Abstract
Down syndrome (DS) results from the triplication of genes located on human chromosome 21 (Hsa21). Though many cognitive and behavioral impairments are associated with DS, sleep disturbances remain poorly understood despite being a reported phenotype in approximately 60% of individuals diagnosed with DS. In this study, sleep and electroencephalography (EEG) oscillations were recorded from aged (12–14 mos.) Dp(16)1Yey/+ mice (Dp16), a mouse model of DS. We observed disrupted sleep demonstrated by increased activity during the dark phase and increased time awake at the expense of NREM sleep compared to wild-type mice. In addition, we found that Dp16 mice display significant differences in relative EEG power distribution among oscillation frequencies in both sleep and awake states. These results in Dp16 mice are consistent with sleep disturbances found in individuals with DS, and the abnormal EEG oscillations in aged Dp16 mice suggest a potential role for GABAergic activity in these sleep and EEG abnormalities. These sleep and EEG data reflect underlying differences in neuronal activity at the network level and thus are causative agents rather than merely symptoms of DS.
Keywords: Down syndrome, EEG, spectral power, Dp16, sleep, beta
INTRODUCTION
Down syndrome (DS) arises from triplication of genes on human chromosome 21 (Hsa21) and is the most common genetic cause of intellectual disability (Desai, 1997; Parker et al., 2010). In addition to the multitude of adverse health outcomes associated with DS, approximately 60% of the DS population displays sleep disturbances (Fan et al., 2017). While in some cases these sleep disruptions result from sleep apnea, previous studies have shown sleep to be independently altered (Nisbet et al., 2015). These sleep alterations consist of increased latency to NREM, sleep fragmentation, and reduced REM (Hamaguchi et al., 1989; Andreou et al., 2002; Fernandez and Edgin, 2013). One way to quantitatively examine changes in sleep structure is by measuring brain activity using electroencephalography (EEG). EEG analyses are advantageous because alterations in EEG activity may serve as a potential biomarker for cognitive impairment in DS (Velikova et al., 2011; Salem et al., 2015). Previous EEG studies have shown that DS individuals have altered EEG oscillations associated with cognitive deficits (Politoff et al., 1996; Śmigielska-Kuzia et al., 2005; Velikova et al., 2011; López-Loeza et al., 2016). However, EEG abnormalities and their role in sleep disturbances are poorly characterized. To date, only a single study has examined EEG activity during sleep in children diagnosed with DS (Śmigielska-Kuzia et al., 2005). In that study, Śmigielska-Kuzia and colleagues found a reduction in alpha power compared to controls during REM sleep. However, it remains unknown if subjects in this study suffered from obstructive sleep apnea (OSA), which is highly comorbid with DS and could serve as a potential confound in this study. Furthermore, whether these EEG abnormalities are present during sleep in adults with DS is currently unknown, as no studies to date have examined EEG activity during sleep in an adult DS population. Therefore, mouse DS models may serve as useful experimental platforms to remove potential confounds present in human DS (i.e. OSA) and delineate the genetic influences of Hsa21 on sleep and EEG activity in DS.
While multiple mouse models of DS are currently available, studies examining sleep and EEG activity in these models are sparse. Hsa21 carries approximately 250 protein-coding genes and one of the most commonly used mouse models of DS, the Ts65Dn mouse, contains a partial trisomy of Hsa21 genes from Zfp295 to Mrp139 on mouse chromosome 16 (Mmu16). This model has been shown to exhibit increased wakefulness and a reduction in NREM sleep that mirrors the clinical population of DS patients (Colas et al., 2008). In the same study, sleep abnormalities were not observed in Ts1Cje mice, another rodent genetic model of DS. This is of key importance because sleep is known to be under heavy genetic control (Franken et al., 2001). The Ts65Dn strain however, contains a trisomy of a ~5.8 Mb subcentromeric region of mouse chromosome 17 which is not syntenic to any region on Hsa21 (Akeson et al., 2001; Li et al., 2007). Therefore, it is possible that genes not present on Hsa21 could impact some of the reported sleep disturbances in the Ts65Dn mouse. To address this issue, the current study sought to examine sleep architecture in Dp(16)1Yey/+ mice (Yu et al., 2010), a recently developed mouse model of DS. The Dp16 mouse is trisomic for ~102 genes on Hsa21 syntenic regions, while not being confounded by additional genes unrelated to DS (Li et al., 2007; Yu et al., 2010).
