Abstract
Whole-genome sequencing clustered Australian Candida auris isolates from sporadic cases within clade III. Case isolates were genomically distinct; however, unexpectedly, those from 1 case comprised 2 groups separated by >60 single nucleotide polymorphisms (SNPs) with no isolate being identical, in contrast to outbreaks where isolates from any 1 individual have differed by <3 SNPs. Multidrug resistance was absent. High within-host genetic heterogeneity should be considered when investigating C. auris infections.
Keywords: Candida auris, drug resistance, whole-genome sequencing
Candida auris is a global multidrug-resistant pathogen with a propensity to cause nosocomial outbreaks [1–6]. Next-generation sequencing analyses have shown that strains of C. auris cluster into 4 predominant, but possibly 5 geographically distinct clades (South Asian [clade I], East Asian [clade II], African [clade III], and South American [clade IV]), with some evidence of phylogeographical mixing of clades [1–3, 7–9]. Short-read whole-genome sequencing (WGS) data demonstrated that there are >10 000 single nucleotide polymorphisms (SNPs) from separate clades, but with lower intraclade diversity (<100 SNP differences) [1]; the low intraclade diversity was most marked in analyses of hospital outbreaks where genomes of outbreak isolates differed by a median of 3 SNPs, and within any 1 patient, by <2 SNPs [1–3, 10]. Whether similar observations emerge in the nonoutbreak setting is uncertain.
Following the first C. auris infections in Australia [11, 12], in 2018/2019, Sydney reported 3 additional cases. Here we used WGS to investigate the genetic relationships and markers of drug resistance of Australian isolates from sporadic cases, including from the first report [11]. To place results in the genomic context, we also sequenced several isolates from India and South Africa and included publicly available C. auris genomes [1, 3, 4, 8]. Phylogenetic SNP analysis was compared with polymerase chain reaction (PCR) fingerprinting [13]. Unexpectedly, mulitple isolates from 1 individual (clade III) had relatively diverse genomes compared with previous observations for this clade [2, 3].
Eighteen clinical isolates, reconfirmed as C. auris by internal transcribed spacers (ITS) sequencing, were studied (Table 1). Of 10 Australian isolates, 9 were from 3 separate cases repatriated to Sydney hospitals from South Africa (Cases 1, 2, 3), with Case 4 repatriated to Perth [11]. Two isolates were from South Africa, and 6 were from India, included as “benchmark” isolates from their respective regions. For Case 1, the incident isolate (strain WM_18.177) was cultured during a prior hospitalization in South Africa but was considered an Australian isolate for this study.
Table 1.
Candida auris Isolates Studied: Geographic Region, Clade, Body Site of Isolation, and in Vitro Susceptibility to 9 Antifungal Agents
| Isolate ID | Country | Clade | Body Site | Date of Isolation | Minimum Inhibitory Concentration, mg/L | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | 5-FC | FLC | ITR | VRC | POS | ANI | MIF | CAS | |||||
