Abstract
Ca2+/calmodulin‐dependent protein kinase II (CaMKII) is a Ser/Thr kinase necessary for long‐term memory formation and other Ca2+‐dependent signaling cascades such as fertilization. Here, we investigated the stability of CaMKIIα using a combination of differential scanning calorimetry (DSC), X‐ray crystallography, and mass photometry (MP). The kinase domain has a low thermal stability (apparent T m = 36°C), which is slightly stabilized by ATP/MgCl2 binding (apparent T m = 40°C) and significantly stabilized by regulatory segment binding (apparent T m = 60°C). We crystallized the kinase domain of CaMKII bound to p‐coumaric acid in the active site. This structure reveals solvent‐exposed hydrophobic residues in the substrate‐binding pocket, which are normally buried in the autoinhibited structure when the regulatory segment is present. This likely accounts for the large stabilization that we observe in DSC measurements comparing the kinase alone with the kinase plus regulatory segment. The hub domain alone is extremely stable (apparent T m ~ 90°C), and the holoenzyme structure has multiple unfolding transitions ranging from ~60°C to 100°C. Using MP, we compared a CaMKIIα holoenzyme with different variable linker regions and determined that the dissociation of both these holoenzymes occurs at a higher concentration (is less stable) compared with the hub domain alone. We conclude that within the context of the holoenzyme structure, the kinase domain is stabilized, whereas the hub domain is destabilized. These data support a model where domains within the holoenzyme interact.
Keywords: CaMKII, differential scanning calorimetry, mass photometry, oligomer dissociation, thermal stability
Abbreviations
- Ca2+/CaM
Ca2+/calmodulin
- CaMKII
Ca2+/calmodulin‐dependent protein kinase II
- cryo‐EM
cryo‐electron microscopy
- DSC
differential scanning calorimetry
- MP
mass photometry
- MW
molecular weight
1. INTRODUCTION
Ca2+/calmodulin‐dependent protein kinase II (CaMKII) is a Ser/Thr kinase that is a central signaling molecule in many systems throughout the body. In humans, CaMKII is encoded by four separate genes: CaMKIIα, β, γ, and δ. CaMKIIα and β are found predominantly in neurons and play a crucial role in long‐term memory formation.1, 2 CaMKIIδ is expressed in cardiomyocytes and contributes to cardiac pacemaking.3, 4 CaMKIIγ is found throughout the body in smooth muscle, skeletal muscle, and elsewhere, and also plays an essential role in eggs during fertilization.5, 6 All CaMKII genes are comprised of four domains: kinase, regulatory segment, variable linker region, and hub (Figure 1a). The kinase domain and regulatory segments are >90% identical between the four genes and the hub domains are >75% identical. CaMKII is activated by Ca2+/calmodulin (Ca2+/CaM) competitively binding to the regulatory segment and dislodging it from the substrate‐binding site, making the pocket accessible. The structure of CaMKII has recently been reviewed. 7
FIGURE 1.
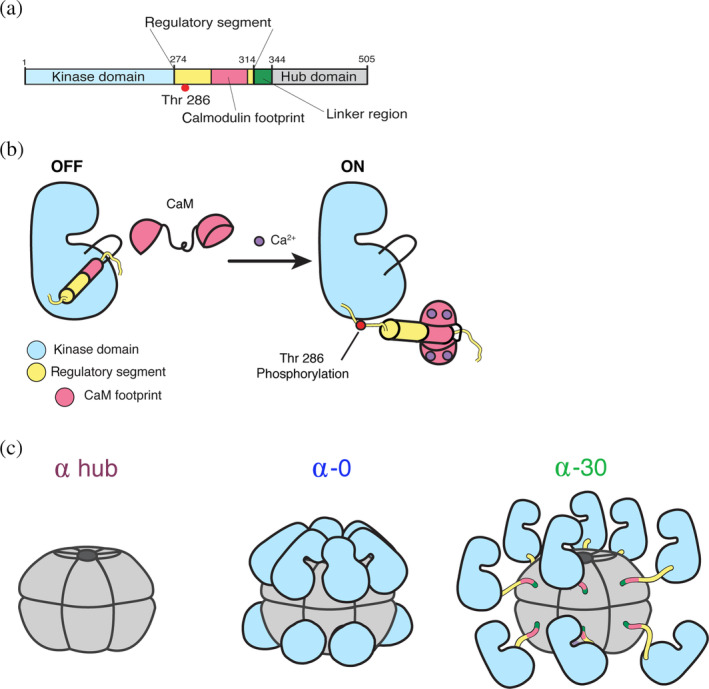
CaMKII architecture. (a) Linear depiction of the CaMKII subunit organization. Domain colors are consistent throughout the figure. (b) Ca2+/calmodulin competitively binds the regulatory segment to expose the substrate‐binding pocket and activate the enzyme. This allows for Thr286 to be trans‐phosphorylated. (c) Cartoon depictions of the hub domain alone, CaMKIIα‐0 (compact conformation), and CaMKIIα‐30 (extended conformation). CaMKII, Ca2+/calmodulin‐dependent protein kinase II
The hub domain of CaMKII is responsible for oligomerization. In this series of experiments, we will focus on human CaMKIIα specifically. The CaMKIIα hub domain alone 8 and the CaMKIIα holoenzyme 9 were both crystallized as dodecameric assemblies. There is agreement in recent studies using both single‐particle negative stain and cryo‐electron microscopy (EM) that autoinhibited CaMKIIα holoenzymes form both dodecamers and tetradecamers.8, 10, 11 There is evidence from the full‐length CaMKIIα (with no variable linker) crystal structure 9 and single‐particle cryo‐EM 11 that the kinase domains directly contact the hub domain.
