Abstract
The placenta utilizes many mechanisms to protect the haploidentical fetus from recognition by the maternal immune system. However, in cases of villitis of unknown etiology (VUE), maternal lymphocytes gain access into the placenta, causing significant health risks for the fetus. Evidence suggests that VUE is a rejection response between the mother and the haploidentical fetus. Therefore, we profiled human leukocyte antigen (HLA), an important predictor of transplant rejection, in VUE using placental tissue from ten patients with VUE and ten gestational age matched controls. Placentas were stained using novel multiplexed immunofluorescence (MxIF) to investigate morphology and HLA classes I and II. Gene expression was evaluated by microarray, and where available, tissue typing of mother/baby pairs was completed to determine HLA type. MxIF demonstrated strong CD8+ T cell infiltration and HLA class I staining both the distal and stem villi of VUE placentas. Compared to controls, VUE cases had significantly higher expression of HLA class II mRNA and pathway analysis demonstrated that 40% of the differentially expressed genes in VUE are related to tissue rejection. The data suggest that VUE resembles a rejection response between the mother and the fetus. It remains unknown what initiates immune recognition and why some mothers appear to be at higher risk for developing this condition than others. Understanding this etiology will be critical for developing effective interventions or prevention strategies during pregnancy.
Electronic supplementary material
The online version of this article (10.1007/s43032-019-00101-9) contains supplementary material, which is available to authorized users.
Keywords: Placenta, HLA, Villitis, Inflammation, Rejection
Introduction
The establishment of maternal immune tolerance to the fetus is a key hallmark of reproduction that is mediated through the trophoblastic cells of the placenta, which interact directly with the maternal system during development; interestingly, many of these mechanisms are mimicked by cancer cells [1, 2]. Trophoblast cells are of fetal origin and play important roles in implantation, exchange of nutrients, gases and waste, as well as vasculature remodeling to provide adequate blood supply to the developing fetus. Additionally, trophoblasts use many mechanisms to keep maternal immune cells from recognizing the haploidentical fetus during pregnancy, including downregulating surface expression of human leukocyte antigen (HLA) [3]. HLA is expressed on all nucleated cells and is utilized by the immune system to determine self from nonself. As half of a fetus’ HLA type is paternally inherited, strict regulation of maternal immune responses to the semi-allogenic fetus is required for a successful pregnancy [4].
Although many mechanisms exist to protect the fetus from maternal recognition, histologic evaluation demonstrates that 7%–33% of placentas will exhibit various degrees of maternal lymphocyte infiltration and villous necrosis without infectious cause [5, 6]. This placental pathology is known as villitis of unknown etiology (VUE), and it can be associated with serious complications including intrauterine growth restriction [7, 8], preterm labor [9], and fetal demise [8, 10]. Additionally, placental lesions are often observed in infants born at term who are diagnosed with neurological impairment [11]. Although VUE is most commonly observed in late third trimester placentas, at time at which maternal immune tolerance is believed to be waning, the biology behind this condition remains poorly understood. Currently, there are two main hypotheses regarding pathogenesis: (1) VUE is the result of an undetected infection or (2) VUE represents a kind of allograft rejection. Another consolidating possibility is that an undetected infection triggers an allograft rejection-like response. The first hypothesis is driven by the fact that some cases originally diagnosed as VUE were later determined to be due to missed infection [12]. However, other studies that target ribosomal 16S DNA have failed to identify any pathogenic source in placentas with VUE [13]. The second hypothesis was proposed in 1993 and suggests that VUE is an inflammatory response that resembles transplant rejection [14]. This immunological rejection theory has gained support based on the fact that VUE is more prevalent in women with autoimmune disease [15, 16], in vitro fertilization pregnancies with egg donation (completely allogeneic fetus) [17, 18], and VUE tends to recur with subsequent pregnancies [10, 19].
The immune system appears to play a central role in placental VUE; therefore, our aim was to further define this biology in a cohort of patients diagnosed with VUE compared to unaffected controls. As HLA is a main driver of rejection responses, our goal was to focus on the expression and compatibility of this antigen in the placenta, mother, and child. Herein, we describe the histological, genetic, and tissue typing results obtained from human placentas diagnosed with VUE.
Methods
Patient Selection
This study was approved by the institutional review board at Mayo Clinic. The primary objective was to compare differences in immune responses between placentas with villitis of unknown etiology (VUE) and controls by imaging, gene expression, and tissue typing. Ten subjects with a positive high-grade VUE diagnosis by Amsterdam guidelines [20] (and no morphologic, clinical, or laboratory evidence of infection) were gestational age matched with ten subjects without any placental pathology. Specifically, any cases with plasma cell villitis, granulomas, or acute villitis/intervillositis/chorioamnionitis were excluded. We also excluded VUE cases that resulted in a demise to minimize potential confounding variables related to this multifactorial outcome. Residual formalin-fixed paraffin-embedded tissue was utilized after routine histopathologic assessment by a pathologist. Only samples that had been in blocks less than 1 year were included to maximize tissue quality.
Immunofluorescence
Tissues were reviewed by a pathologist prior to inclusion for analysis utilizing the multiplexed immunofluorescence (MxIF) method as previously described [21]. Briefly, tissues underwent antigen retrieval and were stained with DAPI so that approximately 20 regions of interest could be selected from each placenta. These representative regions included chorionic fetal vessels, stem villi, distal villi, and maternal decidua. In affected cases, areas of inflamed and non-inflamed villi were selected separately for analysis. A normal tonsil slide was also used as a positive control for each experiment. Tissues then underwent five cycles of staining, bleaching, and imaging on the IN Cell Analyzer 6500HS (GE Healthcare, Marlborough, MA) using antibodies conjugated to either Cy3 or Cy5. The full list of antibodies can be found in supplemental Table 1. Images were then overlaid using Layers Software (GE Healthcare) at 20X magnification.