| Australian isolates | |||||||||||||
| Case 1 | |||||||||||||
| WM_18.181 | Australia | III | Wound swab PICC site | 05.09.2018 | 1 | 0.12 | 256 | 0.25 | 2 | 0.12 | 0.25 | 0.12 | 0.12 |
| WM_18.182 | Australia | III | Sputum | 05.09.2018 | 1 | 0.12 | 256 | 0.25 | 2 | 0.12 | 0.25 | 0.12 | 0.25 |
| WM_18.187 | Australia | III | Axilla swab | 06.09.2018 | 1 | 0.12 | 256 | 0.25 | 2 | 0.12 | 0.5 | 0.12 | 0.25 |
| WM_18.188 | Australia | III | Sputum | 07.09.2018 | 1 | 0.25 | 256 | 0.5 | 4 | 0.25 | 0.25 | 0.06 | 0.12 |
| WM_18.189 | Australia | III | Axilla swab | 11.09.2018 | 1 | 0.12 | 256 | 0.25 | 2 | 0.12 | 0.25 | 0.12 | 0.25 |
| WM_18.177 a | Australia | III | Blood culture | July 2018 | 1 | 0.12 | 256 | 0.5 | 2 | 0.12 | 0.25 | 0.12 | 0.12 |
| WM_18.190 | Australia | III | Groin | 11.09.2018 | 1 | 0.12 | 256 | 0.25 | 2 | 0.12 | 0.25 | 0.12 | 0.25 |
| Case 2 | |||||||||||||
| WM_18.180 | Australia | III | Nose swab | 20.09.2018 | 1 | 0.12 | >256 | 0.25 | 2 | 0.12 | 0.25 | 0.12 | 0.25 |
| Case 3 | |||||||||||||
| WM_18.197 | Australia | III | Throat | 08.11.2018 | 1 | 0.12 | 256 | 0.5 | 2 | 0.12 | 0.25 | 0.12 | 0.25 |
| Case 4b | |||||||||||||
| WM_18.176 | Australia | III | Sternal bone | 2015 | 1 | 0.06 | >256 | 1 | 1 | 0.12 | 0.5 | 0.5 | 0.5 |
| South African comparison isolatesc | |||||||||||||
| WM_18.178 | South Africa | III | Blood culture | 2017 | 0.5 | 0.12 | >256 | 0.5 | 4 | 0.25 | 0.12 | 0.06 | 0.06 |
| WM_18.179 | South Africa | III | Blood culture | 2017 | 1 | 0.12 | 256 | 0.25 | 1 | 0.12 | 0.25 | 0.12 | 0.12 |
| Indian comparison isolatesd | |||||||||||||
| WM_18.173 | India | I | Blood culture | Unknown | 1 | <0.06 | 64 | 0.25 | 1 | 0.03 | 0.25 | 0.12 | >8 |
| WM_18.174 | India | I | Blood culture | Unknown | 2 | <0.06 | 64 | 0.12 | 0.5 | 0.03 | 0.5 | 0.12 | >8 |
| WM_18.175 | India | I | Blood culture | Unknown | 2 | <0.06 | 128 | 0.12 | 0.5 | 0.03 | 0.25 | 0.25 | 0.5 |
| WM_18.194 | India | I | Blood culture | Unknown | 2 | 0.12 | 32 | 0.32 | 0.25 | 0.008 | >8 | >8 | 2 |
| WM_18.195 | India | I | Blood culture | Unknown | 2 | 0.12 | >256 | 0.25 | 1 | 0.12 | 0.25 | 0.12 | 0.12 |
| WM_18.196 | India | I | Blood culture | Unknown | 2 | 0.12 | 256 | 0.25 | 1 | 0.25 | 0.25 | 0.12 | 0.12 |
Abbreviations: 5-FC, 5-flucytosine; AMB, amphotericin B; ANI, anidulafungin; CAS, caspofungin; FLC, fluconazole; ITR, itraconazole; MIC, minimum inhibitory concentration; MIF, micafungin; POS, posaconazole; VRC, voriconazole.
aOriginal (incident) clinical isolate from Case 1, isolated while hospitalized in South Africa.
bClinical isolate obtained from Heath et al. [11].
cClinical isolates from South Africa
dClinical isolates from India.
Antifungal susceptibility was determined using Sensititre YeastOne YO10 (TREK Diagnostics, Cleveland, OH, USA). MICs were interpreted against the proposed US Centers for Disease Control and Prevention (CDC) breakpoints [14] or proposed C. auris–specific Clinical Laboratory Standards Institute epidemiological cutoff values to define wild-type (WT) or non-WT isolates [15]. PCR fingerprinting was also performed [13].
