Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 19 (COVID-19) has rapidly spread to the entire world within a few months. The origin of SARS-CoV-2 has been related to the lineage B Betacoronavirus SARS-CoV and SARS-related coronaviruses found in bats. Early characterizations of the SARS-CoV-2 genome revealed the existence of a distinct four amino acid insert within the spike (S) protein (underlined, SPRRAR↓S), at the S1/S2 site located at the interface between the S1 receptor binding subunit and the S2 fusion subunit. Notably, this insert appears to be a distinguishing feature among SARS-related sequences and introduces a potential cleavage site for the protease furin. Here, we investigate the potential role of this novel S1/S2 cleavage site and present direct biochemical evidence for proteolytic processing by a variety of proteases. We discuss these findings in the context of the origin of SARS-CoV-2, viral stability, and transmission.
Subject Areas: Biochemistry, Virology
Graphical Abstract
Highlights
-
•
SARS-CoV-2 spike protein harbors a distinct four amino acid insertion at the S1/S2 site
-
•
The S1/S2 site can be cleaved by furin-like, trypsin-like, and cathepsin proteases
-
•
The S1/S2 insert likely enhances spike protein cleavage by several proteases in vivo
Biochemistry; Virology
Introduction
Since December 2019, human infections by a novel coronavirus (CoV) named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have rapidly spread globally from an initial outbreak of severe pneumonia centered on Wuhan, Hubei Province, China. The virus is the etiological agent of an infectious respiratory disease termed coronavirus disease 19 (COVID-19). On March 11th 2020, due to its alarming spread and severity across most countries globally the WHO characterized COVID-19 as a pandemic.
Initial genomic characterization and phylogenetic analyses revealed that SARS-CoV-2 is related to the lineage B betacoronavirus SARS-CoV, which caused the 2002–2003 SARS epidemic. Bats are considered as the likely source of SARS-CoV and SARS-CoV-2 viruses, as a rich diversity of SARS-related (or SARS-like) viruses have been identified in several bat species. Notably, a bat coronavirus named BatCoV-RaTG13 has been identified as being most closely related to SARS-CoV-2 on a genomic level with a remarkable 96% sequence identity (Zhou et al., 2020b). To date, the direct precursor of SARS-CoV-2 and the involvement of an intermediate host in the emergence of the human virus remain to be identified, and there has been much attention drawn to the origin of the virus.
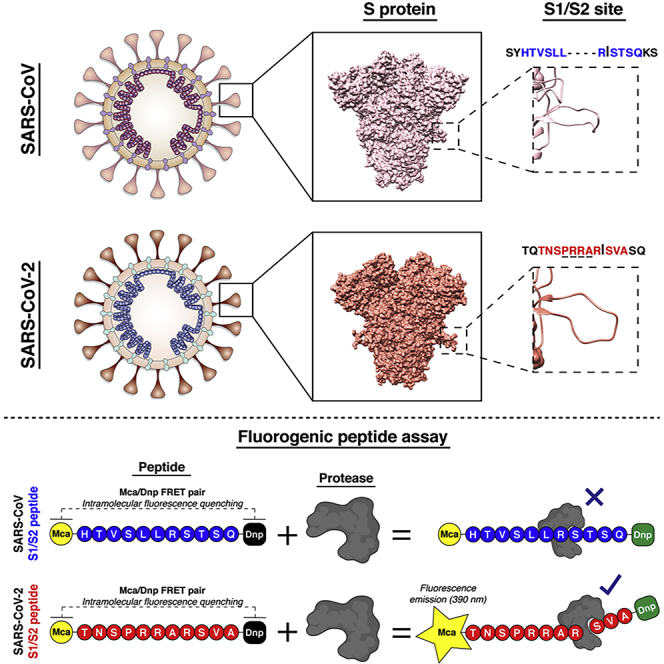
One of the notable features of the SARS-CoV-2 genome is a four amino acid insert (shown underlined below) found at the S1/S2 junction of the SARS-CoV-2 spike protein. Following the release of the SARS-CoV-2 genome sequence, several groups including ourselves identified this insert (underlined, SPRRAR↓S) as a potential cleavage site for the protease furin (Coutard et al., 2020, Hoffmann et al., 2020a, Jaimes et al., 2020, Walls et al., 2020, Wrapp et al., 2020), which is widely used to activate the fusion machinery of viral glycoproteins. This insert has also been referred to as a “polybasic site” or “multibasic site” (Andersen et al., 2020, Hoffmann et al., 2020a). A recent publication highlighted the role of furin in the activation of the SARS-CoV-2 spike protein (Hoffmann et al., 2020a). However, trypsin-like proteases (notably TMPRSS2) and cathepsins have also recently been shown to be functionally important for SARS-CoV-2 infection (Hoffmann et al., 2020b, Ou et al., 2020). Previous studies on SARS-CoV, MERS-CoV, and