Table 1.
Patient demographics
| Control | VUE | p value | |
|---|---|---|---|
| Maternal age (years, range) | 31.3 (27–38) | 30.5 (29–40) | 0.68 |
| Maternal BMI (kg/m2) | 31 (18.8–45.2) | 28.8 (19.8–45.2) | 0.19 |
| Gestational age at birth (weeks, range) |
37 4/7 (34 2/7–40 1/7) |
38 1/7 (34 3/7–39 5/7) |
0.27 |
| Gravida (range) | 2 (1–4) | 4 (2–6) | 0.01 |
| Apgar 1 min | 7 | 8 | 0.45 |
| Apgar 5 min | 9 | 9 | 0.66 |
| Fetal weight (kg) | 3.29 | 3.06 | 0.43 |
|
Fetal sex (% female) |
40 | 80 | * |
| Placental weight for GA (% range) |
50–75 (10–25 to >97) |
50–75 (10–25 to >97) |
0.89 |
Data is presented as the median and range
Abbreviations: BMI, body mass index; GA, gestational age
Microarray
After pathologist review to select areas of interest, ten slides were cut at 10 μm for each sample (n = 20), and areas of least inflamed and most inflamed VUE regions were collected from affected cases by macrodissection of marked areas on the H&E template slide. Similar regions of normal villi were macrodissected from control cases (n = 10). RNA was then extracted from these sections using the RNeasy FFPE kit (Qiagen, Valencia, CA) as per manufacturer’s instructions. RNA concentration and purity was determined by the A260/280 ratio using a NanoDrop 8000 (Thermo Fisher Scientific, Waltham, MA). RNA Integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples with a RNA integrity number (RIN) ≥ 8 were utilized for analysis. Microarray targets were prepared using RNA transcript labeling reagent (Affymetrix, Santa Clara, CA) and then hybridized to the Affymetrix GeneChip U133 plus 2.0 Array (Affymetrix) according to manufacturers’ instructions. Arrays were washed and stained with streptavidin phycoerythrin using the Affymetrix GeneChip protocol and scanned using an Affymetrix GeneChip Scanner 3000 (Affymetrix). Acquisition and initial quantification of array images was conducted using AGCC software (Affymetrix), and subsequent data analysis was performed using Partek Genomics Suite Version 7.0 (Partek, St Louis, MO). Gene IDs were utilized for gene pathway analysis by Kyoto Encyclopedia of Genes and Genomes (KEGG; Kyoto University).
HLA Typing
In order to acquire pure maternal and fetal tissue for tissue typing, we searched for mothers from our original cohort who also had a noncancerous specimen collected in the pathology archive. In total, four women diagnosed with VUE and four who had an unaffected placenta were found to have tissue from a previous biopsy that could be used as the maternal sample. The fetal samples were isolated from the tissue of the umbilical cord. Again, ten slides were cut at 10 μm for each sample (n = 16), and DNA was isolated using the QIAamp DNA FFPE Tissue Kit (Qiagen) per manufacturer’s instruction. DNA concentration and purity was determined by NanoDrop 8000. HLA typing was performed using reverse sequence-specific probe (rSSOP) method (LABType™, One Lambda, Inc. Canoga Park, CA) as per manufacturer’s recommendation. First, target DNA is PCR amplified using a group-specific primer. The PCR product is biotinylated, which allows it to be detected using R-Phycoerythrin conjugated streptavidin (SAPE). The PCR product is denatured and allowed to re-hybridize to complementary DNA probes conjugated to fluorescently coded microspheres. A flow analyzer, the LABScan 3D (Luminex) identifies the fluorescent intensity of PE (phycoerythrin) on each microsphere. The assignment of the HLA typing is based on the reaction pattern compared to patterns associated with published HLA gene sequences. Tissue types tested included class I, HLA-A, HLA-B and HLA-C loci; and class II, HLA-DRB1 and HLA-DQB1 loci.
Data Analysis
For patient demographics, nonparametric categorical data were analyzed with Fisher’s exact test, and nonparametric continuous data were analyzed with Kruskal-Wallis test. Differentially expressed genes in each group were identified using ANOVA test. A gene was considered as differentially expressed if two conditions were met: (1) expression fold change between two groups was ≥ 2 and (2) uncorrected p value was ≤ 0.05. All statistical analysis was performed with GraphPad Prism software version 6.0 (GraphPad Software, San Diego, CA).
Results
Patient Characteristics
Twenty subjects were included in this study, ten of whom had a high-grade VUE diagnosis in the placenta and ten subjects who had a normal placental examination. Cases and controls were intentionally matched pairwise to be within 1 week gestational age of each other. No indication of infection was identified in the VUE cases. Patient demographics are summarized in Table 1. There were no differences in maternal age, body mass index (BMI), gestational age at birth, placental weight, or fetal health (Apgar and weight) between groups; however, the VUE group did have a higher number of prior pregnancies and included more pregnancies with a female fetus.
