the NPC (Figure S1, Table S1). To avoid any specific self‐association and incorporation into native human NPCs,20 we employed isolated FG segments of yeast FG‐Nups as candidates (Figure 2). By tracking nuclear transport of these FG‐containing IDPs (FG‐IDPs) through 10 different NPCs of 10 permeabilized cells by SPEED microscopy with a spatiotemporal resolution of 400 μs and <10 nm, we found that not only small (37‐kDa Nup42 and 11‐kDa Nup116) but also large (62‐kDa Nsp1 and 45‐kDa Nup159) FG segments can passively diffuse through the native NPCs without the presence of any transport receptors (Figure 2a–d). This is in direct contrast to 2x‐GFP and the larger dextrans. Moreover, these large FG‐IDPs completed their passive diffusions through the NPCs with similar diffusion times (1–4 ms) and transport efficiencies (~50%) as those smaller ones, further demonstrating that the molecular weight is unlikely a dominant factor in determining the transport mode and kinetics for IDPs (Tables 2 and S1). Moreover, the 3D nuclear transport routes of these large FG‐IDPs further suggested that they mainly diffuse through the central axial channel of the NPC (the spatial probability of IDPs staying in the channel >70%), which is previously believed to be reserved for the passive diffusion of small folded proteins (Figures 2a,b and S2B, Tables 1 and S1).29, 30, 31 In contrast, the two smaller FG‐IDPs, Nup42 (1–372) and Nup116 (348–458), primarily diffused through the peripheral regions of the pore (the spatial probability of IDPs staying in the periphery >80%). Remarkably, the transport routes for these small IDPs in absence of transport receptors are similar to those obtained for facilitated translocation of large folded protein cargos (Figures 2c,d and S2C,D, Table 1). It appears that the above distinct transport routes obtained for small and large IDPs are exactly opposite to the previous spatial transport routes identified for small and large folded cargo proteins.23, 29, 30, 31, 32
Figure 2.

Two‐dimensional super‐resolution spatial distributions and 3D transport routes of various IDPs with different molecular weights. The molecular weight (i), disordered level (ii), 2D super‐resolution spatial distribution superposed with the 2D physical schematic of NPC in the (X, Y) coordinate plane (iii), and 3D spatial probability density map superposed with the 3D physical schematic of NPC (a cut‐away view) in the (X, R, Ɵ) cylindrical coordinate system for each IDP candidate were provided. In addition, a histogram of spatial probability density for the radial dimension was shown in iv to highlight either a single peak representing a central axial transport route or two peaks representing a peripheral transport route. (a) Nsp1 (1–603) refers an FG segment (AA 1–603) of yeast Nsp1 with a molecular weight of 60 kDa (i). With a high disordered level (ii), Nsp 1 (1–603) was found to mainly diffuse through the NPC's central axial channel, based on its 2D super‐resolution spatial distribution (black dots in iii) and 3D spatial probability density map (red clouds in NPC's axial and radial views shown in iv). N, nucleus; C, cytoplasm. (b–g) Defined similarly as in A, the major information and results for Nup159 (441–881), Nup42 (1–372), Nup116 (348–458), SLD2 (1–453), Coilin (1–576), and CREST (1–402) were presented. IDP, intrinsically disordered protein; NE, nuclear envelope; NPC, nuclear pore complex
To test the possibility that the above characteristics may not be unique to only FG‐IDPs, we additionally tested three non‐FG nuclear IDPs: SLD2, Coilin, and CREST (Video S1). Following the same experimental procedure as the FG‐IDPs, we have determined the spatial pathways for these non‐FG IDPs after collecting data from 10 different NPCs of 10 permeabilized cells, as shown in Figure 2e–g. All tested non‐FG IDPs similarly appeared to passively diffuse through the NPC as complete import and export events were recorded without the presence of transport receptors. However, unlike the two large FG‐IDPs that passively diffuse through the NPC's central axial channel, these large non‐FG IDPs (>40 kDa) took distinct and separate spatial pathways through the NPC. In detail, SLD2 and Coilin, agreeing with large FG‐IDPs, mainly diffused through the central axial channel, but CREST mainly moves in the periphery around the central channel. Again it seems that the IDP's molecule weight is not a determining factor for IDP's transport modes through the NPCs, in addition to its incapability in deciding the nuclear transport routes for IDPs.
Additionally, we further estimated the hydrodynamic radii of these IDP candidates. We first determined which portion of each candidate would likely fold or unfold using the protein modeling software, Phyre2.33 We then calculated the hydrodynamic radii of our candidates for the folded and unfolded regions by pulsed field gradient techniques.34 As shown in Table S2, the estimated IDP's hydrodynamic sizes (3–8 nm in radii and 6–16 nm in diameter) would suggest that only the candidates with smaller sizes (<10 nm in diameter) could potentially diffuse through the NPC's central channel (~ 10 nm in diameter at the narrowest waist). However, our data indicated that some IDP candidates with larger sizes (up to 16 nm in diameter) diffuse through the central channel, while some smaller IDPs (down to 6 nm in diameter) take peripheral transport routes rather than the central channel through the NPCs. The above results again suggest that the IDP's size is not a determining factor for neither its diffusion modes (passive or facilitated) nor transport routes through the NPCs.
2.2. AA composition of IDPs plays critical roles in determining their transport routes through the NPC
Next we sought what features of IDPs distinguish their diffusion pathways through the NPC. The FG‐Nup barrier forms through hydrophobic interactions between FG Nups and generates a highly hydrophobic and slightly positively charged microenvironment for transiting molecules in the NPC.35, 36, 37 Recent evidence further revealed that the FG‐Nup barrier possesses a heterogeneous spatial distribution of FG domains, to which sparsely populated FG domains exist in the central axial channel and more concentrated FG domains stay in the peripheral regions around the central channel.37, 38, 39 As a result, the central axial channel could be more hydrophilic and its periphery may be more hydrophobic in the NPC. Besides the molecular size, previous studies suggested that the charge and hydrophobicity of folded proteins can also affect their translocation through the NPC.35, 40, 41, 42 By defining the charged (K, R, D, and E) and hydrophobic (A, V, L, I, M, P, F, and W) residues at physiological pH 7.4 (Figure S1),43 we found that the IDP candidates that adopt different diffusion routes can have similar percentages of hydrophobic AAs and net charges, but differ in their content ratios of charged (Ch) and hydrophobic (Hy) AAs (Table 1). For example, SLD2 diffusing through the central channel contains 35% of charged AAs and 32% of hydrophobic AAs, generating a Ch/Hy AA ratio of 1.08. However, Nup116 (348–458) moving through the periphery (3% of charged AAs and 28% of hydrophobic AAs) possesses a Ch/Hy ratio of 0.09, which is about 12‐fold lower than that of SLD2. Therefore, we postulate that IDPs that contain high Ch/Hy ratios may diffuse through the NPC's central axial channel, and those with low Ch/Hy ratios could diffuse through the peripheral regions around the central channel.
