Abstract
Motile cilia protrude from cell surfaces and are necessary to create movement of cells and fluids in the body. At the molecular level, cilia contain several dynein molecular motor complexes including outer dynein arms (ODAs) that are attached periodically to the ciliary axoneme, where they hydrolyse ATP to create the force required for bending and motility of the cilium. ODAs are preassembled in the cytoplasm and subsequently trafficked into the cilium by the intraflagellar transport (IFT) system. In the case of the green alga Chlamydomonas reinhardtii, the adaptor protein ODA16 binds to ODAs and directly to the IFT complex component IFT46 to facilitate the ciliary import of ODAs. Here, we purified recombinant human IFT46 and ODA16, determined the high‐resolution crystal structure of the ODA16 protein, and carried out direct interaction studies of IFT46 and ODA16. The human ODA16 C‐terminal 320 residues adopt the fold of an eight‐bladed β‐propeller with high overall structural similarity to the Chlamydomonas ODA16. However, the small 80 residue N‐terminal domain, which in Chlamydomonas ODA16 is located on top of the β‐propeller and is required to form the binding cleft for IFT46, has no visible electron density in case of the human ODA16 structure. Furthermore, size exclusion chromatography and pull‐down experiments failed to detect a direct interaction between human ODA16 and IFT46. These data suggest that additional factors may be required for the ciliary import of ODAs in human cells with motile cilia.
Keywords: cilium, flagella, IFT46, intraflagellar transport, ODA16, outer dynein arms
1. INTRODUCTION
Cilia are conserved eukaryotic organelles that function in signaling, sensory reception, and motility. 1 They provide unicellular organisms with the ability to move in aqueous environments. In humans, motile cilia are found in the embryonic node where they establish the left–right asymmetry required for the correct positioning of inner organs, and in the brain where they propel the cerebrospinal fluid flow.2, 3 They also propel the extra‐cellular mucociliary flow that clears the airways of inhaled pathogens and create the fluid flow that moves the oocyte through the fallopian tubes to reach the uterus. Mutations in ciliary factors can thus result in primary cilia dyskinesia (PCD) with disease phenotypes such as bronchitis and pneumonia due to infections in the airways, and ectopic pregnancy resulting in reduced fertility in females.4, 5, 6, 7, 8, 9 In case of the male reproductive system, one long motile cilium powers the sperm cell and allows it to swim toward the oocyte for fertilization. Autosomal recessive mutations that hamper the functionality of the motile cilium often result in immotile sperm and male infertility.3, 5
The motility of cilia relies on conserved molecular structures including a central microtubule (MT)‐based axoneme onto which inner and outer dynein arms (ODAs) are periodically attached. 10 ODAs are macro‐molecular motors that consist of several heavy, intermediate, and light chains that assemble into large MDa ODA complexes. The dynein heavy chains are 4,000–5,000 residues long ATPases that power the sliding movement of MT doublets resulting in beating of the cilium.11, 12, 13 An elegant study by Lin and Nicastro used cryo‐electron tomography to demonstrate that an asymmetric distribution of dynein motor activity provide a molecular basis for ciliary beating. 14 Around 80% of PCD patients with structural defects in the ciliary axoneme were reported to have a total or partial absence of dynein arms. 3 Specifically, patient mutations in dynein arm heavy chains DNAH5, DNAH9, and DNAH11, intermediate chains DNAI1, DNAI2, the outer doublet MT inner protein TXNDC3 (FAP67 in Chlamydomonas reinhardtii) 15 and dynein light chain 1 DNAL1 were reported to result in PCD.3, 5