other coronaviruses have shown that activation of the spike protein is often a complex process involving multiple cleavage events occurring at distinct sites and with the involvement of several host proteases (Hulswit et al., 2016). Moreover, work on SARS-CoV has also demonstrated the criticality of a cleavage event occurring at another site termed S2’ (Belouzard et al., 2009), found in the S2 fusion domain immediately upstream of the fusion peptide—whose functional role is more directly comparable to that of influenza virus HA cleavage site (Steinhauer, 1999).
Several cellular proteases have been described to participate in the proteolytic activation of the CoV S protein. The proprotein convertase (PC) family of enzymes encloses a group of nine proteases with a diverse range of substrate activity. From this group, furin is ubiquitously expressed in the Golgi apparatus of all cells, but generally only at low levels—with some cell types showing enhanced expression or altered intracellular distribution (Shapiro et al., 1997). Trypsin is a serine endopeptidase that is commonly expressed in digestive and respiratory cells, and it has been described to be highly active as a digestive enzyme in the small intestine (Millet and Whittaker, 2015). Cathepsins, on the other hand, are a diverse group of proteases that typically found in endosomes and lysosomes and are involved in numerous degradative and antigen presenting processes (Millet and Whittaker, 2015). Finally, type II transmembrane serine proteases or TTSP, are another group of enzymes with diverse cellular functions, that have been described to participate in viral activation (Millet and Whittaker, 2015). This group encloses enzymes such as transmembrane protease, serine 2 (TMPRSS2), and matriptase, among others. As seen with other coronaviruses (Le Coupanec et al., 2015, Licitra et al., 2013, Millet and Whittaker, 2014), the role of furin (or other proteases) is potentially highly significant but needs to be carefully interpreted in the context of other aspects of virus infection. The role of furin cleavage as a virulence factor for viruses is rooted in findings on influenza virus, where a “polybasic” or “multibasic” site was found to be a defining feature of highly pathogenic avian influenza (HPAI) (Kawaoka and Webster, 1988), as adopted by the World Organization for Animal Health (OIE). In a subset of avian influenza virus subtypes (i.e. H7 and H5) polymerase slippage inserts a stretch of purine residues at the interface of the HA1 and HA2 subdomains, based on a specific secondary RNA structure found in the H7 and H5 HA gene (Nao et al., 2017). Such polybasic sites typically exist as a stretch of 6-7 arginine (R) and lysine (K) residues (e.g. RKKRKR↓G) that can be cleaved by furin, therefore allowing systemic spread based on the ubiquitous expression of the protease. Without the polybasic cleavage site, infection is restricted based on the localized presence of the trypsin-like protease activating low pathogenicity influenza viruses that are more abundant and usually restrict their infection to the respiratory tract. However, other influenza viruses (notably H9) can modulate cleavage sites in a distinct and less well-recognized way, creating a more defined (but not necessarily “polybasic”) furin cleavage site through recombination and mutation (Lee and Whittaker, 2017), which results in a more virulent form of the virus.
Here we provide context and clarify the role of the novel SARS-CoV-2 S1/S2 cleavage site in virus emergence and infection and perform a direct assessment of the proteases cleaving this site by use of biochemical assays.