To further test the hypothesis, we mutated a majority of (WT) Nup116 (348–458) neutral AAs adjacent to the FG motifs into charged AAs (Figure 3a,b, Table 1). These charge mutations of WT Nup116 (348–458) changed the Ch/Hy ratio of 0.03 to 0.74 for the charged version of Nup116 (348–458) while maintaining the same approximate molecular weight. We found that dramatically increasing the Ch/Hy ratio caused the charged mutant of Nup116 (348–458) to diffuse through the central axial channel, avoiding the peripheral pathway adopted by WT Nup116 (348–458) (Figure 3a,b). This observation was reaffirmed by Nsp1 and its truncates (Nsp1 [1–603], Ch/Hy = 0.73), (Nsp1 [1–172], Ch/Hy = 0.07), and (Nsp1 [173–603], Ch/Hy = 0.94). As shown in Figures 2a and 3c,d, both Nsp1 (1–603) and Nsp1 (173–603) with high Ch/Hy ratios diffused through the central axial channel, while Nsp1 (1–172) with a low Ch/Hy ratio diffused through the periphery, thus confirming the results of Nup116 and its charged mutant. This suggests that the Ch/Hy ratio of IDPs is the dominant role in deciding the spatial transport route through the NPCs, instead of the size.
Figure 3.

IDP's Ch/Hy ratio determines its spatial transport route through the NPC. The 3D spatial probability density map overlaid with the 3D physical schematic of NPC (a cut‐away view) in the (X, R, Ɵ) cylindrical coordinate system (i) and the AA sequence (ii) for each IDP candidate were provided. Additionally, a histogram of spatial probability density for the radial dimension was shown in (ii) to highlight either a single peak representing a central axial transport route or two peaks representing a peripheral transport route. (a, b) WT Nup116 (348–458) and its charge mutant separate their 3D transport routes through the NPC after mutating 31 more neutral AAs adjacent to FG motifs into highly charged residues (k in red and d, e in blue). N, nucleus; C, cytoplasm. (c, d) Two segments of WT Nsp1 (1–603), Nsp1 (1–172), and Nsp1 (173–603), have different charged AAs (k in red and d, e in light blue) and Ch/Hy ratios, which differentiates them into two distinct 3D diffusion routes through the NPCs. AA, amino acid; Ch/Hy, charged/hydrophobic; IDP, intrinsically disordered protein; NPC, nuclear pore complex
2.3. Relationship between Ch/Hy ratio and nuclear transport efficiency of IDPs
The content information of hydrophobic and charged AAs is always the focus in studying the structure and function of IDPs. More than three decades ago, protein's Ch/Hy ratio was firstly used as an informal IDP predictor.44 In recent years, the Ch/Hy ratios of FG Nups have been characterized in understanding the structure and dynamics of these IDPs in the NPCs.39, 45, 46 Here our results for the first time demonstrated that the IDP's Ch/Hy ratio can also significantly influences the spatial transport route of IDPs through the NPCs. Interestingly, we further found that the spatial transport pathways of IDPs through the NPCs seem to be largely a two‐state phenomenon with a Ch/Hy ratio transition point approximately at 0.37, obtained by sigmoidal fitting (Figure S7). As demonstrated, the IDPs with Ch/Hy ratios above this transition point have higher probabilities of diffusing through the NPC's central axial channel with a lower probability to occupy its periphery and vice versa for the IDPs with Ch/Hy ratio below 0.37. Additionally, we further surveyed 80 more human nuclear IDPs and found that ~90% of them have Ch/Hy ratios above 0.37 and the rest with ratios below the transition point (Table S3). The Ch/Hy ratio distribution in these IDPs agrees with previous measurements and estimations.47 The dramatic two‐state scenario observed for NPC transport routes of IDPs might suggest that endogenous nuclear IDPs could be differentiated into two different transport behaviors through the NPCs.
An interesting question is whether the high or the low Ch/Hy ratios in these endogenous human nuclear IDPs could enhance or inhibit the nuclear import efficiency of the protein from cytoplasm into nucleus through the NPC. This question can serve important for a better biological understanding of the activity of endogenous nuclear IDPs as well as for synthetic peptides intended to reach the nucleus of the cell. To answer this question, we further measured nuclear transport (including both import and export) efficiencies for both FG‐IDPs and non‐FG IDP candidates by SPEED microscopy, including SLD2, CREST, Nup116 (348–458), and its charged mutant, as these candidates have highly varying Ch/Hy ratios (Table 2). Nuclear import or export events through the NPCs, classified as being either successful or abortive, describes the complete process that an IDP entering the NPC from the cytoplasmic or nucleoplasmic side, successfully reach the other side or return to the starting side after interacting with the NPC. By comparing the percentage of total events deemed successful events, we can obtain nuclear import or export efficiency. After collecting 200 nuclear import or export events for each IDP candidate from 10 different NPCs of 10 cells, we found that these IDP candidates have almost equal probability for bidirectional moving through the NPC but different Ch/Hy‐dependent import or export efficiencies (Table 2). In detail, WT Nup116 (348–458) with a low Ch/Hy ratio of 0.09 diffusing through the peripheral regions possesses higher import and export efficiencies (~50%) than its charged mutant having a high Ch/Hy ratio of 0.74 moving through the central axial channel (~39%) (p‐value < .01). Similarly, the measured nuclear import efficiencies of other IDPs, CREST (Ch/Hy = 0.30) and SLD2 (Ch/Hy =1.08), confirm the trend (import efficiency of ~56% for CREST and ~36% for SLD2), although they have almost the same export efficiencies (~50%). Thus far, our data indicates that IDPs with lower Ch/Hy ratios corresponding with a NPC transport route through the periphery have more effective nuclear import than those with larger Ch/Hy ratios diffusing through the central axial channel.
3. DISCUSSION
One of our major research goals in this paper is to examine whether the size‐exclusion rule established for nucleocytoplasmic transport of folded proteins would be applicable to IDPs as they transport through the NPCs under physiological conditions. This information can help toward a better understanding of endogenous nuclear IDP structure–function pairing. By understanding the characteristics that determine IDP nuclear translocation, insight into synthetic peptide design can additionally be gained for targeting particles to the nucleus. By using the high‐speed super‐resolution fluorescence SPEED microscopy setup, we tracked real‐time transport processes and mapped 3D transport routes for various IDP candidates through the native NPCs in permeabilized human cells. Our data revealed that both large and small tested IDPs ranging from 11 to 63 kDa can diffuse through the NPC without the addition of transport receptors. This is in stark contrast to data obtained for folded proteins and large dextrans. Moreover, we found that the nucleocytoplasmic transport efficiencies and the spatial transport routes of these IDPs through the NPCs heavily depend on the Ch/Hy ratios in their AA compositions rather than their molecular weights (or hydrodynamic radii). These new findings enabled us to suggest a Ch/Hy‐ratio mechanism for nuclear transport of IDPs through the NPCs (Figure 4, Videos S2 and S3). In detail, differing from the passive and facilitated transport models of NPC transport for folded proteins (Figure 4a), IDPs rely less on size (or molecular weight) and more on their composition of AAs to determine their effectiveness and mode of transport. It must be noted that extremely large IDPs (over 100 kDa) have not yet been tested. It remains unknown if there is a maximum threshold for molecular weight in IDPs that would prevent them from diffusing through the pore unaided.