Individuals with DS have a high risk of developing early-onset Alzheimer’s disease (AD). Alzheimer’s disease is also known to be comorbid with sleep disturbances (Bliwise, 2004a; Roh et al., 2012). In this study, we characterize for the first time, the sleep architecture and EEG activity in aged Dp16 mice. We found sleep and EEG activity were disturbed in aged Dp16 mice and these disturbances are consistent with previous findings in the DS population.
MATERIALS AND METHODS
Animals
Dp(16)1Yey/+ (Dp16) mice on a C57BL/6J background were obtained from Jackson Laboratory (stock #013530). Male Dp16 mice were bred with wild-type (WT) C57BL/6J female mice to generate the Dp16 (N = 10) and WT (N = 9) littermates used for this study. All experiments were performed using male age-matched littermates of 12- to 14-month-old obtained from three independent cohorts. Mice were maintained on a 12-h light/dark cycle at an ambient temperature of 22–24 °C. Food and water were available ad lib. All procedures relating to animal care and treatment conformed to University of Colorado at Boulder Animal Care and Use Committee and NIH guidelines.
Surgery
Head-mount and electrode implantation were performed per manufacturer protocol (Pinnacle Technology, Lawrence, KS). Briefly, animal surgeries were performed under deep isoflurane anesthesia (5% induction followed by 1.5–2% maintenance) in sterile conditions with animal body temperature thermostatically maintained using a heating pad. Under anesthesia, the eyes were covered with a thin layer of artificial tear ointment (Rugby, Livonia, MI), the fur from the head was removed and the skin disinfected with Betadine. A 1.5-cm rostral–caudal incision was made and the periosteum layer removed. The exposed skull was cleaned with ethanol prior to attaching a prefabricated head-mount containing three channels; two EEG and one EMG (Pinnacle Technology, #8201). Each head-mount was affixed to the skull using cyanoacrylate in addition to four stainless steel screws (2.5 mm for anterior placement and 3.0 mm for posterior placement (Pinnacle Technology, #8209)), which also served as electrodes. To ensure that all screws rested on the cerebral cortex for optimal EEG alignment, the front edge of the head-mount was placed 3.0 mm anterior of bregma. Recording leads were coated with silver epoxy (Pinnacle Technology) for a solid connection to the head-mount. To insert the EMG wires, a small pocket was made in the nuchal muscles and the EMG leads were positioned in the muscle cavity. The head-mount was secured with Metabond dental cement (Parkell, NY). All animals received buprenorphine (0.1 mg/kg) post-surgery, were individually housed in recording chambers (20 cm high and 25.4 cm in dia., Pinnacle Technology) and allowed 7–10 days to recover.
Apparatus
Animals were habituated to the recording cable for 3–5 days prior to EEG/EMG recording. Electrophysiological signals were collected from the mice using 6-pin preamplifiers (Pinnacle Technology, #8202). These were connected to swivel commutators (Pinnacle Technology, #8204) connected to a secondary amplifier and data conditioner (DCAS #8206, Pinnacle Technology). EEG and EMG signals were sampled at 2000 Hz. The EEG was low-pass filtered at 100 Hz and high-pass filtered at 0.1 Hz; EMG was low-pass filtered at 6 KHz and high-pass filtered at 3 Hz. All data were collected using Sirenia Sleep Pro (Version 1.7.4, Pinnacle Technology). Animal motor activity was recorded (N = 4–7 mice per group as some locomotor activity data across the 24-h period was lost due to recording loss) by video camera and activity scored using AnyMaze behavioral scanning software (Stoelting USA). EEG, EMG and motor activity for each animal were recorded for two 24-h periods.