The hub domain organizes the kinases into an oligomeric conformation, which confers several unique biophysical properties to the enzyme including highly cooperative and frequency‐dependent activation and activation‐dependent exchange of subunits between holoenzymes. CaMKII activation is highly cooperative, where binding of one Ca2+/CaM potentiates adjacent subunits to bind Ca2+/CaM, likely due to interactions between subunits within the holoenzyme ring.12, 13, 14 It has been shown in vitro that CaMKII has a threshold frequency for activation; meaning that above a specific frequency of Ca2+, CaMKII will turn on; but below this frequency, it will remain off.9, 15, 16 Stochastic kinetic simulations suggest that this frequency response is a product of the rates of 1kinases binding to the hub domain, 2Ca2+/CaM binding to the regulatory segment, and 3Thr286 transphosphorylation. 9 CaMKII activity biosensors, Camui 17 and FRESCA (föster resonance energy transfer (FRET) sensor for CaMKII activity), 18 have been used to monitor CaMKII activity in response to Ca2+ oscillations in live cells. These studies indicate that also in vivo, CaMKII activation is cooperative and varies depending on the frequency of Ca2+ delivered.18, 19 Finally, Ca2+/CaM binding and subsequent phosphorylation of Thr 286 result in autonomous CaMKII activity and induce exchange of subunits between holoenzymes.8, 20 Significant evidence supports a mechanism where the regulatory segment interacts with the hub domain, resulting in the release of a vertical dimer unit, which can then assemble into a nearby CaMKII holoenzyme.8, 20
Structural regulation of CaMKII activity appears to depend on the combination of variable linker and hub identity. In CaMKIIα, completely removing the variable linker region makes the enzyme much harder to activate (i.e., requires more Ca2+/CaM for activation).9, 11 Intriguingly, attaching a CaMKIIα kinase domain to the hub domain of CaMKIIβ with no linker makes the enzyme easier to activate. 11 Little is known about the assembly or disassembly of CaMKII holoenzymes, other than the assertion that the vertical dimer is likely the unit of subunit exchange, and possibly also the unit of assembly. Motivated to further understand how the multiple domains of CaMKII influence one another from a biophysical perspective, herein, we focus on characterizing the stability of the CaMKIIα holoenzyme.
2. RESULTS
2.1. CaMKII protein domain stability
We employed differential scanning calorimetry (DSC) to measure the thermal stability of CaMKIIα domains in order to dissect how each domain affects the overall stability of the holoenzyme (Figures 2 and S1). We separately measured the melting temperatures of the kinase domain, kinase domain with regulatory segment, hub domain, holoenzyme with no linker, and holoenzyme with a 30‐residue linker (CaMKIIα(14, 18)). The nomenclature CaMKIIα(14, 18) designates that this variant of CaMKIIα includes exons 14 and 18, which results in a 30‐residue linker. For simplicity, since we are only using two variants of CaMKIIα, we will refer to these as CaMKIIα‐0 (no linker version) and CaMKIIα‐30 (30‐residue linker). The transcripts of both these variants are found in human hippocampus. 11
FIGURE 2.
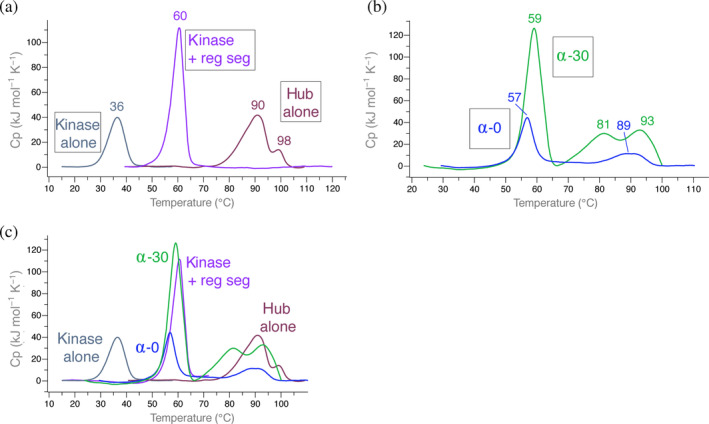
Differential scanning calorimetry of CaMKII domains. Representative traces are shown as well as the average apparent T m rounded to the nearest integer calculated from all replicates (see replicate data sets in Figure S1). (a) From left to right: overlays of CaMKIIα kinase domain without (gray) and with (purple) regulatory segment and the CaMKIIα hub only (burgundy). (b) Overlays of CaMKIIα‐0 (blue) and CaMKIIα‐30 (green). (c) Overlays of all traces for comparison. CaMKII, Ca2+/calmodulin‐dependent protein kinase II
We first measured the kinase domain alone, and the maximum temperature of unfolding was 36°C (Figure 2a). Adding the regulatory segment to the kinase domain stabilized it by a remarkable 24°C (apparent T m = 60°C). The hub domain alone was extremely stable, with the majority of the sample unfolding at 90°C. There is an additional transition that is stabilized by ~8° (apparent T m = 98°C). Fusing the kinase domain to the hub domain with no variable linker region (CaMKIIα‐0) results in a transition very similar to that of the kinase domain plus regulatory segment (apparent T m = 57°C) (Figure 2b). In the presence of a 30‐residue disordered variable linker region (CaMKIIα‐30), there is a small stabilization by 2° (apparent T m = 59°C).