Genomic DNA was extracted using the MasterPure Yeast Kit (Epicentre, Lucigen Corporation, WIS, USA). DNA libraries were prepared (Nextera XT; Illumina, CA, USA) and sequenced on the NextSeq 500 platform (Illumina, CA, USA) [16]. Raw reads were trimmed (Trimmomatic, version 3.6) and taxonomically classified using Kraken, version 1.0. Single nucleotide polymorphisms (SNPs) were called using 3 pipelines where SNPs were defined as substitutions present in at least 90% of reads with minimum coverage above 30. For the Nullarbor and RedDog pipelines (see below), reads were mapped to C. auris strain B11221 (African clade; NCBI GenBank Accession PGLS01000001.1) [17] as the reference. Nullarbor (version e2.0) (https://github.com/tseemann/nullarbor) employed Freebayes (version 1.2.0-dirty) for SNP calling and filtered them using Snippy, version 4.3.5, based on a minimum coverage of 10, a quality of 100, and a read fraction of 0.9 supporting the variant. The analysis was also undertaken using RedDog (https://github.com/katholt/RedDog) employing Bowtie2 [18] to map reads to the reference and BCFtools, version 1.3.1 (https://www.sanger.ac.uk/science/tools/samtools-bcftools-htslib), for SNP calling. In parallel, sequences were examined using the Oxford pipeline [3], which mapped short reads to the reference sequence T26425 (clade III) using Stampy, version 1.0.23, without BWA premapping, with an expected substitution rate of 0.01. SNPs were identified with Samtools4, version 0.1.19, mpileup with extended base-alignment quality flag. A consensus of ≥75% was necessary to support a SNP, and calls were required to be homozygous under a diploid model. Only SNPs supported by ≥5 reads, including 1 in each direction, were accepted. Repetitive regions of the genome were excluded as described by Eyre et al. [3].
Consensus alleles at all SNP variant sites were extracted; SNP sites present in all genomes were concatenated to generate a core SNP alignment for phylogenetic analysis. A maximum likelihood tree was estimated using IQTree, version 1.6.7, at 1000 bootstraps using the SNP matrix of Nullarbor, version 2.0. Sequence reads were deposited in the NCBI Archive (SRA: PRJNA559200). The data set was supplemented with 121 published genome sequences of C. auris (SRAs: PRJEB21518, PRJNA328792, PRJNA328792, PRJEB20230) [1, 3, 8]. The number and location of SNPs called by the 3 pipelines were highly concordant (correlation coefficient r = .999) (Supplementary Tables 1–4).
Genes known for their role for antifungal drug resistance in C. auris and Candida spp. (ERG11, MDR1, CDR1 [azole resistance], FKS1 [echinocandin resistance], and ERG2, ERG3, ERG5, ERG6 [amphotericin B resistance]) [19, 20] were examined for SNPs using the criteria above (CLC Genomics Workbench; Bio, version 7.0, Aarhus, Denmark) [16].
Table 1 summarizes the isolate details including the timeline of Australian isolates. Six of 7 isolates from Case 1 were cultured over a period of 3 weeks during hospitalization. The seventh (incident isolate from South Africa) was forwarded to us for comparative analysis. Isolates cultured from various body sites from Case 1 were available for study.