Electrophysiological signal analysis
EEG recordings for each animal were initially scored using semi-automated cluster analyses based on EEG and EMG activity over 24 hrs. The recordings were divided into 4-s epochs and each epoch was scored as awake (wake; high frequency, low amplitude EEG with high and variable EMG activity), slow-wave sleep (NREM; low frequency, high amplitude EEG and reduced EMG activity) or rapid-eye movement sleep (REM; high frequency, low amplitude EEG with no EMG activity) states. Following the cluster analysis, epochs were analyzed manually to ensure accuracy by two independent evaluators blind to the genotype (Dp16 or WT) using Sirenia Sleep Pro software (Pinnacle Technology). All sleep measures (percent time in each state, bout length, state transitions, and EEG spectral power) were assessed across 24 h. A bout was defined as four consecutively scored epochs in wake, NREM, or REM without transitions. The time in each state, length of bouts, and transitions were quantified using Sirenia Sleep Pro (Pinnacle Technology). Additionally, wake and sleep states were verified with the video recordings. A Fast Fourier transform (FFT), using a Hann window function, of EEG data between 0.5 and 60 Hz from artifact-free epochs produced a spectral analysis in 0.25-Hz bins. These bins were grouped into standard frequency bands including: delta (0.5–4.5 Hz), theta (4.5–8 Hz), alpha (8–13 Hz), beta (13–30 Hz), and gamma (30–60 Hz) for each animal (Sirenia Sleep Pro, Pinnacle Technology). To control for differences in EEG head-mount placement across animals, relative power was calculated by normalizing all bands to the total state-specific power summed across all frequency bins from 0.5 to 60 Hz and expressed as a percent of total power.
Statistical analysis
Statistical analyses were performed using SPSS (IBM Corporation, Armonk, NY). Data are presented as mean ± SEM. The use of parametric tests was determined with the Shapiro–Wilk test for normality and outliers were identified and excluded using Grubb’s method. An alpha level of p < .05 was used to indicate a significant difference from controls. Repeated-measures ANOVA and two-tailed t-tests were used as applicable. For EEG analyses, relative power across the defined bandwidths for each sleep state was compared between groups using t-tests.
RESULTS
Dp16 mice exhibit increased locomotor activity
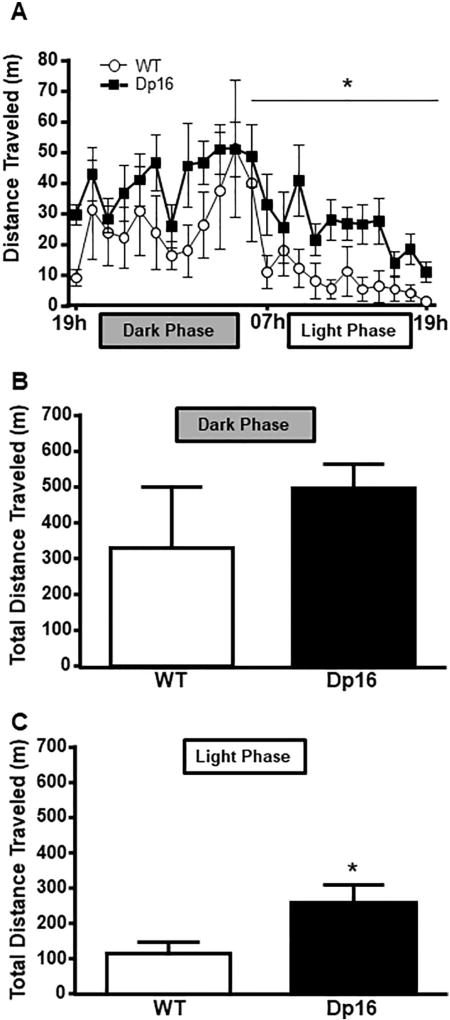
To examine whether sleep and activity patterns are disturbed in an aged mouse model of DS, we assessed the sleep and activity patterns in Dp16 and WT mice across two 24-h periods during the dark (e.g. active phase) and light (e.g. inactive phase) phases. During the dark phase (19 h–07 h) there were no significant differences between the genotypes in distances traveled (F(1,7) = 0.934, p = .366). However, across the 12-h light phase, Dp16 mice displayed significantly more locomotor activity than WT controls (F(1,7) = 78.08, p < .001; Fig. 1A). Total distance was calculated for both groups and no difference was observed during the dark phase (t(12) = 0.97, p = 0.11; Fig. 1B). However, Dp16 mice were significantly more active compared to WT controls during the light phase (Fig. 1C) as measured by total distance traveled (t(12) = 6.28, p = 0.041). This increase in activity during the light phase suggests an altered sleep–wake profile in aged Dp16 mice.