Both CaMKIIα‐0 and CaMKIIα‐30 had additional transitions that are approximately the same maximum temperature of unfolding as the hub domain alone (Figure 2c). These transitions had significantly lower enthalpy (ΔH) compared with the major transition around 56–58°C. CaMKIIα‐30 has an additional transition (apparent T m = 81°C) with a similar ΔH as the most stable transition (apparent T m = 93°C) (Figure 2b).
2.2. Crystal structure of the CaMKII kinase domain
We crystallized the CaMKII kinase domain with an inactivating mutation (D135N) and lacking the regulatory segment (Figure 3a, PDB: http://firstglance.jmol.org/fg.htm?mol=6VZK). In the structure, the ATP‐binding site is occupied with p‐coumaric acid, which was present in the silver bullet additive in our screen. ATP binding stabilizes the kinase domain by 4°C (apparent T m = 40°C) (Figure 3b). The benzene ring of p‐coumaric acid mimics where the adenine base of ATP would be oriented. The carboxylic acid tail of p‐coumaric acid is in close contact with K42 (2.87 Å) and D156 (3.34 Å). This lysine (K42 in CaMKIIα) is highly conserved among protein kinases to facilitate ATP binding, specifically phosphate group orientation.21, 22 The hydroxyl group of p‐coumaric acid is in contact with the backbone of V92 (3.27 and 2.85 Å).
FIGURE 3.
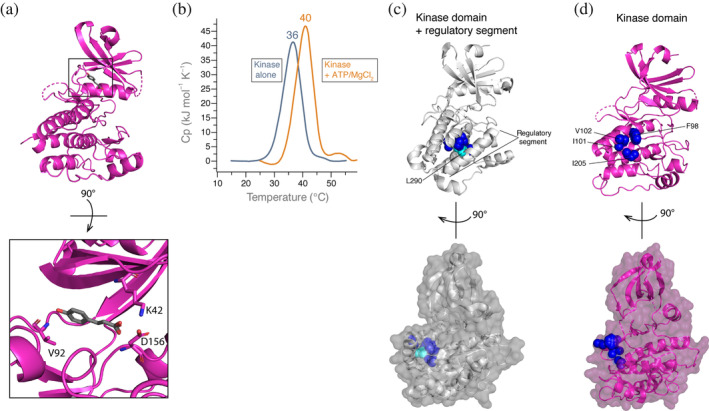
Crystal structure of the CaMKII kinase domain. (a) Kinase domain alone crystallized bound to p‐coumaric acid. Inset (bottom) highlights residues coordinating p‐coumaric acid. (b) DSC comparing kinase alone with no ATP or MgCl2 (gray, shown in Figure 2a) to kinase domain plus 1 mM ATP/10 mM MgCl2 (orange). (c) Crystal structure of kinase domain plus regulatory segment (PDB: http://firstglance.jmol.org/fg.htm?mol=2VZ6). Hydrophobic residues on the kinase domain (dark blue, labeled in 3D) are buried by the regulatory segment, specifically interacting with L290 (cyan). Surface representation is shown below at a 90° rotation. (d) Hydrophobic residues on the kinase domain (dark blue) are solvent exposed. Surface representation is shown below at a 90° rotation. CaMKII, Ca2+/calmodulin‐dependent protein kinase II; DSC, differential scanning calorimetry
In its autoinhibited state, the helical CaMKII regulatory segment occupies the substrate‐binding site (PDB: http://firstglance.jmol.org/fg.htm?mol=2VZ6). 23 This binding pocket on the kinase domain contains several hydrophobic residues, which interact with L290 on regulatory segment, protecting these residues from the solvent 23 (Figure 3c). While F98, I101, and I205 residues are completely buried, V102 is only partially buried as one gamma carbon atom is still exposed. In the absence of the regulatory segment, these hydrophobic residues are solvent exposed, shown in our structure (Figure 3d). Similar to the active conformation (PDB: http://firstglance.jmol.org/fg.htm?mol=2WEL 23 ), the αD helix reorients toward where the regulatory segment was bound (Figure S2). This movement of the αD helix leaves the hydrophobic amino acid patch unoccupied, resulting in complete exposure to the solvent (Figure 3d).
2.3. Analysis of CaMKII dissociation using mass photometry
We employed mass photometry (MP) to analyze the dissociation of CaMKII oligomers. 24 We measured the molecular weight (MW) of single molecules of CaMKII in solution over a concentration range to determine the concentration at which CaMKII oligomers dissociate. We used MP to compare the CaMKIIα hub domain alone to the full‐length holoenzyme with and without a variable linker region (CaMKIIα‐30 and CaMKIIα‐0, respectively) over a subunit concentration range of 100–45 nM (Figure 4; for raw count profiles, see Figures S3–S5). For all replicates, we observed populations of molecules at high‐MW values (200–600 kDa). For most samples, we also observed the clear emergence of a population of molecules at low‐MW values (50–200 kDa).
FIGURE 4.