VUE Demonstrates High Levels of CD8 Infiltration by Multiplex IHC
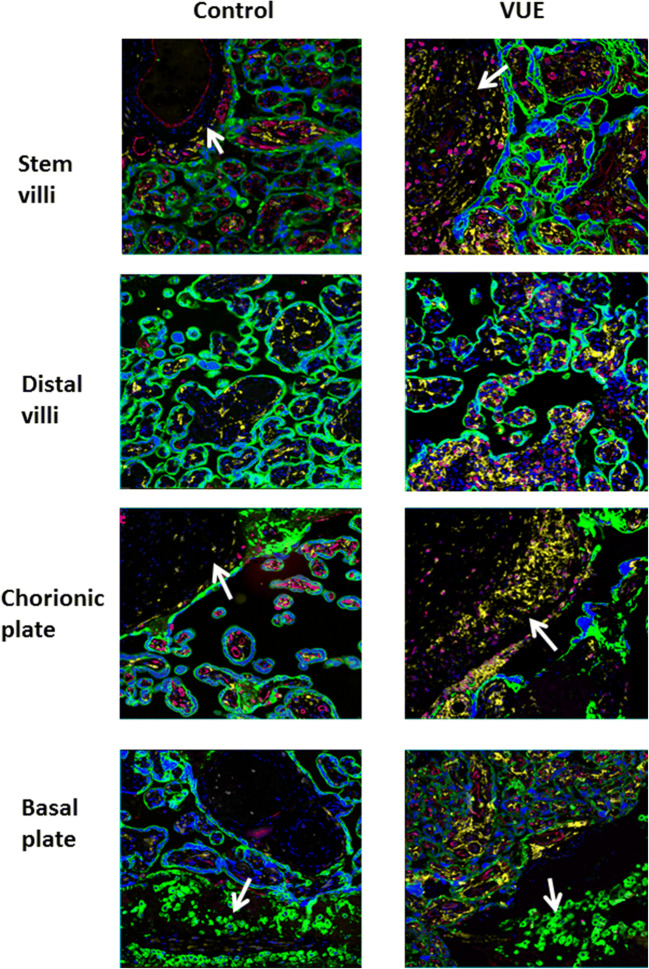
All 20 placentas were stained and images were collected from 5 representative regions of the stem villi, distal villi, chorionic vessels, and decidua basalis. Representative images from one control and one VUE affected placenta can be seen in Fig. 1. There were minimal CD4+ T helper cells (white) detected in any of the interior regions of the placenta; however, strong staining of maternal CD8+ cytotoxic T cells (pink) was observed in VUE placentas. All placentas demonstrated strong CD14+ fetal macrophage (yellow) presence in the villi (green), but these cells were not always found in areas of high CD8 infiltration. CD31+ endothelial cells (red), which line the fetal blood vessels, were much less defined in areas of the placenta with VUE compared to unaffected placenta, confirming the typical villous vascular obliteration also appreciated by light microscopy with H&E staining. No significant immune interactions were seen in the decidua or chorionic vessels.
Fig. 1.
Characterizing VUE morphology in different areas of the placenta compared to unaffected control. Representative 20X image by MxIF staining of the stem villi, distal villi, chorionic plate and decidual plate in VUE versus gestational age matched control placentae. Blue-DAPI, green-cytokeratin-7, red-CD31, yellow-CD14 and pink-CD8
Gene Expression Is Similar in Low and High Areas of Inflammation in VUE
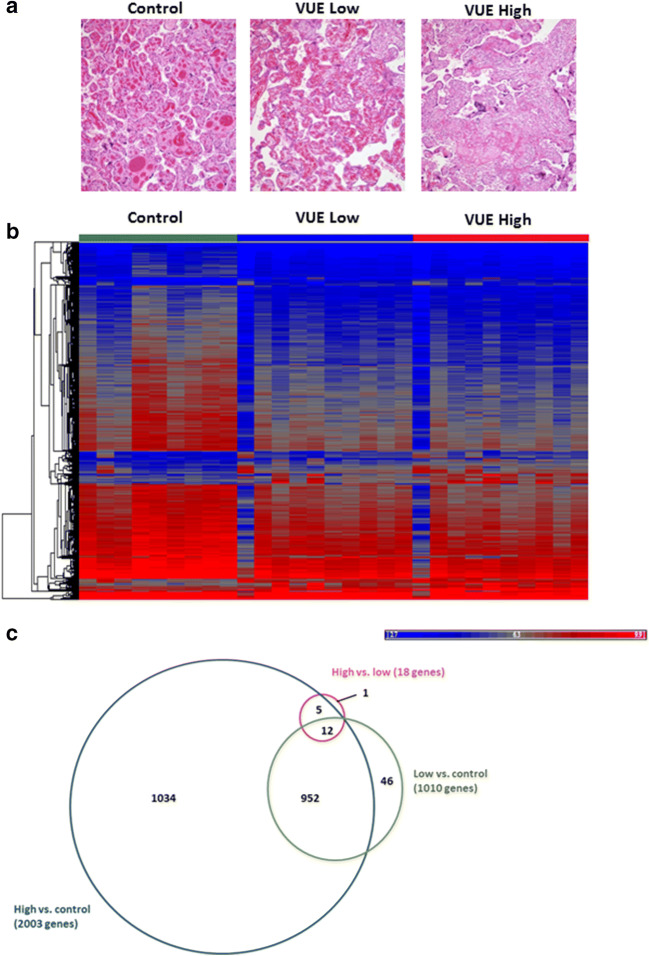
Macrodissection was carried out on each of the VUE cases to capture differences in gene expression between least and most inflamed areas (Fig. 2A). As histologically these less inflamed areas do not appear to be as immunologically active as areas of high lymphocytic infiltration and villous necrosis, we wanted to understand if there may be some indication of pathways still upregulated in an attempt to protect the trophoblast cells from the maternal infiltrating cells. Using microarrays, we measured changes in gene expression between more and less inflamed areas of VUE cases and paired unaffected controls. The complete dataset is available for download from Gene Expression Omnibus (GEO [GSE13085]). The data demonstrated that there is significant downregulation of genes in cases of VUE compared to controls (Fig. 2B). In total, we found 3031 genes with a fold change greater than 2 and 1034 that were differentially expressed between cases of high VUE compared to control placentas (Fig. 3C). Interestingly, there were very few gene expression differences in areas of high inflammation versus low inflammation in VUE. Table 2 indicates the top 15 over and under expressed genes in cases of high VUE infiltration compared to control. In genes involved in T cell trafficking, HLA and HLA regulation were increased in VUE, whereas in genes involved in responses related to stress, vascular function, cell adhesion and transcription control were significantly decreased.
