Figure 1.
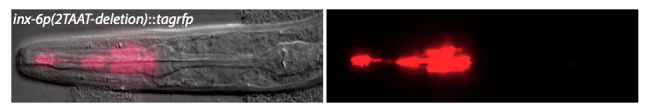
inx-6p(2TAAT-deletion)::tagRFP reporter (otEx5854) is only expressed in the procorpus region of the pharynx.
Description
Among the fluorescence-based co-transformation markers, two of the most commonly used markers are pharyngeal expression of green fluorescent protein (GFP) and red fluorescent protein (RFP) under the myo-2 cis-regulatory element (Frokjaer-Jensen et al., 2008; Tabara et al., 1996). However, bright expression of GFP or RFP in the posterior pharynx, i.e. in the isthmus and in the posterior pharyngeal bulb, restricts expression analysis of reporter genes that express fluorescent proteins with overlapping spectra in the head neurons. This shortcoming particularly limits the choice of fluorescent proteins that can be used when simultaneous imaging of multiple distinct reporter genes is required. We report here generation of a new fluorescence-based co-transformation marker that allows easy identification of transgenic animals based on bright TagRFP expression only in the anterior pharyngeal region (procorpus region) muscles at all developmental and adult stages (Figure 1) and does not interfere with the analysis of reporter gene expression in the head neurons.
This reporter expressed TagRFP under the regulation of 1654 bp cis-regulatory region upstream of the inx-6 locus (Primers used to amplify the cis-regulatory region: upstream: 5’ cgataagattttgacgaatccg 3’ and downstream: 5’ tgtgaacaagctaaggagag 3’). We deleted two conserved putative homeodomain binding sites (TAAT) present within this region. Removal of the first binding site, at 422 bp from the start of the cis-regulatory region (TGTAATAC>TGAC) prevented reporter gene expression in the marginal cells. Deletion of the second binding site, at 521 bp (GATAATTA>GATA) prevented reporter gene expression in AIB interneurons during dauer and L1-diapause stages (Bhattacharya, 2018). We typically used 40ng/ul of circular reporter plasmid (pAB1) in microinjections. For complex arrays (Kelly et al., 1997), we used 6-8ng/ul of linearized pAB1.
Reagents
OH12747 otEx5854[inx-6p(2TAAT-deletion)::tagRFP (8ng/ul); pRF4 (rol-6) (5ng/ul), OP50 gDNA (100ng/ul)]. Will be available at CGC.
pAB1: inx-6p(2TAAT-deletion)::tagRFP::unc-54 3’ UTR; contains 1654 bp region upstream of inx-6 coding region, where two TAAT sites were deleted. Will be available at Addgene.
Acknowledgments
Acknowledgments
Conceptualization, A.B., and O.H.; Methodology, A.B.; Data curation, A.B.; Writing – original draft, A.B.; Revision, A.B.; Funding acquisition, O.H.; Supervision, A.B., and O.H.
Funding
This work was supported by the NIH (R21NS106909) and the Howard Hughes Medical Institute.
References
- Bhattacharya, A., Aghayeva, U., Berghoff, E., Hobert, O. (2018). Plasticity of the electrical connectome of C. elegans. Cell (in press), BioRxiv 406207. doi: https://doi.org/10.1101/406207. [DOI] [PMC free article] [PubMed]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008 Oct 26;40(11):1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997 May 01;146(1):227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Motohashi T, Kohara Y. A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans. Nucleic Acids Res. 1996 Jun 01;24(11):2119–2124. doi: 10.1093/nar/24.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]