Figure 4.
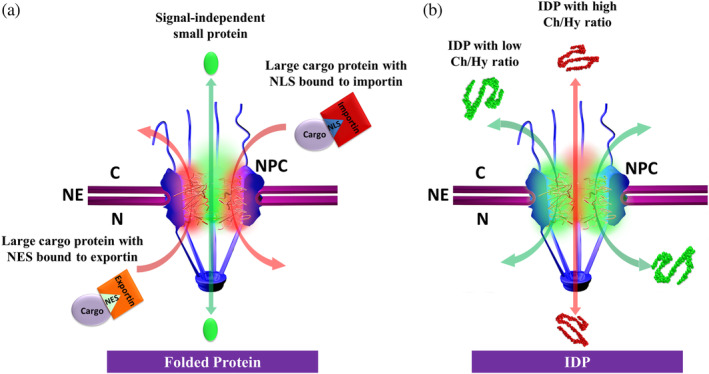
A Ch/Hy ratio model for nucleocytoplasmic transport of IDPs. (a) A size‐exclusion model for folded proteins. Small signal‐independent proteins (<40 kDa) can passively diffuse through the NPC in both directions (green double‐headed arrow); however, larger protein cargos can be repelled by the NPC unless carrying an NLS for import or an NES for export. These signals will be recognized by transport receptors (importins for NLS or exportins for NES) to form complexes and then the complexes transport through the NPC in the mode of facilitated translocation (red single‐headed arrows). Emerging evidence suggests that passive and facilitated transports mainly adopt their spatial routes through the NPC's central axial channel (green regions) and the peripheral areas (red areas), respectively. (b) A Ch/Hy ratio model for IDPs. Both large (>40 kDa) and small (<40 kDa) IDPs can passively diffuse through the NPC (double‐headed arrows). Moreover, the IDPs with high Ch/Hy ratio (> ~0.37) prefer diffusing through the central axial channel (red arrow and area), while the IDPs with smaller Ch/Hy ratio (< ~0.37) mainly diffuse through the peripheral area around the central channel (green arrows and areas). Ch/Hy, charged/hydrophobic; IDP, intrinsically disordered protein; NES, nuclear export signal; NLS, nuclear localization sequence; NPC, nuclear pore complex
Given the heterogeneous micro‐environments formed in the NPC,48 we suggest that these two distinct passive diffusion pathways through the NPC for IDPs could be driven by two different mechanisms. As proven previously,15, 16, 27 the peripheral regions around the central axial channel in the NPC are filled with concentrated disordered FG domains to form a highly hydrophobic micro‐environment. As a result, such hydrophobic circumstances in the NPC would favor transiting molecules with more exposed hydrophobic groups at their surfaces to diffuse in, but may repel those with hydrophilic surfaces. Given the fact that the IDPs with low Ch/Hy ratio contain higher percentage of exposed hydrophobic AAs than those IDPs with high Ch/Hy ratio, the diffusion of the former IDPs through the hydrophobic periphery can be favorably driven by hydrophobic interactions and much easier than the latter ones. In contrast, the central axial channel occupied by few or very sparse FG domains creates an aquatic‐like space, which would more likely favor hydrophilic transiting molecules to diffuse through, such as ions, small soluble proteins, and IDPs with high Ch/Hy ratios. Thus, the passive diffusion of IDPs with high Ch/Hy ratios through the central axial channel could be simply a diffusion process modeled as hydrogen bonding in aquatic narrow channel.15, 16, 27
Moreover, the tested large IDPs can passively diffuse through the NPCs, while large folded proteins with similar molecular weights cannot, might due to their natively disordered conformations. Differing from the structured or globular shape of folded proteins, IDPs do not hold a certain geometry and can remain flexible and dynamic in nature.47 First, the lack of rigidity in shape may allow large IDPs to “fit” into the NPC's narrow central axial channel (~10–14 nm in diameter) or “sneak” into the gaps between FG Nups located at the periphery,18, 27, 49, 50 resulting in effective entrance into the NPC. Second, the lack of secondary structure and dynamic nature of many IDPs allows them to fully expose hydrophobic or hydrophilic interaction groups at the surface to interact with the surroundings. For example, as addressed in the former paragraph, large IPDs with low Ch/Hy ratios could employ exposed hydrophobic AAs to directly interact with FG domains in the peripheral regions in the NPC, enhancing easier entrance into the FG‐Nup barrier and generating higher transport efficiency. These direct hydrophobic interactions between transiting IDPs and FG Nups may explain why transport receptors are not a necessity for transport of nuclear IDPs with low Ch/Hy ratios. Additionally, since some nuclear IDPs accumulate in the nucleus, they might be retained through binding to prevent diffusion out of the nucleus.
Currently it is still challenging, but highly desirable, to obtain high‐resolution (<50 nm) 3D information of structures and dynamic processes in either fixed or live samples. SPEED microscopy is one of the newly developed techniques to determine 3D sub‐diffraction‐limited structural or dynamic information in rotationally symmetric bio‐structures23, 24, 27 without using 3D super‐resolution microscopy or real‐time 3D particle tracking. So far, the method has been successfully applied to achieve 3D structural and functional information for 25–300 nm subcellular organelles that meet the rotational symmetry requirement such as NPC, primary cilium, microtubule and glass nanocapillary.51 To further introduce the method and respond to questions about the reproducibility in obtaining 3D information by 2D to 3D transformation,52, 53 we have provided comprehensive analyses of this method by detailing the mathematical principle, the experimental data and the computational simulations used in our method.51, 52 There are two critical factors in reproducing the accurate 3D super‐resolution information, the single molecule localization error and the number of single molecule locations. Both have been fully discussed.51 The recent successful applications of SPEED microscopy in various systems prove the robustness of achieving accurate and highly reproducible 3D super‐resolution information by following the 2D‐to‐3D transformation procedures we developed.51 Many of our major findings have been subsequently confirmed by other independent studies using different approaches, such as coarse‐grained molecular dynamics simulation and cryo‐electron microscopy.22, 36, 54 Finally, open source code of the 2D to 3D transformation algorithm (MATLAB) and simulations (Python) are also provided online at https://github.com/SamJunod/yanglab.
4. MATERIALS AND METHODS
4.1. Cell culture and transport conditions
A HeLa cell line stably expressing the GFP‐conjugated POM121 was authenticated by the NIH ATCC and monitored to be free of mycoplasma on a routine check before experiments. Freshly split cells were grown overnight on coverslips in DMEM supplemented with 10% FBS. By conjugating GFPs with POM121, the middle plane of the NE can be determined.21 However, multiple‐GFPs‐conjugate‐POM121 NPCs have enhanced transport time and efficiency compared to that of WT NPCs.23 Following the same experimental procedure, the transport time and efficiency for single‐GFP‐POM121 NPCs were measured, and it was found that nuclear transport was not enhanced by a single GFP conjugate. Therefore, experiments were conducted with a single‐GFP‐POM121 cell line, and GFP fluorescence was utilized to localize the location of individual NPC on the NE.