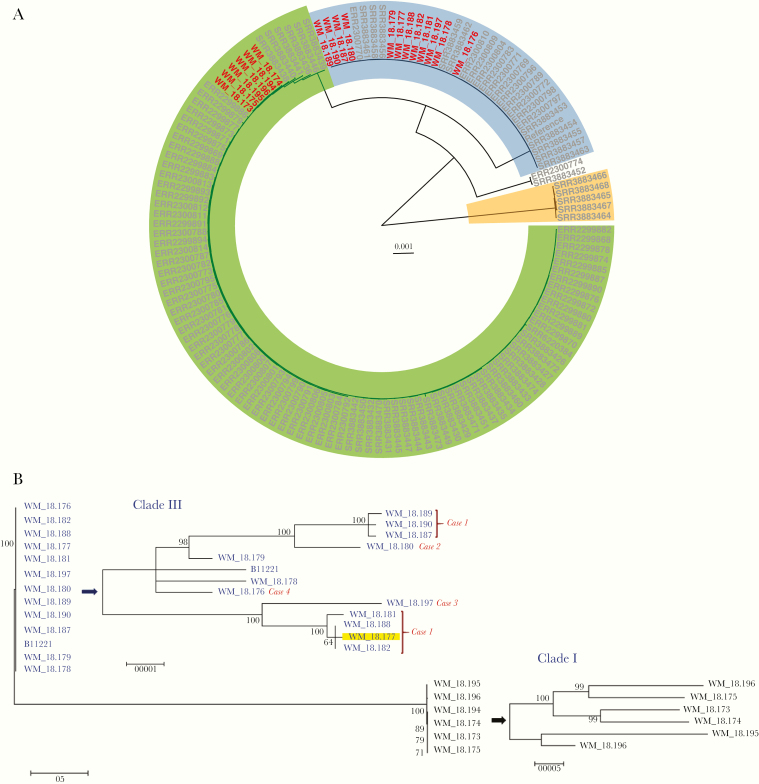
Phylogenetic analysis of the study isolates identified 2 major isolate clusters assigned to clades I and III (Figure 1A). All Australian isolates clustered within clade III (Table 1), whereas the Indian isolates grouped within clade I, separated from clade III by >40 000 SNPs. Genomes of isolates from Case 1 formed 2 distinct “groups,” separated by 60–70 SNPs (Figure 1B; Supplementary Tables 2–4). Three isolates (strains WM_18.181, WM_18.188, and WM_18.182) were in the same subgroup as the incident isolate (highlighted in yellow). Within each subgroup, the SNP difference between isolates was 6–9 SNPs. For example, for isolates from Case 1, strains WM_18.181 and WM_18.182 were separated by 5–8 SNPs. However, others, also from Case 1, differed by >60 SNPs (eg, WM_18.181 and WM_18.190) (Supplementary Tables 2 and 3).
Figure 1.
A, Circular maximum likelihood phylogeny by r package ggtree of Candida auris isolates. Clades are represented by highlighted sections of the tree—clade I (green), clade II (white), clade III (blue), and clade IV (orange)—and generally correspond with geographical regions, namely India/Pakistan, Japan, South Africa, and South America, respectively. “Australian” isolates are depicted by the red tip labels. B, Maximum likelihood phylogeny of Candida auris isolates using MEGA, version 6.0, with expanded subtree views with scales and bootstrap values on the righthand side. Isolate identification (tips) and arrows are colored by clade (clade III: blue; clade I: black). The yellow highlighted tip represents the index isolate from case 1 in South Africa before repatriation to Australia (see the text and Table 1 for further details). For the Australian isolates, the case from which the isolate was cultured is shown on the tree tip.
Isolates from Cases 2, 3, and 4 and 2 other South African isolates (Table 1) were all distinct from each other (>60 SNPs separating their genomes). Of 2 PCR fingerprinting patterns (Figure S1), pattern 1 encompassed the 6 Indian isolates, indistinguishable from each other, and corresponded to genome clade I. Other isolates belonged to pattern 2, also indistinguishable with the exception of 1 strain, and corresponded to clade III.
Isolates had low 5-flucytosine MICs (0.06–0.12 µg/mL) (Table 1). Clade III isolates showed WT MICs to amphotericin B (≤1 µg/mL) with no SNPs in ERG2, ERG3, ERG5, and ERG6; 5/6 Indian isolates had MICs at the proposed breakpoint of 2 µg/mL [14] and harbored an ERG2 SNP at nucleotide position A117C (E39D mutation) (Supplementary Table 5). Isolates universally had high fluconazole MICs (32–>256 µg/mL) with varying susceptibility to other azoles. Two SNPs in ERG11 were present, leading to mutations V125A and F126L in the clade III isolates, while mutations Y132F and K143R were in Indian isolates, as were nonsynonymous SNPs in the MDR1 (T801G and A1225G) and CDR1 (A2127T) genes. Australian and South African isolates had echinocandin MICs of ≤0.5 µg/mL, but a single pan-echinocandin-resistant isolate (strain WM_18.194) from India contained the FKS1 mutation, S639F. No FKS1 mutations was detected in 2 Indian strains with MICs of ≥8 µg/mL to caspofungin only (Tables 1; Supplementary Table 5).