The cell relies on an intracellular transport process known as intraflagellar transport (IFT) to build the cilium and ferry ODAs from their site of assembly in the cytoplasm to their location of action in the axoneme. 16 IFT depends on the 22‐subunit IFT complex that serves as an adaptor for ciliary cargo.17, 18, 19 IFT was shown to be essential for spermiogenesis in mice and male Ift88 Tg737Rpw mutant mice carrying a hypermorphic mutation in the IFT subunit IFT88 are sterile. 20 The sperm count in Ift88 Tg737Rpw mice was 350 times lower than in wild‐type mice and the sperm flagella were completely immotile. 20 Consistent with this notion, disruption of a different IFT gene, Ift20, also resulted in infertile mice. 21 IFT20 is highly abundant in the testes of mice and IFT20 expression is upregulated during spermiogenesis. 21 Additionally, a recently published study demonstrated that the IFT protein IFT25, which is not required for ciliogenesis in somatic cells, 22 is essential for sperm flagella formation and fertility in mice, suggesting sperm‐specific function of certain IFT factors. 23 These studies clearly show that IFT is a required process for proper cilium formation in sperm cells and for fertility of mice. Interestingly, IFT proteins are present during spermiogenesis but are absent in mature sperm, suggesting that IFT is not required for the maintenance of sperm flagella. 20
Important work in the green alga model organism C. reinhardtii (Cr) revealed that the IFT of ODAs requires the IFT complex subunit IFT46 as well the cargo adaptor protein ODA16.24, 25, 26 The zebrafish ODA16 ortholog WDR69 is also required for axonemal dynein assembly and motility in this organism, demonstrating evolutionary conservation. 27 We recently investigated the Cr ODA‐ODA16‐IFT46 interaction network using purified proteins to show that the CrODA16 C‐terminal domain is required for the interaction with ODAs and that the cleft between the N‐ and C‐terminal domains of CrODA16 likely forms the CrIFT46 binding site. 28 Given that both IFT46 and ODA16 are conserved between Cr and Homo sapiens (Hs) (about 60% sequence identity for both proteins), it is a reasonable assumption that the mechanism of ciliary ODA trafficking is also conserved. Additionally, yeast‐2‐hybrid experiments identified a direct interaction between human ODA16 and mouse IFT46. 26 However, as we show here, the HsODA16 structure does not have the pronounced cleft between ODA16 domains and the purified protein does not interact directly with IFT46 in a high affinity complex in vitro. In contrast, the surface of the C‐terminal domain is completely conserved between Cr and Hs ODA16 proteins, suggesting that they interact with ODAs in a similar manner. Our data suggest that additional factors are required to tether ODA‐bound ODA16 to the IFT machinery for ciliary delivery.
2. RESULTS
2.1. Purification and crystal structure determination of HsODA16
To provide insights into the mechanism of IFT of mammalian ODAs, we overexpressed recombinant full‐length HsODA16‐His6 in insect cells (HsODA16 was not soluble when expressed in Escherichia coli) and purified the protein by affinity‐, ion‐exchange and size exclusion chromatography (SEC). Purified HsODA16 was highly soluble and could be concentrated to >30 mg/ml without showing signs of precipitation and was >95% pure judging from the SDS‐PAGE gel (Figure 1a). To verify that we purified full‐length HsODA16 and not a proteolysed fragment, total mass MS on a micrOTOF instrument was carried out, which resulted in an experimental mass that deviated only 0.11 Da from the theoretical mass of the his‐tagged protein (Figure 1b). The purified HsODA16 sample was of sufficient quality to obtain crystals and complete X‐ray diffraction data were collected to a resolution limit of 2.8 Å (Table 1). The crystallographic phase problem was overcome by molecular replacement using the previously determined structure of CrODA16 as a search model. Resulting electron density maps clearly revealed the presence of the β‐propeller domain of HsODA16, while only spurious density was observed for the N‐terminal domain (Figure 2). The structure of the β‐propeller domain was modeled and refined with a Rfree value of 28.4% and good stereochemistry (Table 1).
FIGURE 1.
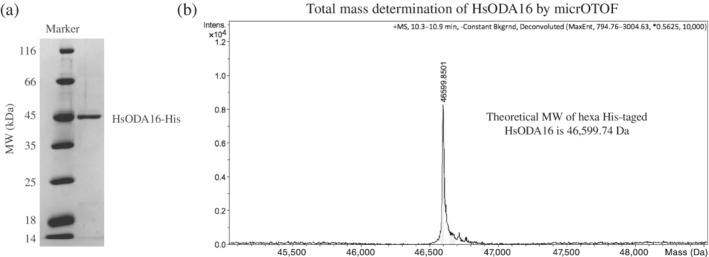
Purification and mass‐spec analysis of HsODA16. (a) SDS‐PAGE showing the purity of HsODA16 after size exclusion chromatography. (b) Total mass determination of purified hexa his‐tagged HsODA16 by mass spectrometry on a micrOTOF
TABLE 1.