For microscopy, flow chambers were constructed with a top coverslip and two lines of silicone grease as spacers. Cells were washed with transport buffer (20 mM HEPES, 110 mM KOAc, 5 mM NaOAc, 2 mM MgOAc, 1 mM EGTA, and pH 7.3), permeabilized for 2 min with 40 μg/ml digitonin in transport buffer, and washed again with transport buffer supplemented with 1.5% polyvinylpyrrolidone (PVP; 360 kDa). PVP was included in all transport buffer solutions after digitonin treatment to prevent osmotic swelling of the nuclei. For single‐molecule measurements of facilitated translocation and passive diffusion of various molecules through the NPCs, 1 nM intrinsic fluorescent or dye‐labeled substrates were used. For the efficient nuclear import of 1 nM labeled NLS‐2xGFP, the transport buffer contained the import cofactors 1 mM GTP, 2 μM Ran, 1 μM NTF2, and 0.5 μM Imp β1 and 0.5 μM Imp α, respectively. More details can be found in previously published methods.23, 25
4.2. Dextran, folded proteins, and labeling
Dye‐labeled dextrans were purchased from Invitrogen. N‐terminal His‐tagged GFP and 2xGFP were expressed in E. coli and purified by Ni‐NTA Superflow (Qiagen), MonoQ, and Superdex 200 (Amersham) chromatography. The transport cofactors used for nuclear import of NLS‐2xGFP were purified as described in previous reports,23, 24 unless specified in the methods. For labeling, the solvent‐accessible cysteines on the proteins were labeled with 20‐fold molar excess Alexa Fluor 647 maleimide dye (Invitrogen) for 2 hr at room temperature in 50 mM sodium phosphate, 150 mM NaCl, and pH 7.5. Reactions were quenched with β‐mercaptoethanol, and the products were dialyzed to remove the free dyes. The labeling ratio is about two dyes per GFP and four dyes per NLS‐2xGFP.
4.3. FG‐domain synthesis, expression, purification, and labeling
Coding sequences for FG‐domains were PCR amplified from S. cerevisiae DNA or synthesized de novo by GenScript and were cloned into pGEX‐2TK in frame with the 3′ end of the glutathione Stransferase (GST) gene. Where indicated, codons encoding six His and one Trp residues were added at the 3′ end. FG‐domains were expressed as GST fusions in E. coli BL21 strain, and glutathione‐coated beads were used to isolate them from bacterial extracts. FG‐domains were released by thrombolysis from their GST tag, and in some cases, nickel‐coated beads were used to recapture the FG‐domain via its C‐terminal His‐tag. These were eluted from beads with 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 250 mM imidazole, 0.1% Tween‐20, and concentrated (Centricon 3) when necessary. All proteins were then labeled with 20‐fold molar excess Alexa Fluor® 647 Succinimidyl Ester (Invitrogen) for 2 hr at room temperature in binding buffer (20 mM HEPES pH 6.8, 150 mM KOAc, 2 mM MgOAc) and the products were dialyzed to remove the free dyes. The labeling ratios are in the range of 3–6.
4.4. Production, expression, purification, and labeling of non‐FG IDPs
Full‐length constructs of yeast SLD2 or human CREST were cloned into a C Terminal GST Vector. Expression was performed in BL121(DE3) E. coli until an OD of 0.6 was reached. Cells were then lysed via homogenization (Avestin B‐15) in Lysis Buffer (1X PBS, 2 M NaCl) and dissolved Protease inhibitor Cocktail (Sigma Aldrich). Cell lysate was then incubated with Glutathione Hicap matrix (Qiagen). After centrifugation, the beads and protein were re‐suspended in PBS‐DTT and placed into a chromatography column. The non‐FG IDPs were then cleaved off the column using Precision Protease (GE Heathcare Lifescience). Purity was confirmed using SDS‐PAGE. For labeling, IDPs were reduced with a 100× molar excess of TCEP for 20 min and then labeled with a 20‐fold molar excess Alexa Fluor® 647 Maleimide (Invitrogen) overnight at 4°C. Products were dialyzed to remove the free dyes. The labeling ratios are in the range of 1–2 dyes per protein.
4.5. Instrumentation
The SPEED microscope includes an Olympus IX81 equipped with a 1.4 NA 100× oil‐immersion apochromatic objective (UPLSAPO 100XO, Olympus), a 35 mW 633 nm He‐Ne laser (Melles Griot), a 120 mW ArKr tunable ion laser (Melles Griot), an on‐chip multiplication gain charge‐coupled device camera (Cascade 128+, Roper Scientific), and the Slidebook software package (Intelligent Imaging Innovations) for data acquisition and processing. An optical chopper (Newport) was used to generate an on–off mode of laser excitation. GFP and Alexa Fluor 647 fluorescence were excited by 488 and 633 nm lasers, respectively. The two lasers were combined by an optical filter (FFF555/646 Di01, Semrock), collimated and focused into an overlapped illumination volume in the focal plane. The green and red fluorescence emissions were collected by the same objective, filtered by a dichroic filter (Di01‐R405/488/561/635‐25x36, Semrock) and an emission filter (NF01‐405/488/561/635‐25X5.0, Semrock) and imaged by an identical CCD camera. The system error of alignment between red and green fluorescence channels is 3.0 ± 0.1 nm, determined by measuring 230 immobile Alexa Fluor 647‐labeled GFP fluorescent molecules on the surface of a coverslip.
4.6. Orientation of a single NPC
The position of the NE was localized by fitting GFP fluorescence of POM121 as follows: the pixel intensities within a row or a column approximately perpendicular to the NE were fit to a Gaussian. The peak position of the Gaussian for a particular set of pixel intensities was considered the NE position for that row and column. The peak positions of a series of such Gaussians were then fit with a second‐degree polynomial, yielding the path of the NE within the entire image.23
Two rules were then used to select a single NPC oriented perpendicularly to the NE on the equator of the nucleus and to the y direction of the Cartesian coordinates (x, y) in the CCD camera: (a) choose a fluorescent NPC on the equator of the nucleus such that the tangent of the NE at the location of this NPC should be parallel to the y direction of the Cartesian coordinates (x, y) in the CCD camera; and (b) examine the ratio of Gaussian widths in the long and short axes of the chosen GFP‐NPC fluorescence spot. The ratio needs to fall between 1.74 and 1.82. Within this range, an illuminated NPC only has a free angle of 1.40 to the perpendicular direction to the NE.23
4.7. Localization precisions of isolated fluorescent spots
The localization precision for fluorescent NPCs, immobile fluorescence molecules and moving fluorescence molecules was defined as how precisely the central point of each detected fluorescent diffraction‐limited spot was determined. For immobile molecules or fluorescent NPCs, the fluorescent spot was fitted to a 2D symmetrical or an elliptical Gaussian function, respectively, and the localization precision was determined by the SD of multiple measurements of the central point. However, for moving molecules, the influence of particle motion during image acquisition should be considered in the determination of localization precision. In detail, the localization precision for moving substrates (σ) was determined by an algorithm of , where F is equal to 2, N is the number of collected photons, a is the effective pixel size of the detector, b is the SD of the background in photons per pixel, and , s 0 is the SD of the point spread function in the focal plane, D is the diffusion coefficient of substrate in the NPC, and Δt is the image acquisition time.23, 24
In our experiments, more than 1,000 signal photons were collected from each targeted molecule. The localization precision for the immobile molecules based on the SD of multiple measurements is approximately 10 ± 1 nm. For moving molecules through the NPCs (captured within ~300 nm in the focal plane of SPEED microscopy as shown in Figure S1), the localization precision is calculated to be <13 nm based on the above equations and the parameters determined experimentally (N > 1,000, a = 240 nm, b ≈ 2, s 0 = 150 ± 50 nm, D is in the range of 0.4–0.8μm2/s for the tested substrates). The approach to determine the diffusion coefficients here follows the published procedure.55 The localization precision of the NPC centroid was determined to be 2 ± 1 nm in permeabilized cell. The system error of alignment between red and green fluorescence channels is 3.0 ± 0.1 nm, determined by measuring 230 immobile Alexa Fluor 647‐labeled GFP fluorescent molecules on the surface of a cover‐slip. Therefore, the overall tracking precision for labeled molecular transport through the GFP‐labeled NPC was estimated to be <13.5 nm in permeabilized cells.