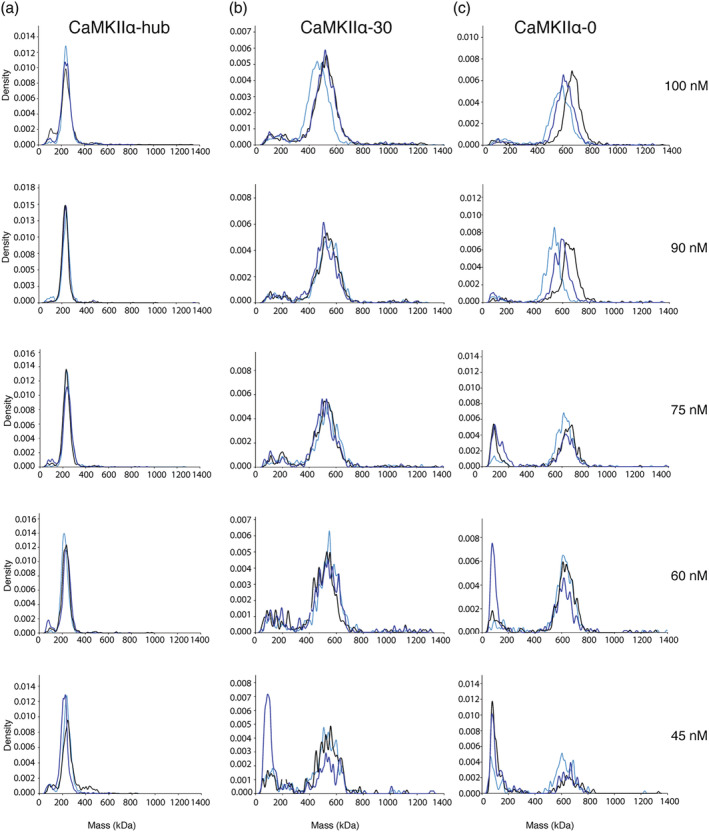
Mass photometry analysis of CaMKII variants. Overlays of three replicates for each concentration measured (100, 90, 75, 60, and 45 nM) are shown for CaMKIIα‐hub (a), CaMKIIα‐30 (b), and CaMKIIα‐0 (c). Density is plotted against mass (kDa). Raw counts were converted to kernel density, which is used to better depict multistep processes. CaMKII, Ca2+/calmodulin‐dependent protein kinase II
The hub domain was the best‐behaved sample across all concentrations tested (Figures 4a and S2). At 100 nM subunit concentration, there was a clear major population of molecules with an average MW of 239 ± 2 kDa, which corresponds roughly to a 16mer (hub subunit = 15.3 kDa). This large oligomeric species was still intact even at 45 nM. The size distribution of large hub oligomers is centered on 16–18 subunits per molecule at these concentrations (Figure S6a). At all concentrations except 90 nM, there was an additional population of molecules at a lower MW (~80–100 kDa). The ratio of molecules in the high‐MW population compared with low‐MW population remained high (22‐fold) across all samples.
Both versions of the CaMKII holoenzyme tested were less stable than the hub domain alone (Figure 4b,c). At concentrations of 90–100 nM, CaMKIIα‐30 had a major population of molecules at a high MW: 519 ± 23 kDa, which roughly corresponds to a 10mer (CaMKIIα‐30 subunit = 53.1 kDa) (Figure S6b). At these same concentrations, CaMKIIα‐0 also had a major population of molecules at a high MW: 606 ± 50 kDa, which roughly corresponds to a 12mer (CaMKIIα‐0 subunit = 50.4 kDa) (Figures 4c and S6c).
CaMKIIα‐30 and CaMKIIα‐0 showed an increase in the number of low‐MW molecules across the dilution series. For CaMKIIα‐30, the low‐MW population was evident at 100 nM, and increased relative to the high‐MW population at the lowest concentrations tested (45 and 60 nM). The majority of the low‐MW species were roughly 95–245 kDa, corresponding to a dimer/pentamer. Similarly for CaMKIIα‐0, the low‐MW population increased at 75 nM and continued to increase at the lowest concentrations tested (60 and 45 nM). The majority of the low‐MW CaMKIIα‐0 species were 77–151 kDa, corresponding to a monomer/dimer/trimer.
From our MP measurements, we summed the total number of molecules within specific MW ranges consistent with the size of larger oligomers (Figure S7; see Section 4 for calculation details). For the hub domain samples, the percentage of high‐MW molecules does not change over the concentration range we tested. This indicates that the K d for the hub domain alone must be lower than that of the holoenzyme. For CaMKIIα‐30 and CaMKIIα‐0, there is a clear increase in the percentage of high‐MW molecules over this concentration range, indicating that we are sampling near the K d.
3. DISCUSSION
It has been known that CaMKII is a large protein assembly since 1983, when it was determined to be a dodecamer. 25 However, still little is known about the assembly and disassembly of CaMKII, including estimates for K d. This has been a difficult question to tackle due to technical limitations in addition to indirect evidence suggesting CaMKII oligomerization must have a very low K d. Our stability measurements combined with MP shed new light on this old problem.
Our DSC data show that the hub domain alone is extremely stable, with the majority unfolding around 90°C (Figure 2). The second transition has a 2.5‐fold lower ΔH and is stabilized to 100°C. This transition is reproducible between all samples, which may represent a final unfolding transition such as monomer unfolding. Further experiments are needed to fully elucidate this. Conversely, the kinase domain is thermally unstable, with the majority unfolding around 36°C. This is similar to that measured for mitogen‐activated protein kinase. 26 Addition of the regulatory segment to the CaMKII kinase domain results in a remarkable 26°C stabilization. Our crystal structure of the CaMKII kinase domain shows several hydrophobic residues (F98, I101, V102, and I205) that are solvent exposed in the absence of the regulatory segment (Figure 3). This is likely an entropic cost that accounts for the stability difference we observe. This hydrophobic pocket is largely responsible for substrate binding; substrate binding (or inhibitors such as CaMKIINtide) would likely result in a similar stability shift, but this will require further testing. The crystals were obtained without any nucleotide added; however, an additive (p‐coumaric acid) was resolved in the ATP‐binding site. As we suspected, and is true for other kinases, 27 ATP/MgCl2 binding stabilizes the kinase domain by 4°C (Figure 3b). Adding MgCl2 without ATP may also have an effect on stability, which will be tested in future studies.