Fig. 2.
Gene expression changes based on VUE severity. (A) Representative H&E stain used for micro dissection of intermediate vs. high VUE in the same tissue sample. (B) Heat map demonstrating up (red) and down (blue) regulation of genes in VUE lesions as a group average with low and high infiltration compared to control (fold change >2; p value ≤0.05). (C) Proportional Venn diagram demonstrating the number of genes shared and different in control, low and high VUE placentae
Fig. 3.
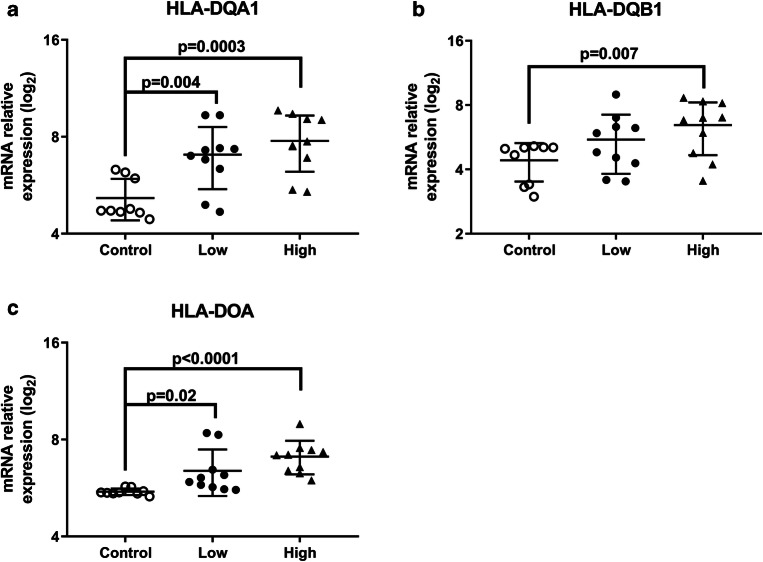
HLA genetic profile in VUE compared to unaffected controls. Relative expression of (A) HLA-DQA1; (B) HLA-DQB1; (C) HLA-DOA in control, low and high VUE (n = 10/group, p value determined by Wilcoxon rank test compared to control)
Table 2.
Top 15 up- and downregulated genes in VUE
| Upregulated Genes | Fold change | Downregulated Genes | Fold change |
|---|---|---|---|
| CXCL10 | 22.65 | HIF3A | −8.06 |
| CXCL9 | 20.87 | SOX7 | −6.70 |
| IGHG1/2 | 6.51 | PPP1CB | −6.02 |
| SERPINA1 | 4.48 | TAC3 | −5.42 |
| CD74 | 4.34 | APOLD1 | −5.31 |
| CCL8 | 4.28 | ID4 | −5.16 |
| HLA-DRA | 4.14 | DNAJB4 | −5.10 |
| HLA-DQB1 | 4.09 | NEXN | −4.95 |
| HLA-DOA | 4.07 | DKK1 | −4.91 |
| STAT1 | 3.97 | ZNF512B | −4.74 |
| GALT | 3.95 | IFNGR1 | −4.68 |
| GBP5 | 3.93 | KDM5D | −4.50 |
| MUC6 | 3.87 | TLK1 | −4.38 |
| CIITA | 3.61 | PTX3 | −4.25 |
| KLRC1/2 | 3.56 | EPPK1 | −4.24 |
Abbreviations: VUE, villitis of unknown etiology
Using these upregulated genes in the high VUE compared to unaffected controls, we utilized Kyoto Encyclopedia of Genes and Genomes (KEGG; Kyoto University) to identify pathways impacted most significantly by VUE. This program searches presence of inputted genes in highly annotated pathways and determines the most relevant pathways based on your input data. Ratio of affected pathways is determined by the number of genes significantly altered in samples divided by the total number of genes known to be involved in the pathway. In VUE, 43% of genes mapped to perturbations in graft vs. host disease and 38% of genes mapped to allograft rejection (Table 3). Other significantly impacted pathways in VUE have a strong immunological basis.
Table 3.
Pathways altered in cases of VUE
| Pathway | Gene count | Reference | Ratio |
|---|---|---|---|
| Graft vs. host disease | 13 | 30 | 0.43 |
| Allo-rejection | 11 | 29 | 0.38 |
| Diabetes type 1 | 12 | 34 | 0.35 |
| Asthma | 8 | 25 | 0.32 |
| Autoimmune thyroid disease | 13 | 45 | 0.29 |
| Antigen presentation | 14 | 56 | 0.25 |
| Rheumatoid arthritis | 20 | 84 | 0.21 |
| Adherens junction | 17 | 71 | 0.24 |
| TGF-β signaling | 17 | 82 | 0.21 |
Abbreviations: TGF, transforming growth factor; VUE, villitis of unknown etiology
VUE Cases Suggest Increased HLA Expression and Mismatch
Our next aim was to better understand the role of HLA in VUE. Microarray data demonstrated that many HLA class II molecules were increased in VUE cases relative to unaffected control. HLA-DQA1 relative expression increased with severity of VUE (5.1 vs. 7.0 vs. 7.8; Fig. 3A), as did HLA-DQB1 (4.4 vs. 5.5 vs. 6.4; Fig. 3B) and HLA-DOA (5.5 vs. 6.4 vs. 7.1; Fig. 3C). Interestingly, gene expression of HLA class I molecules were not significantly different; however, we expect this to be related to the sample and not the biology.