4.8. 2D to 3D transform algorithms and image processing
The detailed transform process used to compute the 3D spatial probability density maps of particles transiting through the NPC was described in our previous publications23, 24, 27 and demonstrated again here in Figure S3. In short, the 3D spatial locations of molecules transiting through the NPC can be considered in either Cartesian (X, Y, Z) or cylindrical (X, R, Ɵ) coordinates. In microscopic imaging, the observed 2D spatial distribution of particle localizations is a projection of its actual 3D spatial locations onto the XY plane. The underlying 3D spatial distributions can be recovered by projection of the measured Cartesian (X, Y) coordinates back onto the simplified cylindrical (X, R, constant) coordinates, based on the expected cylindrically symmetrical distribution along the Ɵ direction of the nuclear pore. More detailed information can be found in Figure S3 and its figure caption. The resulting 3D, surface‐rendered visualizations shown in figures and movies were generated with Amira 5.2 (Visage Imaging).
4.9. Accurate 3D nuclear transport routes through the NPC further validated by Monte Carlo simulations
A Monte Carlo simulation was employed to demonstrate the reproducibility of our method in obtaining accurate 3D super‐resolution information in the setting of rotationally symmetrical biostructures.51 Two critical parameters, the number of acquired single‐molecule locations and the localization error of those acquired locations, are used to determine this reproducibility. The Monte Carlo simulation56, 57 and our use of it to validate our technique, as well as other details, are described elsewhere.51 Briefly, an estimated number for the amount of data points needed to resolve the pathway through the NPC as being either through the central axial channel, or periphery of the NPC was calculated using Monte Carlo simulations. Simulations were conducted using randomly sampled X and Y positions confined to either the axial center of a 200 nm length cylinder, or the periphery of the 50 nm radius within the NPC. Each sampled point was calculated using a probability distribution corresponding to our experimental single molecule localization precision to simulate error. The data was then run through our deconvolution process. Many iterations were run under the necessary conditions for each experiment to determine the minimum amount of data needed to resolve 3D spatial transport route.51 The README file is provided on https://github.com/samjunod/yanglab under the IDP folder, which allows readers to download and test the 2D to 3D Transformation Process.
4.10. Statistical analysis
Experimental measurements are reported as means ± SEs of the mean unless otherwise noted. Binomial Z test for proportions was used for p‐values regarding nuclear transport efficiency.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
W.Y. designed experiments; J.K., S.J., and J.M. performed experiments; J.M. and W.Y. established cell lines and built microscopy equipment; S.J., J.K., and W.Y. conducted data analysis and wrote the manuscript.
Supporting information
Figure S1 | Disordered level, AA composition and Ch/Hy ratio for IDPs
Disorder levels obtained via IUPRED, AA sequences and compositions, and Ch/Hy ratios were shown for both FG‐ and non‐FG IDPs.1 Disordered level score above 0.5 were considered highly disordered. Hydrophobic and charged AA residues were shown in red and blue respectively.
Figure S2 | 3D spatial transport routes for passive and facilitated transports of folded proteins mapped by SPEED Microscopy
(a) A single NPC was illuminated by the inclined point spread function (iPSF) of SPEED microscopy at the NE of a stable HeLa cell line expressing green fluorescent protein (GFP)‐POM121. The iPSF was achieved by shifting an incident laser beam off the center of an objective's back aperture, resulting in an angle of 450 relative to the focal plane and the effective PSF smaller than the average nearest distance between NPCs in X, Y, or Z direction.2, 3 The iPSF was further used to track single‐folded protein molecules or complexes labeled in distinct colors from GFP moving through the NPC. Capturing the trajectories of these transiting molecules (red dots) and superimposing the data over the centroid position of the NPC (blue dot) results in 2D projection (XY plane) of the nuclear transport process. N, nucleus; C, cytoplasm. (b) Collecting hundreds of single‐molecule trajectories for GFPs passively diffusing through the NPC generated a 2D spatial super‐resolution distribution (green dots) of GFPs in the NPC, overlaid with the NPC's 2D schematics (grey). (c) A 2D spatial super‐resolution distribution (red dots) of a facilitated translocation complex of NLS‐2xGFP/Importin β/Importin α in the NPC, overlaid with the NPC's 2D schematics (g).2, 4 (d) A 2D to 3D transform algorithm was utilized to obtain the 3D transport routes of the above 2D passive (b) and facilitated (c) transport data. As shown in the Cylindrical coordinates (x, R, and θ), passive diffusion (green clouds) and facilitated transport (red clouds) of folded proteins possess distinct pathways through the NPC.2 Figure a‐d were adapted from our previous publications with permissions.4
Figure S3 | 2D and 3D nuclear transport data for 2x green fluorescent protein (GFP) and dextrans. 2D super‐resolution locations and 3D spatial probability density maps are shown for 2xGFP (c) and dextrans ranging in size from 10 to 70 kDa (a, b and d). 3D spatial probability density maps (green clouds) are superimposed with wireframe NPC schematic (grey).
Figure S4 | Determination of the central axis of the NPC illuminated by SPEED microscopy. Diagrams are used to demonstrate the detailed steps in our data analyses. (A) The initial single‐molecule tracking data (black points) are plotted around the NPC's marker for the central position (red dot) collected with SPEED. (B) The collected data after filtering by using single‐molecule localization filters (photon count, width of emission PSF). (C) The filtered 2D single‐molecule data shape the spatial location of the NE and a single NPC. Furthermore, the 2D single‐molecule distribution indicates the orientation of NPC, which must fall within a free angle of 1.4° perpendicular to the NE, for the data to be processed. The red dot represents the localized centroid of a fluorescently tagged NPC. Periodically, the localized NPC position does not perfectly overlap with the localized single‐molecule data from the fluorescently tagged INM proteins and must be corrected. (D‐G) Due to the rotational symmetry of the NPC, 2D single‐molecule data is expected to be equally distributed along the central axis in the X and Y dimensions. Histograms are prepared for the X and Y dimensional data, and if the histograms are asymmetrical, as in this example, fine adjustments are made to restore symmetry using the formula . If the peaks are symmetrical, this step is skipped. (H) The 2D single‐molecule data will then undergo the 2D to 3D transformation (Figure S5) to produce the 3D density map.