X‐ray data collection and refinement statistics
| HsODA16 (5NNZ) | |
|---|---|
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50–2.8 (2.97–2.8) |
| Space group | P 21 |
| Unit cell parameters | a = 52.2 Å |
| b = 69.5 Å | |
| c = 102.2 Å | |
| α = 90° | |
| β = 90.0° | |
| γ = 90° | |
| Total reflections | 115,301 (16,503) |
| Unique reflections | 18,102 (2,798) |
| Multiplicity | 6.4 (5.9) |
| Completeness (%) | 99.5 (96.7) |
| Mean I/sigma (I) | 5.7 (1.0) |
| R‐merge | 0.217 (1.35) |
| CC1/2 | 0.973 (0.404) |
| R‐work | 0.226 (0.348) |
| R‐free | 0.284 (0.469) |
| Number of non‐hydrogen atoms | 5,248 |
| Macromolecules | 5,116 |
| Waters | 132 |
| Protein residues | 670 |
| RMSD bonds (Å) | 0.005 |
| RMSD angles (°) | 1.08 |
| Ramachandran favored (%) | 92.9 |
| Ramachandran outliers (%) | 0.60 |
| Clashscore | 18.2 |
| Average B‐factor (Å2) | 29.5 |
| Macromolecules | 30.0 |
| Solvent | 12.2 |
Note: Statistics for the highest‐resolution shell are shown in parentheses.
FIGURE 2.
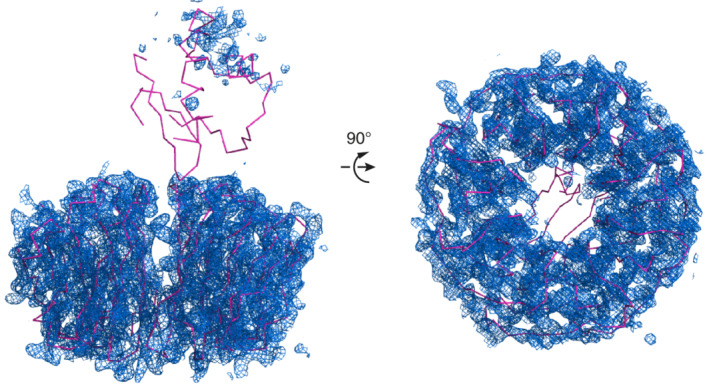
Crystallographic electron density map of HsODA16 after molecular replacement. Two orientations of CrODA16 (shown as pink lines) used in molecular replacement to determine the HsODA16 structure. The 2Fo‐Fc electron density map is shown as a blue meshwork. The map is calculated using phases from the CrODA16 structure (including the N‐terminal domain located on top of the β‐propeller) and is contoured at 1.0 sigma. The crystallographic map displays good electron density for the β‐propeller but only spurious uninterpretable density for the N‐terminal domain
2.2. The structure of HsODA16 reveals a conserved β‐propeller but the N‐terminal domain required for IFT46 association is disordered
The crystallographic map reveals well‐ordered electron density for the eight‐bladed β‐propeller corresponding to residues 81–415 of HsODA16 (Figure 2). The overall structure of the β‐propeller is very similar to that of the previously determined CrODA16 structure as the two models superimpose with a root‐mean‐square‐deviation of 0.8 Å. However, despite the fact that the full‐length HsODA16 was crystallized (Figure 1b), no electron density is observed for the N‐terminal 80 residues (Figure 2), which in case of CrODA16 folded into a small domain consisting of a three‐stranded β‐sheet and two α‐helices positioned on top of the β‐propeller (Figure 3). The N‐terminal domain of HsODA16 shares 46% sequence identity with the CrODA16 counterpart, which suggests that it adopts the same fold. However, the fact that residues 1–80 of HsODA16 were disordered in the crystal indicates that they are not stably attached to the β‐propeller, which is in contrast to what was observed for CrODA16. 28 This structural difference is significant as the cleft formed between the N‐ and C‐terminal domains of CrODA16 was mapped as the binding site for CrIFT46. 28 Our data thus challenge the notion of a direct physical interaction between human ODA16 and IFT46.
FIGURE 3.