Results and Discussion
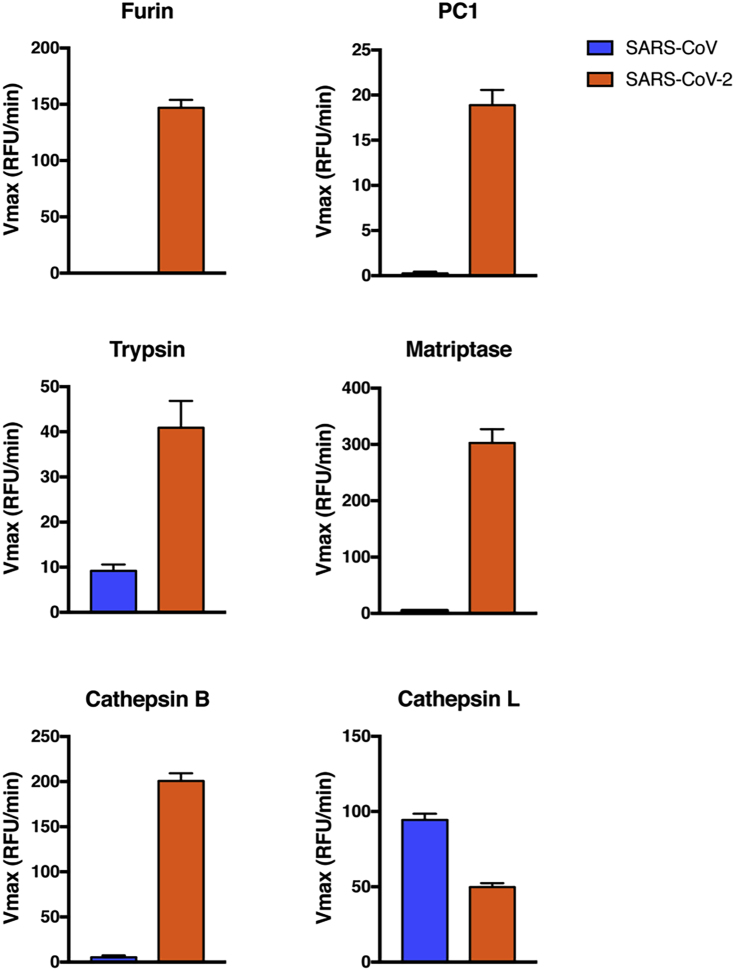
To directly address the proteases cleaving the SARS-CoV-2 S1/S2 site, we used a biochemical peptide cleavage assay (Jaimes et al., 2019), which was previously used to screen emerging influenza viruses (Straus and Whittaker, 2017). This assay was also successfully used to study the functional impact of mutations at coronavirus spike cleavage sites for feline coronavirus and MERS-CoV (Licitra et al., 2013, Millet et al., 2016). The peptide sequences used here were HTVSLLRSTSQ (SARS-CoV S1/S2) and TNSPRRARSVA (SARS-CoV-2 S1/S2). We tested a range of proteases likely to be involved in spike protein processing–the proprotein convertases furin and PC1, trypsin, and the type II transmembrane serine protease (TTSP) matriptase, as well as cathepsins B and L. As predicted, furin cleaves SARS-CoV-2, but not SARS-CoV (Figure 1). However, in addition to furin, other proteases also cleaved SARS-CoV-2 much more readily than SARS-CoV. PC1 showed a similar cleavage pattern but with lower efficiency than for furin. Trypsin cleaved both peptides, but was over 4-fold more efficient for SARS-CoV-2. Both the TTSP matriptase and cathepsin B did not cleave SARS-CoV at all but were highly active on SARS-CoV-2. The only protease that showed more cleavage on SARS-CoV compared with SARS-CoV-2 was cathepsin L. Our data demonstrate that the S1/S2 site of SARS-CoV-2 S is efficiently cleaved by a wide range of proteases, not only furin. The comparative data with SARS-CoV S1/S2 site reveals that the acquisition of the four amino acid insert distinctively broadens the activating protease repertoire of the SARS-CoV-2 S1/S2 cleavage site to all major classes of proteolytic enzymes known to potentially activate coronavirus S proteins.
Figure 1.