Figure S5 | Graphical demonstration of 2D to 3D conversion process A schematic demonstration, with simulated data, of the 2D to 3D transformation algorithms for molecules that diffuse through the NPC. (A) 3D spatial locations of randomly diffusing molecules inside the NPC can be coordinated in a cylindrical coordination system (R, Ɵ, Y) due to the cylindrical rotational symmetry of the NPC. The 3D molecular locations in the NPC (rainbow colored for Z position) are projected onto a 2D plane in a Cartesian coordination system (X and Y, shown as black points) by microscopy imaging (Y and Z shown as gray points). (B) The cross‐sectional view of all the locations (the same as the gray points from the Y and Z dimension). These locations can be grouped into the sub‐regions between concentric rings. Given the high number of randomly distributed molecules in the NPC the spatial density of locations (ρi) in each sub‐region (s(i,j)) between two neighboring rings will be rotationally symmetrical and uniform. These locations can be further projected into 1D along the Y dimension. The locations along the Y dimension can be clustered in a histogram with j columns. The total number of locations in each column (A(i,j)) is equal to , which is further explained in Figure S5. (C) Histograms of 2D projected data from microscopy experiments are identical to B, thereby allowing us to use the aforementioned formula to determine the density of each concentric ring. (D) Using the algorithms, 2D projected data can be used to reconstruct the 3D spatial distribution of protein location on the NPC. More detailed explanation and calculations can be found in Methods and also previous publications.2, 3
Figure S6 | Matrix equation flowchart. (𝑨(𝒊,𝒋)), area of the sub‐region, (i), radial number, (j), axial number, (𝝆𝒊), spatial probability density in each radial ring, (𝑺(𝒊,𝒋)), area of the sub‐region, (𝑵𝒋), number of events, and (∆𝒓), bin size. Equations 1–3 will determine the area of the sub‐region given certain parameters (i.e., i = j). Equation 4 will determine the number of events in the given area. Once the subregion area ((𝒊,)) and events (𝑵𝒋) are known, the spatial probability density in each radial ring (𝝆𝒊) can be calculated.
Figure S7 | A two‐state scenario for IDPs depending on Ch/Hy ratio A plot shown the relationship between Ch/Hy ratio and the probability of occupying either the axial center or periphery of the NPC during transport for the IDP candidates we tested in this paper. A two‐state scenario was revealed with a sigmoidal fitting determining the Ch/Hy ratio of 0.37 as a transition point. IDPs with a high Ch/Hy ratio above 0.37 have a high probability of occupying the axial center of the NPC and vice versa.
Table S1 | Nuclear transport kinetics of IDPs through the NPC In our measurements, more than 200 transport events were collected to determine the transport kinetics for each candidate ordered by decreasing Ch/Hy ratio. The reproducibility of each candidate was determined from the radial peak of the 2D to 3D transformation density map and the number (N) of single‐molecule localizations between the axial region of 70 to −100, cytoplasm to nucleus respectively.5, 6
Table S2 | Estimation of Hydrodynamic Radius of IDPs
Folded and unfolded percentages were estimated using the protein modeling software, Phyre2.7 The hydrodynamic radius (Rh) was calculate using R h = 4.75 N 0.29Å for the folded percentage of the candidates and R h = 2.21 N 0.57Å was used for the unfolded percentage of each candidate. N is the number of residues in the polypeptide chain.8
Table S3 | List of 80 human nuclear IDPs Analysis of 80 IDPs that are localized predominantly within the nucleus. The predicted 3D transport route through the NPC is given for each protein based upon the Ch/Hy ratio obtained. UNIPROT ID is provided for each for easy identification.
Video S1 │ Single IDP molecule interacting with a single NPC.
This video shows a typical transport event of a IDP through the NPC in a eukaryotic cell (CREST is used as an example). Pixels are 240‐nm square, each frame was acquired in 2 ms, and the playback speed is 1,000X slower than real‐time. The compartment on the left side of the NPC (the green fluorescent spot) is the Nucleoplasm(N), and the right side is the Cytoplasm (C).
Video S2 │ 3D diffusion route for IDPs diffusing through the NPC's central axial channel.
This video shows a cut‐away view of the 3D diffusion route of SLD2 through the central axial channel of the NPC (red cloud; brighter color indicates higher spatial density) superimposed on the NPC architecture (grey wire frame). Bar: 20 nm.
Video S3 │ 3D diffusion route for IDPs diffusing through the peripheral regions around the central axial channel in the NPC.
This video shows a cut‐away view of the 3D diffusion route of CREST through the periphery of the NPC (Green cloud; brighter color indicates higher spatial density) superimposed on the NPC architecture (grey wire frame). Bar: 20 nm.
ACKNOWLEDGMENTS
We greatly thank Vladimir Uversky (University of South Florida, Tampa, USA) for providing helpful information on IDP characteristics to aid in our protein selections. We thank M. Rexach (University of California, Santa Cruz, USA) for providing isolated yeast FG domains. We thank Michael D. Hebert (University of Mississippi Medical Center, Jackson, USA) for providing purified Coilin protein. The project was supported by grants from the US National Institutes of Health (NIH GM094041, GM097037, GM116204, and GM122552 to W.Y.).
Junod SL, Kelich JM, Ma J, Yang W. Nucleocytoplasmic transport of intrinsically disordered proteins studied by high‐speed super‐resolution microscopy. Protein Science. 2020;29:1459–1472. 10.1002/pro.3845
Samuel L. Junod and Joseph M. Kelich contributed equally to this study.