CaMKII holoenzyme stability falls in the middle of these two domains, indicating significant interactions and influence between domains within the oligomer. For CaMKIIα‐30 and CaMKIIα‐0, the first large transition around 56°C (~40–60 kJ/mol) is likely the kinase domains unfolding. Included in this transition may also be dissociation of the hub domain into dimers. The second and third unfolding transitions, which produced a smaller heat release (~10–15 kJ/mol), may indicate the unfolding of hub dimers or intact hub domains with unfolded kinase tails. Interestingly, the first transition is comparable between CaMKIIα‐30 and CaMKIIα‐0, whereas the right‐shifted transitions are not exactly overlapping (Figure 2c). This may indicate that the 30‐residue variable linker region additionally destabilizes the hub domain to induce unfolding of some hub domains at a lower temperature (transition 2, 81°C).
Structures of the human hub domains have revealed both sixfold and sevenfold symmetries, forming dodecamers or tetradecamers.8, 23, 28 More recently, CaMKII hub‐like domains from green algae have been shown to assemble into 16–20mers. 29 In our MP measurements with human CaMKIIα hub, the high‐MW population roughly corresponds to a 16mer. It is clear that this oligomeric complex is very stable since it does not dissociate significantly at 45 nM (Figures 4a and S7). Though, it is clear that at 45 nM those additional populations are increasing in the sample, suggesting that we are approaching the K d. Further experiments are needed to determine whether these hubs are actually forming 16mers at these concentrations and also to determine the K d.
Structures of human CaMKII holoenzymes have also revealed both sixfold and sevenfold symmetries.8, 9, 10, 11 The crystal structure of CaMKIIα‐0 showed a dodecamer with all the kinases docked onto the hub domain. 9 Cryo‐EM studies of CaMKIIα‐30 showed that both stoichiometries are adopted, in addition to populations where the kinase is docked onto the hub domain in different orientations. 11 In our MP measurements of CaMKIIα‐0, the high‐MW population corresponds to 12mer/14mer, which is consistent with existing data. For CaMKIIα‐30, we observe a high‐MW population corresponding to 10mer/12mer. Decameric CaMKII assemblies have been observed in solution‐based anisotropy measurements 30 and negative stain EM. 31 Since CaMKIIα‐30 has a disordered region and likely leads to a myriad of possible conformations for all kinase domains, this flexibility may affect the contrast used to calculate MW in the interferometric scattering microscopy method. It is also unknown how dilution may affect oligomerization, since all structural studies are done at significantly higher concentrations by necessity. Regulation and implication of CaMKII stoichiometry are still largely not understood and will be the focus of future studies.
As CaMKII dissociates into smaller oligomers, interfaces that are normally buried must become solvent exposed. The solubility of these species is likely low. We suspect that the low‐MW molecules we measured are underrepresenting what is present in the solution due to adsorption to other surfaces or aggregation. In order to calculate the K d, it will be necessary to screen for conditions that stabilize these dissociated species but do not interfere with the contrast optics of the MP setup. In this way, an accurate number of low‐ and high‐MW molecules can be calculated at equilibrium. However, our measurements indicate that CaMKII dissociation may be cooperative. This is most evident in the CaMKIIα‐0 dilution series where the high‐MW population clearly starts to shift to 1–2 low‐MW populations (Figure 4c). Perhaps as CaMKII dissociates, it preferentially forms dimers and tetramers, as has been predicted from subunit exchange experiments, discussed below. The MP data are not as clear for CaMKIIα‐30, which may either be because the dissociation kinetics are different or because the variable linker region further destabilizes the released subunits, resulting in noisier measurements. As mentioned above, there is a hint of hub destabilization seen in the DSC measurement of CaMKIIα‐30, where there is a transition at a lower apparent T m (81°C) compared to CaMKIIα‐0. Stabilizing these species will help us to clarify this.
Based on our measurements, we have roughly estimated K d values of ~10–20 nM for CaMKII oligomerization. These new data have clear implications on understanding the mechanism of CaMKII subunit exchange.8, 20, 32 The concentration of holoenzymes in dendritic spines is estimated to be 100 μM, 33 indicating that complete dissociation of oligomers on a reasonable timescale in spines is unlikely. As previously discussed, the proposed mechanism for subunit exchange relies on holoenzyme activation by Ca2+/CaM followed by induced release of a vertical dimer by interaction between the regulatory segment and hub domain. Future MP experiments measuring active CaMKIIα will indicate whether activation also affects dissociation, and thereby potentially facilitates subunit exchange.
Importantly, subunit exchange measurements were largely performed using single‐molecule fluorescence measurements. 20 The concentration of CaMKII used during the exchange experiments was ~5 μM. In order to make the single‐molecule measurement, the sample was diluted to ~2 nM, affixed to a glass surface, and imaged. Given that this dilution is slightly below the apparent K d, the total number of exchange events may have been underestimated due to some dissociation under these conditions. However, in order to further dissect this, future MP experiments will be geared toward taking time points under equilibrium conditions to determine the rate of dissociation.
Both the DSC and MP measurements suggest that the kinase and hub domain influence one another and do not behave like beads on a string. This is in line with structural studies showing interactions between the kinases and the hub domain.9, 11 Additionally, when comparing kinase activity between CaMKIIα and CaMKIIβ with no linker, there are significant differences in EC50 value for Ca2+/CaM, indicating that different structures are being adopted. 11 It is logical that these two covalently linked domains have evolved ways to interact. 34
It will be interesting to test the remaining three CaMKII variants in terms of temperature stability and dissociation parameters. There are sufficient differences between the hub domain sequences (>80% identical) that may provide changes in oligomerization kinetics and stability. This may have further implication on understanding mixed oligomers, which are known to exist in the brain 35 and likely exist in other tissues as well.