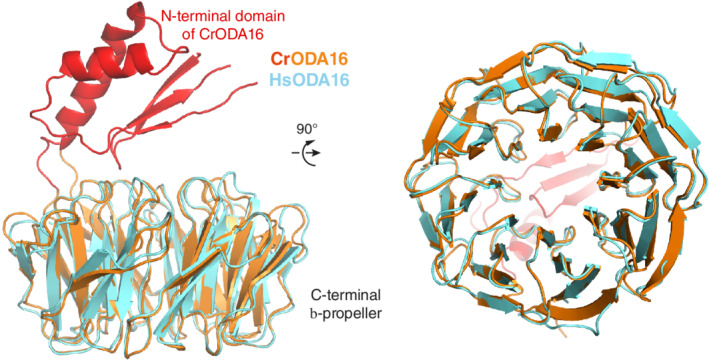
Structural superpositioning of CrODA16 and HsODA16. (Left) Crystal structure of HsODA16 shown as a cartoon and colored cyan. Comparison with the CrODA16 structure (shown in red and orange) reveals that both proteins consist of an eight‐bladed β‐propeller but HsODA16 does not have the small N‐terminal domain positioned on top of the β‐propeller. (Right) After rotating 90° around the horizontal axis, the C‐terminal faces of the Cr‐ and Hs‐ODA16 proteins are structurally highly conserved
2.3. HsODA16 contains a conserved acidic surface area that likely interacts with ODAs
In Chlamydomonas, the association with ODAs by ODA16 was shown to be mediated by the C‐terminal β‐propeller of ODA16. 28 ODAs likely associate with a highly acidic patch found on the C‐terminal face of the β‐propeller (Figure 4). 28 To assess if a similar acidic surface patch is found on HsODA16, we superposed the CrODA16 and HsODA16 structures as shown in Figures 3 and 4. All eight blades of the β‐propeller as well as the connecting loops are in very similar positions in the two structures. Interestingly, 10 of the residues that line the entrance to the central cavity of the β‐propeller are identical in CrODA16 and HsODA16 and form a highly conserved surface patch on the C‐terminal face of ODA16 (Figure 4a,c). This surface patch likely has an evolutionarily conserved role in the recognition of ODAs for ciliary trafficking. Of the 10 identical residues, six are glutamic or aspartic acids that form a prominently negatively charged surface area (Figure 4b). The remaining four conserved residues are aromatic/hydrophobic and three of these residues (Y112, F155, and M197) form a smaller mainly hydrophobic surface patch (Figure 4a,b). Given the highly conserved properties of the surface area of the C‐terminal face of the β‐propeller, it appears likely that ODA16 associates with ODAs using a similar mechanism in mammalian cells as in Chlamydomonas.
FIGURE 4.
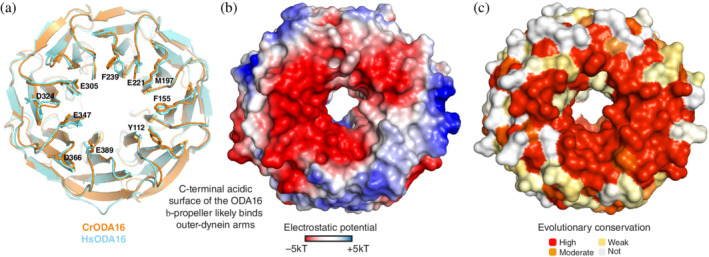
A highly conserved acidic surface patch on the C‐terminal face of ODA16. (a) Superpositioning of Cr‐ and Hs‐ODA16 structures in cartoon representation showing the C‐terminal face of the β‐propeller with 10 completely conserved surface residues displayed as sticks and numbered according to the HsODA16 sequence. The 10 conserved residues likely constitute a common binding site for ODAs. (b) surface representation of ODA16 colored according to electrostatic potential (negatively charged areas are colored red and positively charged areas are colored blue according to the color bar below the figure). The C‐terminal face of ODA16 is mainly negatively charged. (c) Surface representation of the mapping of amino acid conservation onto the ODA16 structure. (a–c) show ODA16 in the same orientation
2.4. HsODA16 does not bind directly to HsIFT46 in vitro
Given the structural analysis presented above, we set out to test if HsODA16 interacts directly with HsIFT46. HsIFT46 Isoform 1 was recombinantly expressed in E. coli, purified and incubated with HsODA16 purified from insect cells before subjecting the protein mix to SEC (Figure 5). The result of this experiment revealed that HsIFT46 and HsODA16 eluted from SEC as two separate peaks, which suggests that no complex was formed under the experimental conditions (Figure 5). As the elution peaks for HsODA16 and HsIFT46 