Funding information Foundation for the National Institutes of Health, Grant/Award Numbers: GM116204, GM122552; US National Institutes of Health, Grant/Award Numbers: GM097037, GM094041; University of Mississippi; University of California, Santa Cruz; University of South Florida
REFERENCES
- 1. Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. [DOI] [PubMed] [Google Scholar]
- 2. Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 3. Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell‐signaling and cancer‐associated proteins. J Mol Biol. 2002;323:573–584. [DOI] [PubMed] [Google Scholar]
- 4. Frege T, Uversky VN. Intrinsically disordered proteins in the nucleus of human cells. Biochem Biophys Rep. 2015;1:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Love JJ, Li X, Chung J, Dyson HJ, Wright PE. The LEF‐1 high‐mobility group domain undergoes a disorder‐to‐order transition upon formation of a complex with cognate DNA. Biochemistry. 2004;43:8725–8734. [DOI] [PubMed] [Google Scholar]
- 6. Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. [DOI] [PubMed] [Google Scholar]
- 7. Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. [DOI] [PubMed] [Google Scholar]
- 8. Velma V, Broome HJ, Hebert MD. Regulated specific proteolysis of the Cajal body marker protein coilin. Chromosoma. 2012;121:629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aizawa H, Hu S‐C, Bobb K, et al. Dendrite development regulated by CREST, a calcium‐regulated transcriptional activator. Science. 2004;303:197–202. [DOI] [PubMed] [Google Scholar]
- 10. Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kabachinski G, Schwartz TU. The nuclear pore complex—Structure and function at a glance. J Cell Sci. 2015;128:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paulillo SM, Phillips EM, Köser J, et al. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol. 2005;351:784–798. [DOI] [PubMed] [Google Scholar]
- 13. Fahrenkrog B, Aebi U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. [DOI] [PubMed] [Google Scholar]
- 14. Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. [DOI] [PubMed] [Google Scholar]
- 15. Terry LJ, Wente SR. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. [DOI] [PubMed] [Google Scholar]
- 18. Sakiyama Y, Mazur A, Kapinos LE, Lim RYH. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high‐speed atomic force microscopy. Nat Nanotechnol. 2016;11:719–723. [DOI] [PubMed] [Google Scholar]
- 19. Rexach M, Blobel G. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. [DOI] [PubMed] [Google Scholar]
- 20. Griffis ER, Altan N, Lippincott‐Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription‐dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cardarelli F, Lanzano L, Gratton E. Capturing directed molecular motion in the nuclear pore complex of live cells. Proc Natl Acad Sci U S A. 2012;109:9863–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. Structure and gating of the nuclear pore complex. Nat Commun. 2015;6:7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma J, Yang W. Three‐dimensional distribution of transient interactions in the nuclear pore complex obtained from single‐molecule snapshots. Proc Natl Acad Sci U S A. 2010;107:7305–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma J, Goryaynov A, Sarma A, Yang W. Self‐regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci U S A. 2012;109:7326–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson GL, Dean BS, Wang G, Dean DA. Nuclear import of plasmid DNA in digitonin‐permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J Biol Chem. 1999;274:22025–22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma J, Goryaynov A, Yang W. Super‐resolution 3D tomography of interactions and competition in the nuclear pore complex. Nat Struct Mol Biol. 2016;23:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hough LE, Dutta K, Sparks S, et al. The molecular mechanism of nuclear transport revealed by atomic‐scale measurements. Elife. 2015;4:e10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feldherr CM, Akin D. The location of the transport gate in the nuclear pore complex. J Cell Sci. 1997;110:3065–3070. [DOI] [PubMed] [Google Scholar]
- 30. Keminer O, Peters R. Permeability of single nuclear pores. Biophysik. 1999;J77:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naim B, Brumfeld V, Kapon R, Kiss V, Nevo R, Reich Z. Passive and facilitated transport in nuclear pore complexes is largely uncoupled. J Biol Chem. 2007;282:3881–3888. [DOI] [PubMed] [Google Scholar]
- 32. Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. [DOI] [PubMed] [Google Scholar]
- 35. Tagliazucchi M, Peleg O, Kröger M, Rabin Y, Szleifer I. Effect of charge, hydrophobicity, and sequence of nucleoporins on the translocation of model particles through the nuclear pore complex. Proc Natl Acad Sci U S A. 2013;110:3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghavami A, Veenhoff LM, van der Giessen E, Onck PR. Probing the disordered domain of the nuclear pore complex through coarse‐grained molecular dynamics simulations. Biophys J. 2014;107:1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Naim B, Zbaida D, Dagan S, Kapon R, Reich Z. Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 2009;28:2697–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atkinson CE, Mattheyses AL, Kampmann M, Simon SM. Conserved spatial organization of FG domains in the nuclear pore complex. Biophys J. 2013;104:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghavami A, van der Giessen E, Onck PR. Energetics of transport through the nuclear pore complex. PLoS One. 2016;11:e0148876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Colwell LJ, Brenner MP, Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLoS Comput Biol. 2010;6:e1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feldherr C, Akin D, Littlewood T, Stewart M. The molecular mechanism of translocation through the nuclear pore complex is highly conserved. J Cell Sci. 2002;115:2997–3005. [DOI] [PubMed] [Google Scholar]
- 42. Stewart M, Baker RP, Bayliss R, et al. Molecular mechanism of translocation through nuclear pore complexes during nuclear protein import. FEBS Lett. 2001;498:145–149. [DOI] [PubMed] [Google Scholar]
- 43. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. [DOI] [PubMed] [Google Scholar]
- 44. Williams RJP. The conformation properties of proteins in solution. Biol Rev. 1979;54:389–437. [DOI] [PubMed] [Google Scholar]
- 45. Milles S, Lemke EA. Single molecule study of the intrinsically disordered FG‐repeat nucleoporin 153. Biophys J. 2011;101:1710–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamada J, Phillips JL, Patel S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uversky VN, Dunker AK. Understanding protein non‐folding. Biochim Biophys Acta, Proteins Proteomics. 2010;1804:1231–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. [DOI] [PubMed] [Google Scholar]
- 49. Popken P, Ghavami A, Onck PR, Poolman B, Veenhoff LM. Size‐dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol Biol Cell. 2015;26:1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ruba A, Luo W, Kelich J, Tingey M, Yang W. 3D tracking‐free approach for obtaining 3D super‐resolution information in rotationally symmetric biostructures. J Phys Chem B. 2019;123:5107–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruba A, Kelich J, Ma J, Yang W. Reply to ‘deconstructing transport‐distribution reconstruction in the nuclear‐pore complex’. Nat Struct Mol Biol. 2018;25:1062–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Junod SL, Ruba A, Kelich JM, Yang W. Nuclear export of mRNA molecules studied by SPEED microscopy. Methods. 2019;153:46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo‐electron tomography. Structure. 2012;20:998–1006. [DOI] [PubMed] [Google Scholar]
- 55. Kahms M, Lehrich P, Hüve J, Sanetra N, Peters R. Binding site distribution of nuclear transport receptors and transport complexes in single nuclear pore complexes. Traffic. 2009;10:1228–1242. [DOI] [PubMed] [Google Scholar]
- 56. Mahadevan S. In: Cruse TA, editor. Monte Carlo Simulation: Reliability‐based mechanical design. New York, NY: Marcel Dekker, 1996; p. 123–146. [Google Scholar]
- 57. Mooney ZC. Monte Carlo Simulation. Thousand Oaks, CA: Sage Publications, 1997. [Google Scholar]
- 58. Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 | Disordered level, AA composition and Ch/Hy ratio for IDPs
Disorder levels obtained via IUPRED, AA sequences and compositions, and Ch/Hy ratios were shown for both FG‐ and non‐FG IDPs.1 Disordered level score above 0.5 were considered highly disordered. Hydrophobic and charged AA residues were shown in red and blue respectively.