4. MATERIALS AND METHODS
4.1. Molecular biology
The wild type (WT) CaMKIIα kinase, CaMKIIα kinase + regulatory segment, and both CaMKIIα holoenzymes (with and without linker) were cloned into a vector containing an N‐terminal His‐SUMO tag. The CaMKIIα hub was cloned into a pET‐28 vector with an N‐terminal 6xHis‐precission tag using Gibson assembly.
4.2. Protein purification
WT CaMKIIα kinase domain (residues 7–274, with and without D135N) and CaMKIIα kinase + regulatory segment (residues 7–314) were grown and co‐expressed in Escherichia coli BL21 with λ phosphatase. The cells were induced at 18°C with 1 mM isopropyl β‐d‐1‐thiogalactopyranoside and grown overnight. The cells were then suspended using buffer A (25 mM tris, pH 8.5, 150 mM KCl, 40 mM imidazole, and 10% glycerol), commercially available protease inhibitors (0.5 mM benzamidine, 0.2 mM 4‐benzenesulfonyl fluoride hydrochloride, 0.1 mg/ml trypsin inhibitor, 0.005 mM leupeptin, 1 μg/ml aproptinin, and 1 μg/ml pepstatin) plus 1 μg/ml DNAse, and then lysed. Purification was done at 4°C utilizing an ÄKTA Pure system (GE, Piscataway, NJ). Clarified lysate was loaded onto a Ni‐NTA column and eluted with an imidazole gradient, desalted into buffer C (25 mM tris pH 8.8, 50 mM KCl, 40 mM imidazole, 2 mM tris(2‐carboxyethyl)phosphine (TCEP), and 10% glycerol), and cleaved overnight with Ulp1 at 4°C. Following a subtractive step, the sample was then bound to a HiTrap Q FF column (GE) and eluded with a KCl gradient. Samples were concentrated and further purified and desalted using a Superdex 75 column into DSC buffer (25 mM tris pH 8, 150 mM KCl, 1 mM TCEP, and 10% glycerol). The pure protein was then frozen and stored at −80°C.
CaMKIIα‐0 (7–475, Δ324–354) and CaMKIIα‐30 (7–475) were co‐expressed with λ phosphatase using Rosetta (DE‐3)pLysS cells. They were purified as previously described. Holoenzymes were purified using a Superose 6 column (GE). All purified holoenzymes eluted consistently (retention volume 12 ml). Retention volumes were consistent with standards as well as other samples which we have verified as holoenzymes using cryo‐EM.
The CaMKIIα hub domain (residues 315–475) was expressed in BL21 cells and purified as described except a different buffer C (25 mM tris pH 8.8, 150 mM KCl, 1 mM ethylenediaminetetraacetic acid, 2 mM dithiothreitol, and 10% glycerol) to facilitate Prescission protease cleavage overnight at 4°C. Samples were eluted from the Superose 6 column into DSC buffer. All purified hubs eluted consistently (retention volume 14 ml). Pure protein samples were frozen and stored at −80°C.
4.3. Differential scanning calorimetry
All protein samples were diluted to 0.5 mg/ml in DSC buffer (25 mM tris pH 8, 150 mM KCl, 1 mM TCEP, and 10% glycerol). DSC measurements were performed on a MicroCal Automated PEAQ‐DSC instrument (Malvern Panalytical, Westborough, MA). For the CaMKIIα kinase + ATP/MgCl2, ATP (1 mM) and MgCl2 (10 mM) were used and added to the buffer blank as well. Unless otherwise indicated, after a 5‐min prescan equilibration step, samples were scanned from 10 to 120°C at a scan rate of 90°C/hr with no feedback. Data were analyzed using the MicroCal PEAQ‐DSC software, and baseline‐subtracted data were fit to a non‐two‐state fitting model to obtain apparent T m values.