partly overlap, the SEC profile shown in Figure 5 does not, however, rule out a low affinity interaction between HsODA16 and HsIFT46. The SEC elution profile is consistent with soluble and folded proteins as no elution peak was observed at the void volume, demonstrating that none of the proteins was aggregated (Figure 5). HsODA16 appears to be a globular protein as it elutes with an apparent mass of 42 kDa, which is very close to the theoretical mass of the his‐tagged protein of 46.6 kDa (Figure 5). Purified HsIFT46 on the other hand appears to have an elongated shape as it elutes in SEC with an apparent mass of 73 kDa (theoretical mass of 34 kDa for HsIFT46 Isoform 1). The fact that no complex formation was observed for HsODA16 and HsIFT46 in SEC is in sharp contrast to what was observed for CrIFT46 and CrODA16, which eluted as a stoichiometric complex during SEC. 28 Interaction studies using various salt concentrations demonstrated that the CrODA16‐IFT46 complex was stable at 200–300 mM NaCl, partly dissociated at 500–750 mM NaCl and almost completely broken apart at 1,000 mM NaCl. 28 The SEC experiment for HsODA16 and HsIFT46 shown in Figure 5 was carried out in a buffer containing 250 mM NaCl, a condition where the Chlamydomonas proteins formed a stoichiometric complex. 28 The binding region of CrIFT46 for CrODA16 was mapped to the N‐terminal 147 residues of CrIFT46 and the dissociation constant for the CrODA16‐IFT46 complex was measured to be ~200 nM. 28 Based on these observations, we conclude that HsIFT46 Isoform 1 does not interact strongly with HsODA16.
FIGURE 5.
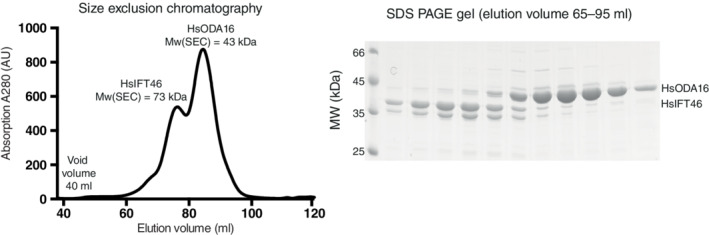
Co‐purification of Hs ODA16 and IFT46. (Left) Elution profile of pre‐incubated HsODA16 and HsIFT46 from a Hiload Superdex 200 SEC column. (Right) Coomassie stained SDS gel of the peak fractions from the SEC run. SEC, size exclusion chromatography
2.5. HsODA16 does not interact with HsIFT46 Isoforms 1 or 2 in pull‐downs
Database searches using Uniprot reveal that human IFT46 is expressed as two different isoforms (Isoform 1, Q9NQC8‐1 and Isoform 2, Q9NQC8‐2). As seen in Figure 6a, the amino acid sequences of the two HsIFT46 isoforms of differ mainly in the N‐terminal region that is responsible for ODA16 association in Chlamydomonas. The SEC experiment shown in Figure 5 was carried out with HsIFT46 Isoform 1 overexpressed in E. coli. Generally, proteins that form stable complexes in SEC interact with at least single digit μM affinity. To investigate if HsODA16 and HsIFT46 form a weak, lower affinity complex, we carried out pull‐down experiments using both isoforms of HsIFT46. Surprisingly, HsIFT46 Isoform 2 was not well expressed in E. coli or insect cells but could only be expressed in mammalian cells. We thus decided to express both HsIFT46 isoforms as GFP‐tagged constructs in HEK293 cells. This mammalian expression system may also allow for posttranslational modifications missing on the E. coli produced protein. The results of this experiment demonstrated no detectable direct interaction between GFP‐HsIFT46 and HsODA16‐His6 in buffer conditions containing 150 mM NaCl (Figure 6b). As this type of pull‐down experiment routinely detects affinities in the double digit μM range, 29 we conclude that a putative HsODA16‐HsIFT46 complex, if it exists at all, is at least two orders of magnitude weaker than the Chlamydomonas IFT46‐ODA16 complex.