Fig. 1.
Dp16 mice are more active during the light phase of a 24-hour period. (A) Locomotor activity over 24 h. (B) Total distance traveled during the dark phase (1900 h–0700 h). (C) Total distance traveled during the inactive phase (0700 h–1900 h). Locomotor activity in Dp16 mice is significantly increased compared to WT. Data shown: mean ± SEM. *p < .05. N = 4–7 mice/per time point/group (note: each group contained 7 mice, but some data were lost during recording).
Altered sleep–wake profiles in Dp16 mice
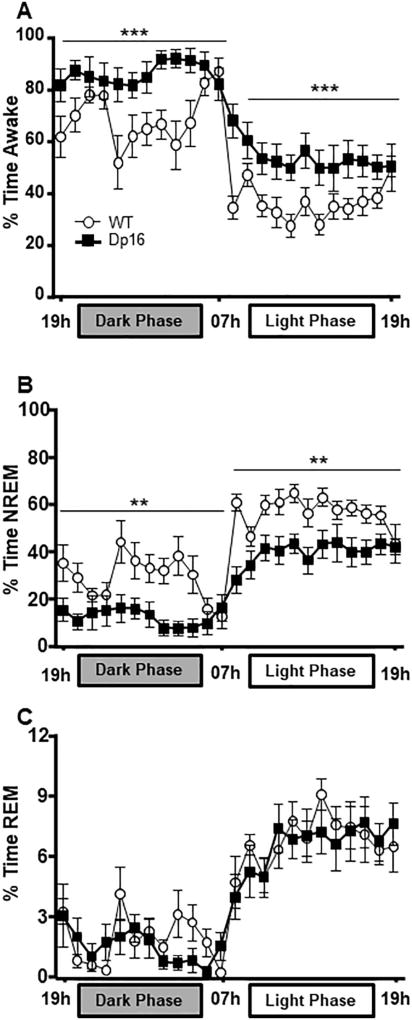
To determine whether sleep architecture was altered in aged Dp16 mice, we examined the percentage of time spent in wake, NREM, and REM states across 24 h by measuring cortical EEG activity in both aged Dp16 and WT mice (N = 8–10 per group). Dp16 mice spent more time awake in the dark phase (F(1,16) = 10.779, p = 0.005) and light phase (F(1,16) = 31.462, p < 0.001) compared to their WT littermates (Fig. 2A). Dp16 mice also spent less time in NREM during the dark phase (F(1,16) = 12.124, p = .0.003 and light phase (F(1,16) = 36.377, p < 0.001) compared to controls (Fig. 2B). No significant difference was observed between the two groups in REM during either period (Dark phase: F(1,16) = 0.487, p = 0.495, Light Phase: F(1,16) = 0.094, p = 0.763; Fig. 2C). The disturbed wake/sleep pattern in Dp16 mice could be explained by: (i) Dp16 mice have longer NREM bouts and therefore require less total time in sleep or (ii) Dp16 mice have difficulties maintaining sleep and therefore have increased transitions from sleep to wake.
Fig. 2.