4.4. Crystallization, data collection, and structure determination
Crystals of CaMKII kinase domain (residues 7–274) with an inactivating mutation (D135N) were grown at 4°C using hanging drop vapor diffusion. An additional mutation in this structure is Q223K, which was acquired during the cloning process. This residue is solvent exposed on the αG helix, and comparing this structure to previous structures with Q223 shows no effect on the fold or orientation. Crystals were obtained in the following condition: 8% polyethylene glycol 6000, 50 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES), pH 7.0, and H10 condition of the Silver Bullets screen (1:10 ratio) containing: 0.16 wt%/vol% 3‐aminobenzene‐sulfonic acid, 0.16% 5‐sulfosalicylic acid dihydrate, 0.16 wt%/vol% p‐coumaric acid, 0.16% pierazine‐N,N′‐bis(2‐ethanesulfonic acid), 0.16% terephthalic acid, 0.16% vanillic acid, and buffer 0.02 M HEPES pH 6.8. The crystal was cryo‐protected in 20% (vol/vol) ethylene glycol prior to being frozen in liquid nitrogen. Diffraction data were collected at a wavelength of 1.5418 Å using a Rigaku MicroMax‐007 HF X‐ray source, which was coupled to a Rigaku VariMax HF optic system (UMass Amherst, Amherst, MA). The X‐ray data were collected at 100 K. Data sets were integrated, merged, and scaled using HKL‐2000 (HKL research, Inc). 36 The structure was solved by molecular replacement with Phaser 37 using the coordinates of the kinase domain as an initial search model (PDB ID: http://firstglance.jmol.org/fg.htm?mol=3SOA 9 ). Model building was performed using Coot. 38 Coot and refinement were performed with REFMAC (CCP4 Suite). 39
4.5. Mass photometry
All MP experiments were carried out using an OneMP mass photometer (Refeyn LTD, Oxford, UK) at room temperature and used Acquire MP software for data collection. Coverslips (cat #630–2105) were cleaned by rinsing with a series of H2O, ethanol, and isopropanol (high performance liquid chromatography (HPLC) grade), and then dried under a clean stream of nitrogen. Gaskets (catalog #CW‐50R‐1.0) were cleaned using H2O and ethanol, and then dried under nitrogen. The gasket was then sealed onto the center of the glass coverslip. The coverslips were stored in a sterile container until used. Ten microliters of buffer (25 mM tris, 150 mM KCl, and 1 mM TCEP at pH 8.0) was added to the coverslip and focused. Images were collected in an area of 3 × 10 μm at a frame rate of 1 kHz. Protein stock solutions (2 μM) were diluted to 200 nM (25 mM tris, 150 mM KCl, and 1 mM TCEP at pH 8.0) right before the experiment. Proteins were further diluted to achieve the following concentrations in the measurements: 100 nM, 90 nM, 75 nM, 60 nM, and 45 nM. Three replicates were performed at each concentration. Collection time was varied to acquire the same number of binding events over the dilution series. Since the technical limit of the instrument at low MW is 50 kDa, we will not be able to fully resolve monomeric hub subunits (15.3 kDa). 40
4.5.1. MP data analysis
DiscoverMP software (Refeyn LTD, Oxford, UK) was used for all analyses using standard settings. Each replicate was analyzed individually. Only molecules that bound to the glass surface were counted. Populations were automatically fit with a Gaussian distribution. 24 For these populations, the mean Gaussian values from replicates were averaged to calculate an average MW; these values are reported ± the SD between three replicates. Some low‐MW populations could not be fitted for a Gaussian distribution due to insufficient sampling of multiple populations. In order to quantify the relative number of low‐ versus high‐MW species in the total number of molecules, we established MW cutoffs in line with replicates where low‐ and high‐MW populations were Gaussian distributed.
The MW ranges for low‐ and high‐MW species used are as follows—CaMKIIα‐hub: low = 14–161 kDa; CaMKIIα‐0: low = 50–400 kDa, high = 400–800 kDa; and CaMKIIα‐30: low = 50–300 kDa, high = 400–800 kDa. These ranges were applied to the following samples—CaMKIIα‐hub: low = 90, 75, 60, 45 nM; CaMKIIα‐0: low = 100, 90, 60 nM, and one replicate of 45 nM, high = 45 nM; and CaMKIIα‐30: low = 90, 75, 60 nM, 45 nM, high = 45 nM. For all other samples, the number of molecules was defined by the Gaussian fit.
AUTHOR CONTRIBUTIONS
Ana Torres‐Ocampo: Conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing‐original draft; writing‐review and editing. Can Özden: Data curation; methodology; writing‐review and editing. Alexandra Hommer: Data curation; formal analysis; writing‐review and editing. Anne Gardella: Data curation; formal analysis; writing‐review and editing. Emily Lapinskas: Investigation; methodology. Alfred Samkutty: Data curation; methodology; writing‐review and editing. Edward Esposito: Data curation; formal analysis; writing‐review and editing. Scott Garman: Formal analysis; visualization; writing‐review and editing. Margaret Stratton: Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; visualization; writing‐original draft; writing‐review and editing.
Supporting information
Figure S1 DSC replicates
Figure S2. Reorientation of the αD helix
Figure S3. Raw MP data: CaMKIIα hub
Figure S4. Raw MP data: CaMKIIα‐30
Figure S5. Raw MP data: CaMKIIα‐0
Figure S6. Size estimation of CaMKII variants from mass photometry
Figure S7. CaMKIIα oligomer dissociation.
ACKNOWLEDGMENTS
We would like to thank Dr. Matthias Langhorst and Dr. Sofia Ferreira for their technical assistance and helpful discussion of mass photometry data and also thank Dr. Roman Sloutsky for helpful discussions and manuscript editing.