Figure S2 | 3D spatial transport routes for passive and facilitated transports of folded proteins mapped by SPEED Microscopy
(a) A single NPC was illuminated by the inclined point spread function (iPSF) of SPEED microscopy at the NE of a stable HeLa cell line expressing green fluorescent protein (GFP)‐POM121. The iPSF was achieved by shifting an incident laser beam off the center of an objective's back aperture, resulting in an angle of 450 relative to the focal plane and the effective PSF smaller than the average nearest distance between NPCs in X, Y, or Z direction.2, 3 The iPSF was further used to track single‐folded protein molecules or complexes labeled in distinct colors from GFP moving through the NPC. Capturing the trajectories of these transiting molecules (red dots) and superimposing the data over the centroid position of the NPC (blue dot) results in 2D projection (XY plane) of the nuclear transport process. N, nucleus; C, cytoplasm. (b) Collecting hundreds of single‐molecule trajectories for GFPs passively diffusing through the NPC generated a 2D spatial super‐resolution distribution (green dots) of GFPs in the NPC, overlaid with the NPC's 2D schematics (grey). (c) A 2D spatial super‐resolution distribution (red dots) of a facilitated translocation complex of NLS‐2xGFP/Importin β/Importin α in the NPC, overlaid with the NPC's 2D schematics (g).2, 4 (d) A 2D to 3D transform algorithm was utilized to obtain the 3D transport routes of the above 2D passive (b) and facilitated (c) transport data. As shown in the Cylindrical coordinates (x, R, and θ), passive diffusion (green clouds) and facilitated transport (red clouds) of folded proteins possess distinct pathways through the NPC.2 Figure a‐d were adapted from our previous publications with permissions.4
Figure S3 | 2D and 3D nuclear transport data for 2x green fluorescent protein (GFP) and dextrans. 2D super‐resolution locations and 3D spatial probability density maps are shown for 2xGFP (c) and dextrans ranging in size from 10 to 70 kDa (a, b and d). 3D spatial probability density maps (green clouds) are superimposed with wireframe NPC schematic (grey).
Figure S4 | Determination of the central axis of the NPC illuminated by SPEED microscopy. Diagrams are used to demonstrate the detailed steps in our data analyses. (A) The initial single‐molecule tracking data (black points) are plotted around the NPC's marker for the central position (red dot) collected with SPEED. (B) The collected data after filtering by using single‐molecule localization filters (photon count, width of emission PSF). (C) The filtered 2D single‐molecule data shape the spatial location of the NE and a single NPC. Furthermore, the 2D single‐molecule distribution indicates the orientation of NPC, which must fall within a free angle of 1.4° perpendicular to the NE, for the data to be processed. The red dot represents the localized centroid of a fluorescently tagged NPC. Periodically, the localized NPC position does not perfectly overlap with the localized single‐molecule data from the fluorescently tagged INM proteins and must be corrected. (D‐G) Due to the rotational symmetry of the NPC, 2D single‐molecule data is expected to be equally distributed along the central axis in the X and Y dimensions. Histograms are prepared for the X and Y dimensional data, and if the histograms are asymmetrical, as in this example, fine adjustments are made to restore symmetry using the formula . If the peaks are symmetrical, this step is skipped. (H) The 2D single‐molecule data will then undergo the 2D to 3D transformation (Figure S5) to produce the 3D density map.
Figure S5 | Graphical demonstration of 2D to 3D conversion process A schematic demonstration, with simulated data, of the 2D to 3D transformation algorithms for molecules that diffuse through the NPC. (A) 3D spatial locations of randomly diffusing molecules inside the NPC can be coordinated in a cylindrical coordination system (R, Ɵ, Y) due to the cylindrical rotational symmetry of the NPC. The 3D molecular locations in the NPC (rainbow colored for Z position) are projected onto a 2D plane in a Cartesian coordination system (X and Y, shown as black points) by microscopy imaging (Y and Z shown as gray points). (B) The cross‐sectional view of all the locations (the same as the gray points from the Y and Z dimension). These locations can be grouped into the sub‐regions between concentric rings. Given the high number of randomly distributed molecules in the NPC the spatial density of locations (ρi) in each sub‐region (s(i,j)) between two neighboring rings will be rotationally symmetrical and uniform. These locations can be further projected into 1D along the Y dimension. The locations along the Y dimension can be clustered in a histogram with j columns. The total number of locations in each column (A(i,j)) is equal to , which is further explained in Figure S5. (C) Histograms of 2D projected data from microscopy experiments are identical to B, thereby allowing us to use the aforementioned formula to determine the density of each concentric ring. (D) Using the algorithms, 2D projected data can be used to reconstruct the 3D spatial distribution of protein location on the NPC. More detailed explanation and calculations can be found in Methods and also previous publications.2, 3
Figure S6 | Matrix equation flowchart. (𝑨(𝒊,𝒋)), area of the sub‐region, (i), radial number, (j), axial number, (𝝆𝒊), spatial probability density in each radial ring, (𝑺(𝒊,𝒋)), area of the sub‐region, (𝑵𝒋), number of events, and (∆𝒓), bin size. Equations 1–3 will determine the area of the sub‐region given certain parameters (i.e., i = j). Equation 4 will determine the number of events in the given area. Once the subregion area ((𝒊,)) and events (𝑵𝒋) are known, the spatial probability density in each radial ring (𝝆𝒊) can be calculated.
Figure S7 | A two‐state scenario for IDPs depending on Ch/Hy ratio A plot shown the relationship between Ch/Hy ratio and the probability of occupying either the axial center or periphery of the NPC during transport for the IDP candidates we tested in this paper. A two‐state scenario was revealed with a sigmoidal fitting determining the Ch/Hy ratio of 0.37 as a transition point. IDPs with a high Ch/Hy ratio above 0.37 have a high probability of occupying the axial center of the NPC and vice versa.
Table S1 | Nuclear transport kinetics of IDPs through the NPC In our measurements, more than 200 transport events were collected to determine the transport kinetics for each candidate ordered by decreasing Ch/Hy ratio. The reproducibility of each candidate was determined from the radial peak of the 2D to 3D transformation density map and the number (N) of single‐molecule localizations between the axial region of 70 to −100, cytoplasm to nucleus respectively.5, 6
Table S2 | Estimation of Hydrodynamic Radius of IDPs
Folded and unfolded percentages were estimated using the protein modeling software, Phyre2.7 The hydrodynamic radius (Rh) was calculate using R h = 4.75 N 0.29Å for the folded percentage of the candidates and R h = 2.21 N 0.57Å was used for the unfolded percentage of each candidate. N is the number of residues in the polypeptide chain.8
Table S3 | List of 80 human nuclear IDPs Analysis of 80 IDPs that are localized predominantly within the nucleus. The predicted 3D transport route through the NPC is given for each protein based upon the Ch/Hy ratio obtained. UNIPROT ID is provided for each for easy identification.
Video S1 │ Single IDP molecule interacting with a single NPC.
This video shows a typical transport event of a IDP through the NPC in a eukaryotic cell (CREST is used as an example). Pixels are 240‐nm square, each frame was acquired in 2 ms, and the playback speed is 1,000X slower than real‐time. The compartment on the left side of the NPC (the green fluorescent spot) is the Nucleoplasm(N), and the right side is the Cytoplasm (C).
Video S2 │ 3D diffusion route for IDPs diffusing through the NPC's central axial channel.
This video shows a cut‐away view of the 3D diffusion route of SLD2 through the central axial channel of the NPC (red cloud; brighter color indicates higher spatial density) superimposed on the NPC architecture (grey wire frame). Bar: 20 nm.
Video S3 │ 3D diffusion route for IDPs diffusing through the peripheral regions around the central axial channel in the NPC.
This video shows a cut‐away view of the 3D diffusion route of CREST through the periphery of the NPC (Green cloud; brighter color indicates higher spatial density) superimposed on the NPC architecture (grey wire frame). Bar: 20 nm.