We then identified eight mothers from our cohort (four controls and four VUE) who had other tissue specimens that could be utilized for determining their HLA type. Class I and II HLA typing was compared for similarity between mother and fetus. Of the five loci measured, only results for DRB1 and DQB1 could be reliably measured in our sample. As demonstrated in Table 4, we observed that 1/3 DRB1 and DQB1loci were mismatched in the control cohort, while 4/4 DRB1 and 3/4 DQB1 loci were mismatched in the VUE group.
Table 4.
HLA DRB1 and DQB1 tissue typing in mother/baby pairs
| DRB1 Locus | DQB1 Locus | |||
|---|---|---|---|---|
| Fetus | Mom | Fetus | Mom | |
| Control | DR7 DR12 | DQ2 DQ7 | ||
| Control | DR4 DR4 | DR4 DR4 | DQ8 DQ8 | DQ8 DQ8 |
| Control | DR17 DR13 | DR17 DR13 | DQ2 DQ6 | DQ2 DQ6 |
| Control | DR13DR15 | DR7 DR15 | DQ6DQ6 | DQ2 DQ6 |
| VUE | DR4 DR9 | DR9 DR13 | DQ7 DQ9 | DQ9 DQ6 |
| VUE | DR4DR14 | DR4 DR4 | DQ7DQ8 | DQ8 DQ8 |
| VUE | DR13DR15 | DR13 DR13 | DQ6 DQ6 | DQ6 DQ6 |
| VUE | DR17 DR11 | DR11 DR16 | DQ2 DQ7 | DQ7 DQ5 |
*Bold denotes a mismatch between the mom and fetus
Intense HLA Class I Staining in VUE
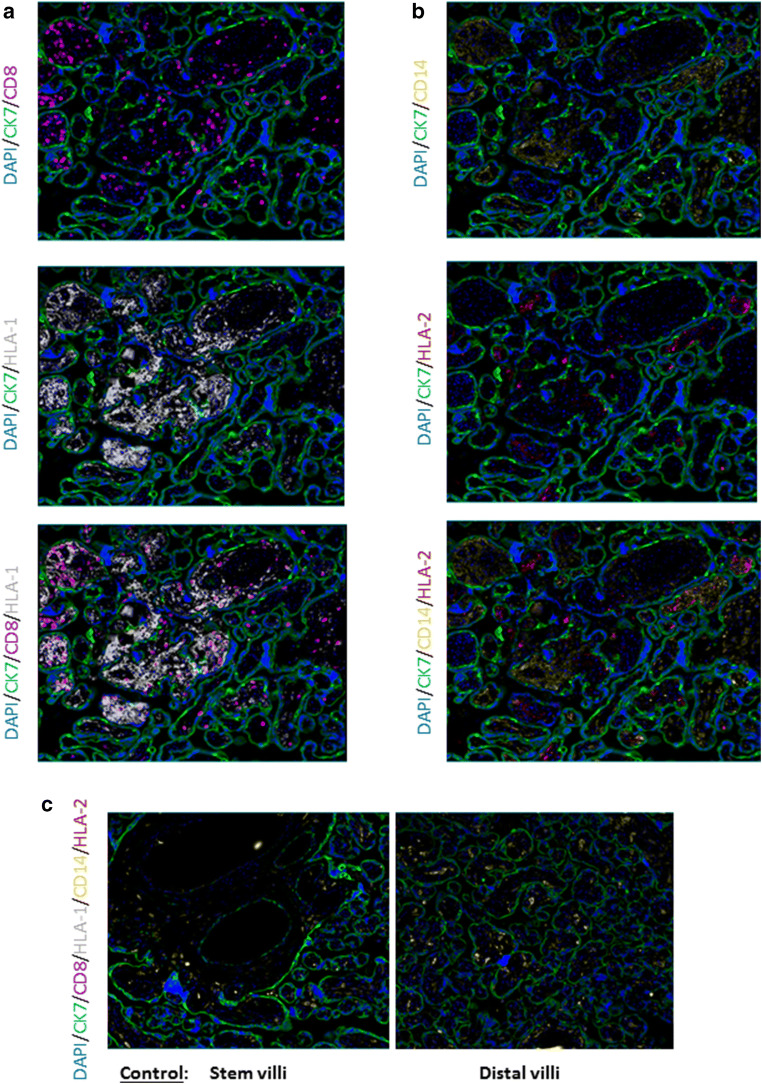
Lastly, we wanted to identify areas of HLA class I and II staining in relation to maternal and fetal immune cells utilizing MxIF. In VUE, areas with high infiltration of maternal CD8+ cells corresponded with areas having strong HLA class I staining (Fig. 4A). On the contrary, HLA class II staining was sparse and did not necessarily align with CD14+ fetal macrophages (Fig. 4B). This indicates that another type of immune cell may be playing an important, but undiscovered antigen presentation role in VUE pathology. HLA class I and II staining was not detectable in both the stem and distal villi of the control placentas (Fig. 4C). Together, these images suggest that VUE is associated with significant increases in HLA class I-positive cell infiltration to the placenta but a much lower level of HLA class II trafficking.
Fig. 4.