FIGURE 6.
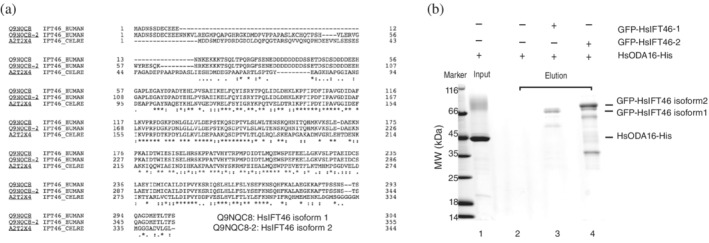
(a) Sequence alignment of the two isoforms of HsIFT46 with the CrIFT46 sequence. (*) completely conserved residues, (:) conservative substitution, and (.) somewhat conservative substitution that does not interchange hydrophobic and non‐hydrophobic residues. (b) GFP‐binder affinity pull‐down experiment of HsODA16 using GFP‐tagged HsIFT46 produced in HEK293 cells. Lanes: (1) 20% of HsODA16 input, (2) elution of GFP‐binder beads incubated with HsODA16 (negative control), (3) elution of GFP‐HsIFT46 Isoform 1 incubated with a 10× molar excess of HsODA16, and (4) elution of HsIFT46 Isoform 2 incubated with a 10× molar excess of HsODA16
3. DISCUSSION
We previously demonstrated that ODA16 of Chlamydomonas interacts with ODAs via the C‐terminal β‐propeller and couples to IFT46 via a pronounced cleft formed by the ODA16 β‐propeller and the small 80‐residue N‐terminal domain. 28 Curiously, the crystal structure of human ODA16 presented here demonstrates the absence of such an IFT46‐binding cleft. As we show by mass spectrometry that the N‐terminal 80 residues are present in the crystallized construct, we conclude that the N‐terminal domain of HsODA16 is not stably attached to the C‐terminal β‐propeller. Given these structural results, one possibility is that the N‐terminal domain of HsODA16 is merely flexible but becomes fixed upon association with HsIFT46. However, we do not observe a stable HsODA16‐IFT46 complex in SEC (Figure 5) suggesting that this hypothesis is not correct. Additionally, both identified isoforms of HsIFT46 harboring N‐terminal GFP‐tag were expressed in human cells and tested for association with C‐terminally hexa‐histidine tagged HsODA16 in affinity pull‐down experiments. We note that the presence of the N‐terminal GFP‐tag on HsIFT46 could interfere with the HsODA16 interaction. However, this was not the case for the Chlamydomonas system where the presence of the GST‐tag at the N‐terminus of the CrIFT46 did not hinder the association with CrODA16 in our previous pull‐down experiments. 28 The fact that mouse IFT46 interacted with human ODA16 in yeast‐2‐hybrid experiments could indicate a low affinity interaction or a false positive, perhaps as a result of the unspecified N‐terminal linker on human IFT46 generated by 5′ UTR sequences. 26 Based on our interaction data, we conclude that human ODA16 and IFT46 do not associate directly with each other in a high affinity complex in vitro.
Despite the evolutionary conservation of ODA16 and IFT46 between Chlamydomonas and human, only the Chlamydomonas but not the human proteins interact directly to form a stable heterodimeric complex. This finding suggests that the coupling of ODA16 to the IFT system, and thus the trafficking of ODAs, is different in the two species. It is possible that other IFT proteins, yet uncharacterized factors or posttranslational modifications in sperm cells are required to anchor the ODA‐ODA16 complex to the ciliary trafficking machinery. It is unlikely that association between HsODA16 and HsIFT46 is isoform specific as neither of the two HsIFT46 isoforms reported in the uniprot database interacts with HsODA16 (Figure 6). However, the fact that we do observe that the C‐terminal interface of the ODA16 β‐propeller is highly conserved between the human and Chlamydomonas structures suggests that the recognition of ODAs by ODA16 likely follows an evolutionarily conserved mechanism. Future experiments will aim at identifying factors required for the ciliary import of ODAs in human cells expressing motile cilia.