Dp16 mice spent more time awake at the expense of NREM. (A) Percent time awake over 24 h. Dp16 mice spend more time awake in both dark and light phases. (B) Percent time in NREM over 24 h. Dp16 mice spend less time in NREM during both phases than WT controls. (C) Percent time in and in REM across 24 h. Data parsed into 1 h bins. (N = 8–10 mice per group). Data shown: mean ± SEM. *p < .05; **p < .01; ***p < .001.
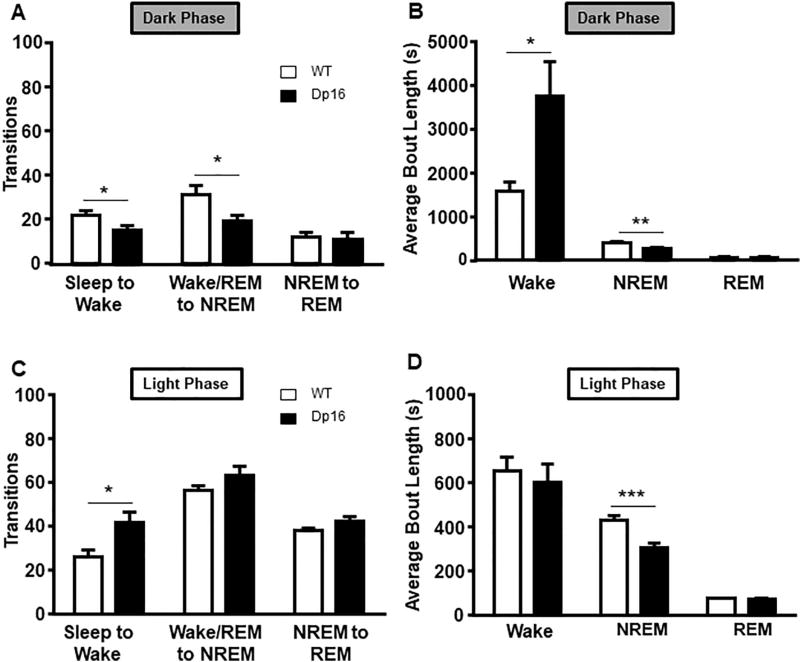
To distinguish between these phenotypes, the number of transitions and the average length of bouts were analyzed. These data revealed phase-specific differences. During the dark phase, significant decreases in the number of wake (t(16) = 2.18, p = 0.043) and NREM transitions (t(16) = 2.54, p = 0.021) in Dp16 mice were observed (Fig. 3A). This reduction in transitions during the dark phase is explained by the increase in wake bout length (t(16) = 2.28, p = 0.036) and decreased NREM bout length (t(16) = 3.18, p = 0.005) (Fig. 3B). During the light phase, the number of transitions to wake increased (t(16) = 2.47, p = 0.025) in Dp16 mice (Fig. 3C), while NREM bout length remained decreased (t(16) = 3.84, p < 0.001) (Fig. 3D). We also analyzed the latency to first NREM bout during the light phase, but found no significant differences between the groups (data not shown). The amount of REM sleep did not differ from WT animals. In summary, the lack of a significant difference in the number of transitions to NREM paired with an increase in transitions to wake during the light phase indicates that aged Dp16 mice have difficulty maintaining NREM sleep.
Fig. 3.
Dp16 mice show disturbed sleep pattern measured in number of transitions and average bout lengths. (A) During the dark phase, Dp16 mice have significantly fewer transitions to wake and NREM states. (B) Wake bout length during the dark phase is significantly increased in Dp16 mice, while NREM average bout length was reduced. (C) Compared to WT mice, Dp16 mice have significantly more transitions to wake paired with a reduction in NREM bout length (D) during the light phase. Data shown: mean ± SEM. *p < .05; **p < .01; ***p < .001. N = 8–10 mice per group.