HLA class I and II expression in VUE affected placentae. Representative 20× image by MxIF staining. (A) HLA class I and maternal CD8+ T cell localization in the lesion. (B) HLA class II and fetal CD14+ macrophages in VUE. (C) HLA class I and II staining in the stem and distal villi of a control placenta
Discussion
Classically, the hallmarks of VUE include the infiltration of maternal CD8+ lymphocytes into the villi, activation of fetal macrophages, and destruction of chorionic villi. Although the etiology of this phenotype is weakly understood, the initiation of chronic inflammatory lesions in the placenta is hypothesized to be either through infection, immunologic rejection, or the interplay of both phenomena. Data presented here support the growing body of evidence that VUE is an allograft rejection between the mother and the fetus. Similar to others, we showed an infiltration of CD8+ cells in the stem and distal villi of placentas diagnosed with VUE that is not observed in unaffected, gestational age matched controls. Interestingly, we did not observe infiltration of CD4+ T helper cells into our cohort of placentas. Other groups have observed CD4+ cells in the lesions [22, 23]; however, the numbers of these reported cells were much lower than CD8+ cells. CD4+ cells are the main regulator of adaptive immunity following an interaction with a pathogenic antigen. It is possible that due to the lack of infectious stimuli, CD4 cells do not receive a signal to traffic into the villi; however, their role in regulating responses systemically may be more important than locally. Future studies that also evaluate blood from mothers and neonates with VUE are warranted. CD14+ Hofbauer cells are of fetal origin and have important roles in placental development, including vasculogenesis and angiogenesis [24]. Hofbauer cells have been shown to express Ki67, a marker of activation and proliferation, during VUE [23]. Hofbauer cells were prominent in our cases, but were not consistently seen in areas of cytotoxic T cell infiltration. We were limited in markers for this first round of staining, but future studies should be expanded to additional cell types and include activation and inflammatory targets to better understand the status of these fetal immune cells.
Expression analysis demonstrated that genes involved in allograft and graft-versus-host disease pathways were significantly altered in VUE compared to unaffected controls. As the genes involved with these two pathways have a high level of overlap, it remains unclear whether initiation of VUE originates from the mother or the fetus. One could hypothesize that maternal CD8+ T cells could be presented with antigen from a fetal cell or placental debris in circulation, initiating expansion and chemokine trafficking. Conversely, due to the intricate sharing of blood during human pregnancy, fetal macrophages could become activated by circulating cytokines, causing them to release oxygen radicals and indirectly recruit other immune cells, including maternal immune cells, into the placenta. Support for maternal origin comes from in situ hybridization experiments demonstrating that the infiltrating lymphocytes are of maternal origin using placentas from a male fetus [14, 25, 26]. Studies in di-zygotic twins who are exposed to the same maternal environment showed marked infiltration of T cells into the placenta of the smaller fetus, whereas the larger twin had no T cell infiltration [27]. This suggests that the fetal cells of the placenta, and not the mother, initiate this inflammatory phenotype.
We found an increase in HLA class II gene expression which led us to tissue typing of maternal versus fetal tissue. Tissue typing measures cell surface expression of HLA which is found on the surface of a majority of cells in the body. This is crucial for determining a best match between a donor and recipient during a transplant procedure. In pregnancy, half of the fetus’ HLA type is paternally inherited; therefore, the mother would have up to 50% mismatch with the fetus. Antibodies against fetal HLA and maternal HLA have been detected in the sera of women and neonates with VUE [28]. C4d, a marker of antibody-mediated rejection in transplant biopsies, was detected in the syncytiotrophoblast of a VUE inflamed placenta [29], but C4d staining was not observed in cases of infectious villitis [30]. Our results suggested a higher percentage of HLA mismatch between mom and fetus in VUE cases compared to controls; however, this observation was made at low resolution using a small cohort of mom/baby pairs. Additionally, our HLA typing was incomplete, particularly for class I, due to poor DNA quality. Therefore, we aim to follow up with larger studies using fresh/frozen tissue to better understand HLA mismatch and tissue rejection in VUE.
Lesions seen in VUE pathology demonstrate differences in HLA expression based on location and cell type. HLA class II expression on Hofbauer cells increases with gestational age [23], which may explain why VUE tends to occur later in pregnancy. In VUE, HLA class II was observed on some of the CD14+ Hofbauer cells but not all of them. As placental macrophages and their functions are highly heterogeneous [24], in depth phenotyping of these cells during VUE is warranted. Nonclassical HLA class I molecules are found on the extra-villous trophoblast, but not the villous trophoblast [3]. In our study, many maternal CD8+ T cells co-expressed HLA class I in placentas from VUE cases, which was not observed in unaffected controls. There are a range of mechanisms by which this VUE response could be mediated: (1) up-regulation of class I and II expression, (2) increased presentation of antigen, (3) increased T cell priming, (4) amplified proinflammatory cytokine milieu, and (5) altered T cell trafficking. These mechanisms have been expertly reviewed elsewhere [31].
The mechanism(s) that traffic lymphocytes into the placenta are still being discovered. In a cohort of fetal demise cases, researchers found chronic inflammatory lesions in over 55% of the placentas and elevated CXCL10 concentrations in amniotic fluid [32]. However, it should be noted that this was a small cohort (n = 40) and systematic studies of the association between fetal demise and VUE are confounded by the classification system utilized, the small number of published studies, differences in study design and lack of in depth pathological review, making the true relationship challenging to define [22, 33, 34]. CXCL10 is secreted in response to proinflammatory cytokines and is effective at attracting T cells to sites of infection/inflammation [35]. Similar to our results, mRNA and protein levels of chemokines, including CXCL10 and CXCL9, were significantly higher in the placenta and the maternal and fetal plasma in VUE [26]. Interestingly, in cases of chorioamnionitis, chemokines were not upregulated in maternal plasma, demonstrating another unique difference between these placental pathologies [26]. Co-culture systems are being developed to address questions related to mechanism and pathophysiology of VUE. In a model of infectious villitis, lipopolysaccharide increased ICAM-1 expression on the trophoblast which allowed peripheral monocyte binding to the placenta [36]. Using tissue explants, Derricott et al. developed a co-culture with CD4 and CD8 T cells which more closely mimics the inflammatory state of VUE [37, 38]. As normal human placentation is significantly more invasive than observed in other species [39], in vitro models are currently our best option for understanding this inflammatory condition.