4. MATERIALS AND METHODS
4.1. Protein purification and interaction studies
The coding sequences for human IFT46 (uniprot accession code: Isoform 1, Q9NQC8‐1 and Isoform 2, Q9NQC8‐2) and ODA16 (uniprot accession code: Q8N136) were amplified from the Megaman DNA library and cloned with various N‐ and C‐terminal affinity tags (specified below). HsODA16 with a C‐terminal hexa‐histidine tag was expressed from recombinant baculovirus in HighFive cells as previously described for CrODA16. 28 Purification of HsODA16 was carried out as for CrODA16 but 250 mM instead of 500 mM NaCl was used in lysis and SEC buffers. 28 HsIFT46 Isoform 1 was expressed with a cleavable N‐terminal hexa‐histidine tag in E. coli and purified as previously described for CrIFT46. 30 Additionally, N‐terminally GFP‐tagged HsIFT46 (Isoforms 1 and 2) was produced in HEK293 cells and purified using the GFP‐nanobody coupled to sepharose beads (“GFP‐binder beads”). For the SEC experiment shown in Figure 5, N‐terminally GFP‐tagged IFT46 was incubated with 20% molar excess of C‐terminally hexa‐histidine tagged HsODA16 for 1 hr at 4°C and injected onto a HiLoad Superdex 200 column in a buffer containing 10 mM Hepes pH 7.5, 250 mM NaCl, 5% glycerol and 1 mM DTT. The affinity pull‐down experiment shown in Figure 6b was carried out as previously described with 150 mM NaCl in the binding and washing buffer.31, 32 Bound proteins were eluted from the GFP‐binder beads using 0.1 M citric acid and visualized by SDS‐PAGE.
4.2. HsODA16 crystallization and structure determination
Full‐length HsODA16 was crystallized using the hanging drop vapor diffusion method at 18°C by mixing 200 nl of purified HsODA16 at 26 mg/ml with 200 nl of the precipitant solution containing 50 mM Tris pH 8.0 and 50 mM NaOxalate. Crystals were cryo‐protected by the addition of 30% glycerol and flash‐cooled in liquid nitrogen. X‐ray diffraction data were collected at 2.8 Å resolution at the Swiss Light Source (SLS; Villigen, Switzerland) at the PXII beamline on a Pilatus 6M detector and indexed with the XDS package 33 before scaling with Aimless as part of the CCP4 package. 34 The crystals appeared to belong to a primitive orthorhombic lattice during indexing, but subsequent structure determination revealed the lattice to be monoclinic primitive with a beta angle close to 90°. The structure of HsODA16 was determined by molecular replacement using the CrODA16 structure (PDB code 5MZH) as a model and refined in the program Phenix. 35 The asymmetric unit contained a total of two chains; chain A was not well ordered and had high B‐factors whereas chain B was well ordered with low B‐factors. Chain B was consequently used for the structure representation shown in the figures (non‐crystallographic symmetry restraints were used in refinement). See Table 1 for data collection and refinement statistics.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Jiaolong Wang: Formal analysis; investigation; methodology; visualization; writing‐original draft; writing‐review and editing. Michael Taschner: Formal analysis; investigation; methodology; project administration. Narcis Petriman: Funding acquisition; project administration; supervision; writing‐review and editing. Marie Andersen: Investigation; methodology. Jerome Basquin: Data curation; formal analysis; investigation; methodology. Sagar Bhogaraju: Investigation; methodology. Melanie Vetter: Investigation; methodology. Stefanie Wachter: Investigation; methodology. Anna Lorentzen: Investigation; methodology. Esben Lorentzen: Conceptualization; data curation; funding acquisition; project administration; supervision; validation; writing‐original draft; writing‐review and editing.
ACKNOWLEDGMENTS
We would like to thank the crystallization facility at the Max‐Planck Institute of Biochemistry (MPIB) for invaluable help with crystallization screening, and the staff at the Swiss Light Source (SLS) at the Paul‐Scherrer‐Institute (PSI) in Villigen, Switzerland, for help with crystal data collection. Furthermore, we are grateful to Lissy Weyher and Dr Stephan Übel in the MPIB core facility for mass spectrometry. We thank the Novo Nordisk Foundation (Grant number NNF15OC0014164) and Carlsberg Foundation (Grant number CF19‐0253) for funding this project. Structural coordinates have been deposited at PDB, accession code 5NNZ.