Proteolytic Cleavage Assay of Peptides Derived from SARS-CoV and SARS-CoV-2 S1/S2 Spike Protein Sites
Fluorogenic peptide mimetics of the S1/S2 spike cleavage sites of SARS-CoV (HTVSLLRSTSQ) and SARS-CoV-2 (TNSPRRARSVA) were incubated with the following recombinant proteases (indicated on top of graphs): furin, PC1, trypsin, matriptase, cathepsin B, and cathepsin L. The increase in fluorescence due to proteolytic processing was measured using a fluorometer enabling calculation of the velocity of each cleavage reactions (Vmax). The assays were performed in triplicates, with results representing Vmax averages obtained from three independent experiments (n = 3). Error bars indicate SD.
The COVID-19 pandemic represents a global public health emergency. The origin of the causative agent of the disease, SARS-CoV-2, remains a mystery. Previous analyses have revealed distinctive features within the genome of SARS-CoV-2. In particular, a four amino acid insert was found within the S1/S2 site of the S glycoprotein. This finding has garnered interest because of the possible introduction of a furin recognition motif at the S1/S2 site. Our study aimed at clarifying the functional role of the S1/S2 cleavage site for SARS-CoV-2. We provide direct biochemical evidence that the site S1/S2 is recognized and cleaved by furin. However, other proteases such as PC1, another member of the PC family of proteases, trypsin-like proteases, and cathepsins can all efficiently recognize and cleave SARS-CoV-2 S1/S2 cleavage site. These data confirm earlier findings showing the SARS-CoV-2 entry was dependent, at least in part, on lysosomal cathepsins and the cell-surface-expressed TTSPs such as TMPRSS2 (Hoffmann et al., 2020b). Interestingly, the comparative assessment we present with SARS-CoV shows that the S1/S2 insertion SARS-CoV-2 has acquired substantially expands its proteolytic activation profile, with potential activation from a wide variety of proteases. Further experiments will include the evaluation of the protease cleavage of the complete trimeric form of the SARS-CoV-2 S protein.
Another feature of the S1/S2 junction that has been noted for SARS-CoV-2 is the presence of a leading proline (P) residue. We favor a structural interpretation for the role of this residue, whereby the presence of the leading proline—and the turn it creates—impacts the cleavability of the S1/S2 junction by directly affecting the accessibility of the cleavage loop with the active site of the protease. It has also been suggested that the leading proline residue allows the addition of O-linked glycans to neighboring residues (Andersen et al., 2020), with the suggestion that these glycans act to positively influence virulence by creating a mucin-like domain that shields critical epitopes from the immune system. Although this is possible, such glycan addition would also be predicted to shield the cleavage site from its activating protease, so negatively influencing virulence. Such a scenario (in this case via N-linked glycans) has been noted for influenza HA (Tse et al., 2014). Not all algorithms predict O-glycans in this region, and it remains to be determined whether the precited glycosylation sites are in fact utilized.
We favor a model whereby the novel S1/S2 insert allows furin cleavage but also reflects a more enhanced exposure of a critical cleavage site to TTSPs such as TMPRSS2 and matriptase, as well as cathepsin B, an activation mechanism that is not necessarily through the action of furin itself. Overall, the novel S1/S2 insert is likely to enhance spike protein cleavage by multiple proteases beyond that for other SARS-like viruses (e.g. containing the sequence SR↓S). The role of the fusion peptide-proximal S2′ site remains to be evaluated, but its sequence (PDPSKPSKR↓SFIEDLLF) is not distinctly different from other SARS-like viruses (Jaimes et al., 2020). We also propose that the SARS-CoV-2 S1/S2 cleavage site likely arose by mutation/recombination as for influenza H9 viruses, rather than by polymerase slippage as for the more widely appreciated polybasic sites found in H7 and H5 HPAI, and that the term “polybasic site” is a misnomer for SARS-CoV-2. This concept has recently been reinforced by the finding of a bat coronavirus (BatCoV-RmYN02) closely related to SARS-CoV-2 that has an extended S1/S2 cleavage loop lacking any additional basic residues (SPAAR↓S) (Zhou et al., 2020a). BatCoV-RmYN02 represents a good candidate to be involved in the emergence of SARS-CoV-2, lacking a furin cleavage site and likely utilizing a distinct receptor but providing evidence for a natural origin of SARS-CoV-2.