To conclude, the pathophysiology of VUE resembles an immunologic rejection response with the cause and initiator (fetal vs. maternal) currently unknown. VUE is a common placental pathology diagnosed after birth and is known to be associated with increased fetal morbidity and mortality. Therefore, the development of methods to predict VUE risk would be clinically beneficial. Additionally, use of models that can be utilized to understand the mechanisms behind these immunological lesions may lead to novel ways to diagnose and potentially prevent VUE and its associated comorbidities in the future.
Electronic supplementary material
(DOCX 13 kb)
Acknowledgments
The authors would like to acknowledge Wendy Nevala, MS, for her excellent training in the MxIF technique.
Funding Information
Financial support for this project was provided by Mayo Clinic Division of Anatomic Pathology (SEK) and NICHD K12 HD065987 (EALE).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elizabeth Ann L. Enninga, Email: Enninga.elizabethann@mayo.edu
Alexey A. Leontovich, Email: Leontovich.alexey@mayo.edu
Bohdana Fedyshyn, Email: Fedyshyn.bohdana@mayo.edu.
Laurie Wakefield, Email: Wakefield.laurie@mayo.edu.
Manish Gandhi, Email: Gandihi.manish@mayo.edu.
Svetomir N. Markovic, Email: Markovic.svetomir@mayo.edu
Rodrigo Ruano, Email: Ruano.rodrigo@mayo.edu.
Sarah E. Kerr, Email: kerr.sarah@mayo.edu
References
- 1.Holtan SG, Creedon DJ, Haluska P, Markovic SN. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin Proc. 2009;84(11):985–1000. doi: 10.1016/S0025-6196(11)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enninga EA, Holtan SG, Creedon DJ, Dronca RS, Nevala WK, Ognjanovic S, Markovic SN. Immunomodulatory effects of sex hormones: requirements for pregnancy and relevance in melanoma. Mayo Clin Proc. 2014;89(4):520–535. doi: 10.1016/j.mayocp.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redman CW, McMichael AJ, Stirrat GM, Sunderland CA, Ting A. Class 1 major histocompatibility complex antigens on human extra-villous trophoblast. Immunology. 1984;52(3):457–468. [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt JS, Orr HT. HLA and maternal-fetal recognition. FASEB J. 1992;6(6):2344–2348. doi: 10.1096/fasebj.6.6.1544544. [DOI] [PubMed] [Google Scholar]
- 5.Boog G. Chronic villitis of unknown etiology. Eur J Obstet Gynecol Reprod Biol. 2008;136(1):9–15. doi: 10.1016/j.ejogrb.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Redline RW. Villitis of unknown etiology: noninfectious chronic villitis in the placenta. Hum Pathol. 2007;38(10):1439–1446. doi: 10.1016/j.humpath.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 7.Becroft DM, Thompson JM, Mitchell EA. Placental villitis of unknown origin: epidemiologic associations. Am J Obstet Gynecol. 2005;192(1):264–271. doi: 10.1016/j.ajog.2004.06.062. [DOI] [PubMed] [Google Scholar]
- 8.Knox WF, Fox H. Villitis of unknown aetiology: its incidence and significance in placentae from a British population. Placenta. 1984;5(5):395–402. doi: 10.1016/S0143-4004(84)80019-3. [DOI] [PubMed] [Google Scholar]
- 9.Salafia CM, Vogel CA, Vintzileos AM, Bantham KF, Pezzullo J, Silberman L. Placental pathologic findings in preterm birth. Am J Obstet Gynecol. 1991;165(4 Pt 1):934–938. doi: 10.1016/0002-9378(91)90443-U. [DOI] [PubMed] [Google Scholar]
- 10.Redline RW, Abramowsky CR. Clinical and pathologic aspects of recurrent placental villitis. Hum Pathol. 1985;16(7):727–731. doi: 10.1016/S0046-8177(85)80159-3. [DOI] [PubMed] [Google Scholar]
- 11.Redline RW. Severe fetal placental vascular lesions in term infants with neurologic impairment. Am J Obstet Gynecol. 2005;192(2):452–457. doi: 10.1016/j.ajog.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Garcia AG, Basso NG, Fonseca ME, Zuardi JA, Outanni HN. Enterovirus associated placental morphology: a light, virological, electron microscopic and immunohistologic study. Placenta. 1991;12(5):533–547. doi: 10.1016/0143-4004(91)90029-F. [DOI] [PubMed] [Google Scholar]
- 13.Ernst LM, Crouch J, Rinder H, Howe JG. Bacterial etiology for chronic villitis is not supported by polymerase chain reaction for 16S rRNA DNA. Pediatr Dev Pathol. 2005;8(6):647–653. doi: 10.1007/s10024-005-0412-1. [DOI] [PubMed] [Google Scholar]
- 14.Redline RW, Patterson P. Villitis of unknown etiology is associated with major infiltration of fetal tissue by maternal inflammatory cells. Am J Pathol. 1993;143(2):473–479. [PMC free article] [PubMed] [Google Scholar]
- 15.Labarrere CA, Catoggio LJ, Mullen EG, Althabe OH. Placental lesions in maternal autoimmune diseases. Am J Reprod Immunol Microbiol. 1986;12(3):78–86. doi: 10.1111/j.1600-0897.1986.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 16.Magid MS, Kaplan C, Sammaritano LR, Peterson M, Druzin ML, Lockshin MD. Placental pathology in systemic lupus erythematosus: a prospective study. Am J Obstet Gynecol. 1998;179(1):226–234. doi: 10.1016/S0002-9378(98)70277-7. [DOI] [PubMed] [Google Scholar]
- 17.Styer AK, Parker HJ, Roberts DJ, Palmer-Toy D, Toth TL, Ecker JL. Placental villitis of unclear etiology during ovum donor in vitro fertilization pregnancy. Am J Obstet Gynecol. 2003;189(4):1184–1186. doi: 10.1067/S0002-9378(03)00577-5. [DOI] [PubMed] [Google Scholar]