Wang J, Taschner M, Petriman NA, et al. Purification and crystal structure of human ODA16: Implications for ciliary import of outer dynein arms by the intraflagellar transport machinery. Protein Science. 2020;29:1502–1510. 10.1002/pro.3864
Jiaolong Wang and Michael Taschner contributed equally to this study.
Funding information Carlsbergfondet, Grant/Award Number: CF19‐0253; H2020 European Research Council, Grant/Award Number: 888322; Novo Nordisk Fonden, Grant/Award Number: NNF15OC0014164
REFERENCES
- 1. Ishikawa H, Marshall WF. Ciliogenesis: Building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. [DOI] [PubMed] [Google Scholar]
- 2. Fliegauf M, Benzing T, Omran H. When cilia go bad: Cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. [DOI] [PubMed] [Google Scholar]
- 3. Ibañez‐Tallon I, Heintz N, Omran H. To beat or not to beat: Roles of cilia in development and disease. Human Mol Genet. 2003;12:R27–R35. [DOI] [PubMed] [Google Scholar]
- 4. Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. [DOI] [PubMed] [Google Scholar]
- 5. Zariwala MA, Omran H, Ferkol TW. The emerging genetics of primary ciliary dyskinesia. Proc Am Thorac Soc. 2011;8:430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horani A, Ferkol TW, Dutcher SK, Brody SL. Genetics and biology of primary ciliary dyskinesia. Paediat Resp Rev. 2016;18:18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirra V, Werner C, Santamaria F. Primary ciliary dyskinesia: An update on clinical aspects, genetics, diagnosis, and future treatment strategies. Front Pediat. 2017;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poprzeczko M, Bicka M, Farahat H, et al. Rare human diseases: Model organisms in deciphering the molecular basis of primary ciliary dyskinesia. Cell. 2019;8:E1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Afzelius B. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. [DOI] [PubMed] [Google Scholar]
- 10. King SM. Axonemal dynein arms. Cold Spring Harb Perspect Biol. 2016;8:a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibbons IR. Studies on the protein components of cilia from Tetrahymena pyriformis . Proc Natl Acad Sci USA. 1963;50:1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein‐2 primed for its power stroke. Nature. 2015;518:435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mizuno N, Taschner M, Engel BD, Lorentzen E. Structural studies of ciliary components. J Mol Biol. 2012;422:163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin J, Nicastro D. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science. 2018;360:eaar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, Zhang R. Structure of the decorated ciliary doublet microtubule. Cell. 2019;179:909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. [DOI] [PubMed] [Google Scholar]
- 17. Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016;8:a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhogaraju S, Engel BD, Lorentzen E. Intraflagellar transport complex structure and cargo interactions. Cilia. 2013;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. San Agustin JT, Pazour GJ, Witman GB. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. MBoC. 2015;26:4358–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z, Li W, Zhang Y, et al. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. MBoC. 2016;27:3705–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keady BT, Samtani R, Tobita K, et al. IFT25 links the signal‐dependent movement of hedgehog components to intraflagellar transport. Dev Cell. 2012;22:940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H, Li W, Zhang Y, et al. IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation. Biol Reprod. 2017;96:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahmed NT, Mitchell DR. ODA16p, a chlamydomonas flagellar protein needed for dynein assembly. MBoC. 2005;16:5004–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao C, Wang G, Amack JD, Mitchell DR. Oda16/Wdr69 is essential for axonemal dynein assembly and ciliary motility during zebrafish embryogenesis. Dev Dynam. 2010;239:2190–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taschner M, Mourão A, Awasthi M, Basquin J, Lorentzen E. Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J Biol Chem. 2017;292:7462–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vetter M, Stehle R, Basquin C, Lorentzen E. Structure of Rab11–FIP3–Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11. Nat Struct Mol Biol. 2015;22:695–702. [DOI] [PubMed] [Google Scholar]
- 30. Taschner M, Bhogaraju S, Vetter M, Morawetz M, Lorentzen E. Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex IFT52 binds directly to four other IFT‐B subunits. J Biol Chem. 2011;286:26344–26352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taschner M, Kotsis F, Braeuer P, Kuehn EW, Lorentzen E. Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J Cell Biol. 2014;207:269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taschner M, Weber K, Mourão A, et al. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin‐binding IFT‐B2 complex. EMBO J. 2016;35:773–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: A comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]