Our work underscores the importance of biochemical validation of newly identified coronavirus spike glycoprotein cleavage sites to gain a better understanding of the potential functional implications of proteolytic activation. Further investigation is required to identify precursor sequences that led to the emergence of SARS-CoV-2 and to shed light on the evolutionary mechanism(s) by which the virus acquired its distinct S1/S2 site.
Limitations of the Study
Fluorogenic peptides mimicking the SARS-CoV and SARS-CoV-2 S1/S2 regions were used to evaluate cleavability by recombinant cellular proteases. Each peptide is 11 amino acids long and harbors the specific S1/S2 sequence for each virus, and cleavage of the peptide by a protease is driven by both the specific amino acid sequence and the conformational structure of the protein. One of the limitations of the fluorogenic peptide assay is the structural conformation of the peptide, which may not resemble the original conformation of the full-length protein. This could result in altered cleavability by the cellular protease. Future experiments using purified full-length SARS-CoV and SARS-CoV-2 spike protein are necessary to validate the results obtained with our S1/S2 peptides.
Resource Availability
Lead Contact
Gary R. Whittaker, grw7@cornell.edu. Address: 930 Camous Rd., Ithaca, NY 14853.
Materials Availability
The fluorogenic peptides were synthetized by a commercial laboratory and its availability in our laboratory is limited.
Data and Code Availability
No custom code, software, or algorithm were used in this research.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Ruth Collins and all member of the Whittaker and Daniel labs at Cornell University for comments and discussion. Work in the author's laboratory is supported by the National Institutes of Health (research grant R01AI35270).
Author Contributions
Conceptualization, J.A.J., J.K.M., and G.R.W.; Methodology, J.A.J. and J.K.M.; Investigation, J.A.J.; Writing—Original Draft, J.K.M. and G.R.W.; Writing—Review & Editing, J.A.J., J.K.M., and G.R.W.; Visualization, J.A.J. and J.K.M.; Supervision, G.R.W.; Funding acquisition, G.R.W.
Declaration of Interests
The authors manifest no conflict of interest.
Published: June 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101212.
Supplemental Information
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U S A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;28:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Andre N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020 doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Millet J.K., Goldstein M.E., Whittaker G.R., Straus M.R. A fluorogenic peptide cleavage assay to screen for proteolytic activity: applications for coronavirus spike protein activation. J. Vis. Exp. 2019;143:e58892. doi: 10.3791/58892. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R.G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U S A. 1988;85:324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Coupanec A., Desforges M., Meessen-Pinard M., Dube M., Day R., Seidah N.G., Talbot P.J. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11:e1005261. doi: 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Whittaker G.R. Use of AAScatterPlot tool for monitoring the evolution of the hemagglutinin cleavage site in H9 avian influenza viruses. Bioinformatics. 2017;33:2431–2435. doi: 10.1093/bioinformatics/btx203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19:1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J., Whittaker G. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Goldstein M.E., Labitt R.N., Hsu H.-L., Daniel S., Whittaker G.R. A camel-derived MERS-CoV with a variant spike protein cleavage site and distinct fusion activation properties. Emerg. Microbes Infect. 2016;5:e126. doi: 10.1038/emi.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci.U S A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio. 2017;8 doi: 10.1128/mBio.02298-16. e02298–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J., Sciaky N., Lee J., Bosshart H., Angeletti R.H., Bonifacino J.S. Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 1997;45:3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- Straus M.R., Whittaker G.R. A peptide-based approach to evaluate the adaptability of influenza A virus to humans based on its hemagglutinin proteolytic cleavage site. PLoS One. 2017;12:e0174827. doi: 10.1371/journal.pone.0174827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse L.V., Hamilton A.M., Friling T., Whittaker G.R. A novel activation mechanism of avian influenza virus H9N2 by furin. J. Virol. 2014;88:1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No custom code, software, or algorithm were used in this research.