- 18.Schonkeren D, Swings G, Roberts D, Claas F, de Heer E, Scherjon S. Pregnancy close to the edge: an immunosuppressive infiltrate in the chorionic plate of placentas from uncomplicated egg cell donation. PLoS One. 2012;7(3):e32347. doi: 10.1371/journal.pone.0032347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labarrere C, Althabe O. Chronic villitis of unknown aetiology in recurrent intrauterine fetal growth retardation. Placenta. 1987;8(2):167–173. doi: 10.1016/0143-4004(87)90019-1. [DOI] [PubMed] [Google Scholar]
- 20.Khong TY, Mooney EE, Ariel I, et al. Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch Pathol Lab Med. 2016;140(7):698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch C, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derricott H, Jones RL, Greenwood SL, Batra G, Evans MJ, Heazell AE. Characterizing Villitis of unknown etiology and inflammation in stillbirth. Am J Pathol. 2016;186(4):952–961. doi: 10.1016/j.ajpath.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Kim JS, Romero R, Kim MR, Kim YM, Friel L, Espinoza J, Kim CJ. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. 2008;52(4):457–464. doi: 10.1111/j.1365-2559.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes L, Golos TG. Hofbauer cells: their role in healthy and complicated pregnancy. Front Immunol. 2018;9:2628. doi: 10.3389/fimmu.2018.02628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myerson D, Parkin RK, Benirschke K, Tschetter CN, Hyde SR. The pathogenesis of villitis of unknown etiology: analysis with a new conjoint immunohistochemistry-in situ hybridization procedure to identify specific maternal and fetal cells. Pediatr Dev Pathol. 2006;9(4):257–265. doi: 10.2350/08-05-0103.1. [DOI] [PubMed] [Google Scholar]
- 26.Kim MJ, Romero R, Kim CJ, et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. 2009;182(6):3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusuf K, Kliman HJ. The fetus, not the mother, elicits maternal immunologic rejection: lessons from discordant dizygotic twin placentas. J Perinat Med. 2008;36(4):291–296. doi: 10.1515/JPM.2008.054. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Romero R, Xu Y, Kim JS, Topping V, Yoo W, Kusanovic JP, Chaiworapongsa T, Hassan SS, Yoon BH, Kim CJ. A signature of maternal anti-fetal rejection in spontaneous preterm birth: chronic chorioamnionitis, anti-human leukocyte antigen antibodies, and C4d. PLoS One. 2011;6(2):e16806. doi: 10.1371/journal.pone.0016806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudzinski E, Gilroy M, Newbill C, Morgan T. Positive C4d immunostaining of placental villous syncytiotrophoblasts supports host-versus-graft rejection in villitis of unknown etiology. Pediatr Dev Pathol. 2013;16(1):7–13. doi: 10.2350/12-05-1195-OA.1. [DOI] [PubMed] [Google Scholar]
- 30.AL K, Kim YW, Shim JY, et al. Distinct patterns of C4d immunoreactivity in placentas with villitis of unknown etiology, cytomegaloviral placentitis, and infarct. Placenta. 2013;34(5):432–435. doi: 10.1016/j.placenta.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Tamblyn JA, Lissauer DM, Powell R, Cox P, Kilby MD. The immunological basis of villitis of unknown etiology - review. Placenta. 2013;34(10):846–855. doi: 10.1016/j.placenta.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Lannaman K, Romero R, Chaiworapongsa T, Kim YM, Korzeniewski SJ, Maymon E, Gomez-Lopez N, Panaitescu B, Hassan SS, Yeo L, Yoon BH, Jai Kim C, Erez O. Fetal death: an extreme manifestation of maternal anti-fetal rejection. J Perinat Med. 2017;45(7):851–868. doi: 10.1515/jpm-2017-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derricott H, Jones RL, Heazell AE. Investigating the association of villitis of unknown etiology with stillbirth and fetal growth restriction - a systematic review. Placenta. 2013;34(10):856–862. doi: 10.1016/j.placenta.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Ptacek I, Sebire NJ, Man JA, Brownbill P, Heazell AE. Systematic review of placental pathology reported in association with stillbirth. Placenta. 2014;35(8):552–562. doi: 10.1016/j.placenta.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168(7):3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 36.Xiao J, Garcia-Lloret M, Winkler-Lowen B, Miller R, Simpson K, Guilbert LJ. ICAM-1-mediated adhesion of peripheral blood monocytes to the maternal surface of placental syncytiotrophoblasts: implications for placental villitis. Am J Pathol. 1997;150(5):1845–1860. [PMC free article] [PubMed] [Google Scholar]
- 37.Derricott H, Jones RL, Heazell AEP, Greenwood SL. Co-culture of placental explants with isolated CD4 and CD8 T cells: a functional model to define the consequences of placental inflammation. 2015.
- 38.Derricott H, Heazell AEP, Greenwood SL, Jones RL. A novel in vitro model of villitis of unknown etiology demonstrates altered placental hormone and cytokine profile. Am J Reprod Immunol. 2017;78(5). [DOI] [PubMed]
- 39.Grigsby PL. Animal models to study placental development and function throughout Normal and dysfunctional human pregnancy. Semin Reprod Med. 2016;34(1):11–16. doi: 10.1055/s-0035-1570031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 13 kb)