The value of WGS for investigation of nosocomial C. auris infections has been documented, yet there are few data on its application to describe the genomic diversity of isolates outside the outbreak setting. Here we present the phylogenetic relatedness based on WGS of 10 Australian isolates from 4 sporadic cases in the wider geographical context. There was no distinct Australian clade as noted for other Australian isolates [11, 12] but rather independent importation events with strains reflecting likely geographic origin of acquisition.
Interestingly, in the nonoutbreak setting herein, whereas isolates from different cases had genomes separated by >60 SNPs, as found elsewhere [1–3] for clade III isolates, those from Case 1 were separated by an unexpectedly higher number of SNPs, with no isolate being identical. This finding contrasts with those from nosocomial C. auris outbreaks where intraclade diversity between isolates from any 1 individual was low (<3 SNPs) [1, 2, 21] although 1 study showed less spatiotemporal clustering [4]. One likely explanation for our findings, although unproven, is that at least 2 distinct populations were acquired while in the hospital in South Africa (a group of 4 and another group of 3 isolates) (Figure 1B). Regardless, the results indicate the presence of genetically heterogeneous strain populations colonizing the host simultaneously where such heterogeneity may enhance fitness [4, 22]. The sampling of >1 body site and/or on >1 occasion to detect C. auris may assist studies addressing this question. We included 2 de novo South African isolates as a “benchmark” for clade classification. Despite the lower discriminatory power of PCR fingerprinting, it may be useful for rapid clade identification in laboratories without WGS technology, as different clades are associated with different drug resistance patterns, which impact empiric antifungal therapy.
Importantly, despite the absence of interpretive MICs, resistance to ≥2 drug classes was not evident among Australian isolates compared with 20%–44% resistance rates elsewhere [1, 6]. Multidrug resistance may be location- or clade-specific. As before, isolates exhibited reduced fluconazole susceptibility and varying susceptibility to other azoles [1–3, 6]. Reassuring, given that echinocandins are first-line agents for treating C. auris infections [23], strains had low echinocandin MICs and WT MICs to amphotericin B [2–4, 20]. In parallel, no Australian isolate harbored FKS1 mutations, for example, involving S639 substitutions [7, 24, 25]. This amino acid is homologous to that at position S645 in C. albicans, where amino acid changes confer high-level echinocandin resistance [26]. Conversely, the pan-echinocandin-resistant clade I strain contained the S639F mutation, as have other pan-echinocandin-resistant isolates. The mutations S639P and S639Y, linked to echinocandin resistance, were absent in our isolates [20, 24].
WGS analysis also informed ERG11 mutations, homologous to those linked to azole resistance in C. albicans [1, 26]. That K143R and Y132F substitutions were present in Indian but not Australian and South African isolates is consistent with the notion these are clade I–specific markers of resistance [1, 17]. Conversely, the F126L and V125A mutations (clade III–specific) [1, 20] were present in Australian and South African isolates only. The association of novel SNPs in MDR1 and CDR1 in the Indian isolates with azole resistance is uncertain, as is the link to amphotericin B resistance with the mutation E39D.
The limitations of our study include the small number of Australian isolates due to the sporadic nature of infection, and in the absence of an outbreak, the environment was not sampled. The unusually high within-host genetic diversity was documented only in 1 case, as multiple isolates from the other patients were not available, limiting wider clinical relevance. Detailed study of patient data and investigation of resistance mechanisms to azoles were outside of the study scope.