Torres‐Ocampo AP, Özden C, Hommer A, et al. Characterization of CaMKIIα holoenzyme stability. Protein Science. 2020;29:1524–1534. 10.1002/pro.3869
Funding information National Institute of General Medical Sciences, Grant/Award Number: R01123157; National Institutes of Health, Grant/Award Number: T32 GM008515
REFERENCES
- 1. Rossetti T, Banerjee S, Kim C, et al. Memory erasure experiments indicate a critical role of CaMKII in memory storage. Neuron. 2017;96:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herring BE, Nicoll RA. Long‐term potentiation: From CaMKII to AMPA receptor trafficking. Annu Rev Physiol. 2016;78:351–365. [DOI] [PubMed] [Google Scholar]
- 3. Backs J, Backs T, Neef S, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106:2342–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang T, Kohlhaas M, Backs J, et al. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem. 2007;282:35078–35087. [DOI] [PubMed] [Google Scholar]
- 5. Escoffier J, Lee HC, Yassine S, et al. Homozygous mutation of PLCZ1 leads to defective human oocyte activation and infertility that is not rescued by the WW‐binding protein PAWP. Hum Mol Genet. 2016;25:878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoon SY, Jellerette T, Salicioni AM, et al. Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J Clin Invest. 2008;118:3671–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhattacharyya M, Karandur D, Kuriyan J. Structural insights into the regulation of Ca(2+)/calmodulin‐dependent protein kinase II (CaMKII). Cold Spring Harb Perspect Biol. 2019;a035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhattacharyya M, Stratton MM, Going CC, et al. Molecular mechanism of activation‐triggered subunit exchange in Ca2+/calmodulin‐dependent protein kinase II. Elife. 2016;5:e13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chao LH, Stratton MM, Lee IH, et al. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin‐dependent kinase II holoenzyme. Cell. 2011;146:732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Myers JB, Zaegel V, Coultrap SJ, Miller AP, Bayer KU, Reichow SL. The CaMKII holoenzyme structure in activation‐competent conformations. Nat Commun. 2017;8:15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sloutsky R, Dziedzic N, Dunn MJ, et al. Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub‐dependent regulation. BioRxiv; 721589; 2020. [DOI] [PMC free article] [PubMed]
- 12. Gaertner TR, Kolodziej SJ, Wang D, et al. Comparative analyses of the three‐dimensional structures and enzymatic properties of alpha, beta, gamma and delta isoforms of Ca2+−calmodulin‐dependent protein kinase II. J Biol Chem. 2004;279:12484–12494. [DOI] [PubMed] [Google Scholar]
- 13. Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. [DOI] [PubMed] [Google Scholar]
- 14. Chao LH, Pellicena P, Deindl S, Barclay LA, Schulman H, Kuriyan J. Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat Struct Mol Biol. 2010;17:264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. [DOI] [PubMed] [Google Scholar]
- 16. Bayer KU, De Koninck P, Schulman H. Alternative splicing modulates the frequency‐dependent response of CaMKII to Ca(2+) oscillations. EMBO J. 2002;21:3590–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takao K, Okamoto K, Nakagawa T, et al. Visualization of synaptic Ca2+/calmodulin‐dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ardestani G, West MC, Maresca TJ, Fissore RA, Stratton MM. FRET‐based sensor for CaMKII activity (FRESCA): A useful tool for assessing CaMKII activity in response to Ca(2+) oscillations in live cells. J Biol Chem. 2019;294:11876–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang JY, Nakahata Y, Hayano Y, Yasuda R. Mechanisms of Ca(2+)/calmodulin‐dependent kinase II activation in single dendritic spines. Nat Commun. 2019;10:2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stratton M, Lee IH, Bhattacharyya M, et al. Activation‐triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. Elife. 2014;3:e01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein‐kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 22. Knighton DR, Zheng JH, Teneyck LF, et al. Crystal‐structure of the catalytic subunit of cyclic adenosine‐monophosphate dependent protein‐kinase. Science. 1991;253:407–414. [DOI] [PubMed] [Google Scholar]
- 23. Rellos P, Pike AC, Niesen FH, et al. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8:e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young G, Hundt N, Cole D, et al. Quantitative mass imaging of single biological macromolecules. Science. 2018;360:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett MK, Erondu NE, Kennedy MB. Purification and characterization of a calmodulin‐dependent protein kinase that is highly concentrated in brain. J Biol Chem. 1983;258:12735–12744. [PubMed] [Google Scholar]
- 26. Krishna SN, Luan CH, Mishra RK, et al. A fluorescence‐based thermal shift assay identifies inhibitors of mitogen activated protein kinase kinase 4. PLoS One. 2013;8:e81504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bauer MS, Baumann F, Daday C, et al. Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc Natl Acad Sci U S A. 2019;116:6766–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosenberg OS, Deindl S, Comolli LR, et al. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006;273:682–694. [DOI] [PubMed] [Google Scholar]
- 29. McSpadden ED, Xia Z, Chi CC, et al. Variation in assembly stoichiometry in non‐metazoan homologs of the hub domain of Ca(2+)/calmodulin‐dependent protein kinase II. Protein Sci. 2019;28:1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarkar P, Davis KA, Puhl HL, Veetil JV, Nguyen TA, Vogel SS. Deciphering CaMKII multimerization using fluorescence correlation spectroscopy and homo‐FRET analysis. Biophys J. 2017;112:1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanaseki T, Ikeuchi Y, Sugiura H, Yamauchi T. Structural features of Ca2+/calmodulin‐dependent protein kinase II revealed by electron microscopy. J Cell Biol. 1991;115:1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh D, Bhalla US. Subunit exchange enhances information retention by CaMKII in dendritic spines. Elife. 2018;7:e41412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otmakhov N, Lisman J. Measuring CaMKII concentration in dendritic spines. J Neurosci Methods. 2012;203:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. [DOI] [PubMed] [Google Scholar]
- 35. Shen K, Teruel MN, Subramanian K, Meyer T. CaMKII beta functions as an F‐actin targeting module that localizes CaMKII alpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. [DOI] [PubMed] [Google Scholar]
- 36. Otwinowski Z, Minor W. Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. [DOI] [PubMed] [Google Scholar]
- 37. McCoy AJ, Grosse‐Kunstleve RW, Storoni LC, Read RJ. Likelihood‐enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. [DOI] [PubMed] [Google Scholar]
- 38. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 39. Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. [DOI] [PubMed] [Google Scholar]
- 40. Wu D, Piszczek G. Measuring the affinity of protein‐protein interactions on a single‐molecule level by mass photometry. Anal Biochem. 2020;592:113575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 DSC replicates
Figure S2. Reorientation of the αD helix
Figure S3. Raw MP data: CaMKIIα hub
Figure S4. Raw MP data: CaMKIIα‐30
Figure S5. Raw MP data: CaMKIIα‐0
Figure S6. Size estimation of CaMKII variants from mass photometry
Figure S7. CaMKIIα oligomer dissociation.