In conclusion, analysis of C. auris genomes in a nonoutbreak setting demonstrated that WGS added value by differentiating independent importations from recent transmission events. Within-host diversity of C. auris suggests concurrent colonization of the host by heterogeneous populations. These findings improve our understanding of colonization/infection by C. auris and assist in interpretation of growing genomic data for this pathogen.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We wish to thank Drs. Ranjeeta Menon, Elena Martinez, and Nathan Bachmann, Centre for Infectious Diseases and Microbiology-Public Health, The University of Sydney, for their advice on the bioinformatics analysis. We also thank Ms. Sunita Gupta, Postgraduate Institute of Medical Education and Research, Chandigarh, India, and Ms. Ruth Mpembe, National Institute for Communicable Diseases, Johannesburg, South Africa, for forwarding isolates from their countries. The authors also thank the Sydney Informatics Hub of the University of Sydney for providing the high-performance computing resources for comparative genomics.
Author contributions. Study conception: S.C.A.C., V.S., C.B., B.H., S.V.H., W.M. Data generation: C.B., Q.W., S.V.H., C.H., B.H., S.C.A.C., K.M., A.K.G. Data analysis: C.B., Q.W., S.V.H., S.C.A.C., D.E., C.H., V.S., L.I., W.M. Wrote the manuscript: S.C.A.C., C.B., Q.W., S.V.H., V.S. Editing and approving final manuscript: all authors.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017; 64:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow NA, Gade L, Tsay SV, et al. ; US Candida auris Investigation Team. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 2018; 18:1377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eyre DW, Sheppard AE, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 2018; 379:1322–31. [DOI] [PubMed] [Google Scholar]
- 4. Rhodes J, Abdolrasouli A, Farrer RA, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 2018; 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Welsh RM, Bentz ML, Shams A, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 2017; 55:2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osei Sekyere J. Candida auris: a systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2018; 7:e00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chow NA, Munoz JF, Gade L, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 2020; 11. doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sekizuka T, Iguchi S, Umeyama T, et al. Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS One 2019; 14:e0223433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis 2019; 25:1780–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma C, Kumar N, Pandey R, et al. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 2016; 13:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heath CH, Dyer JR, Pang S, et al. Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015. Emerg Infect Dis 2019; 25:192–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lane CR, Seemann T, Worth LJ, et al. Incursion of Candida auris into Australia, 2018. Emerg Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer W, Latouche GN, Daniel HM, et al. Identification of pathogenic yeasts of the imperfect genus Candida by polymerase chain reaction fingerprinting. Electrophoresis 1997; 18:1548–59. [DOI] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention. Candida auris - antifungal susceptibility testing and interpretation 2017. Available at: https://wwwcdcgov/fungal/candida-auris/c-auris-antifungal. Updated 21 December 2018. Accessed January 2020.
- 15. Arendrup MC, Prakash A, Meletiadis J, et al. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 2017; 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biswas C, Chen SC, Halliday C, et al. Whole genome sequencing of Candida glabrata for detection of markers of antifungal drug resistance. J Vis Exp 2017; ( 130). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muñoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 2018; 9:5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Healey KR, Kordalewska M, Jimenez Ortigosa C, et al. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 2018; 62:e01427–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chowdhary A, Prakash A, Sharma C, et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 2018; 73:891–9. [DOI] [PubMed] [Google Scholar]
- 21. Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 2016; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holland SL, Reader T, Dyer PS, Avery SV. Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environ Microbiol 2014; 16:1729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsay S, Kallen A, Jackson BR, et al. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin Infect Dis 2018; 66:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kordalewska M, Lee A, Park S, et al. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother 2018; 62:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perlin DS. Mechanisms of echinocandin antifungal drug resistance. Ann N Y Acad Sci 2015; 1354:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flowers SA, Colón B, Whaley SG, et al. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 2015; 59:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.