Altered EEG spectral power in Dp16 mice
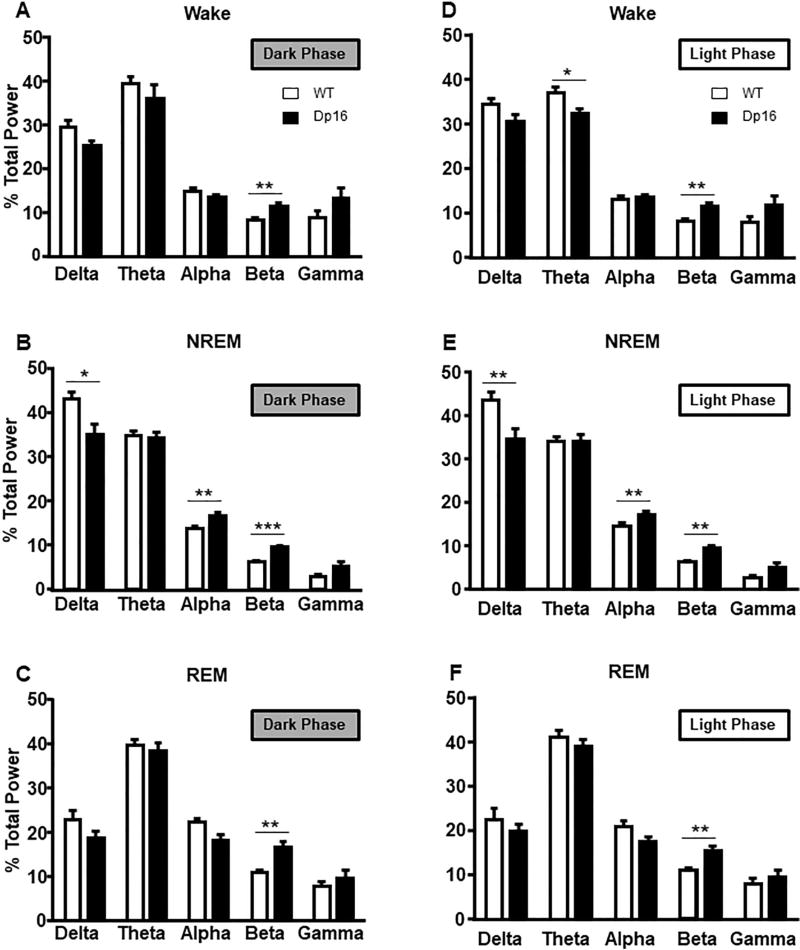
Next, we determined the power distribution in EEG oscillations during the dark and light phases for wake, NREM, and REM states across frequency bands (delta, theta, alpha, beta, and gamma). Analysis of the spectral power during the dark phase showed that beta power (13–30 Hz) was significantly increased in Dp16 mice during wake (t(17) = 3.04, p = 0.007), NREM (t(17) = 4.50, p < 0.001) and REM (t(17) = 3.84, p = 0.001) (Fig. 4A–C). Dp16 mice also showed significantly decreased delta power (t(17) = 2.59, p = 0.018) and increased alpha power (t(17) = 2.64, p = 0.017) during NREM (Fig. 4B). During the light phase, beta power in wake (t(17) = 3.66, p < 0.001), NREM (t(17) = 4.77, p < 0.001) and REM (t(17) = 3.53, p = 0.002) remains increased in Dp16 mice (Fig. 4D–F). Additionally, we found that Dp16 mice have reduced theta power (4.5–8 Hz) during wake states (t(17) = 2.24, p = 0.038) and reduced delta power during NREM (t(17) = 2.85, p = 0.01) (Fig. 4D, E). This reduction in theta was not present during the dark phase. Finally, to further assess EEG activity in Dp16 mice, we graphed relative spectral power across the full frequency spectra over 24 h (Fig. 5; Dark phase (A–C), Light Phase (D–F)). Taken together, these data show that EEG oscillations are altered in aged Dp16 mice, and these alterations are distinctly different during dark and light phases.
Fig. 4.
Dp16 mice have shifts in EEG power that vary across behavioral states during the dark phase (panels A–C) and light phase (panels D–F). Spectral power within delta (0.5–4 Hz), theta (4.5–8 Hz), alpha (8.5–14 Hz), beta (15–30 Hz), and gamma (30–60 Hz) expressed as a percentage of mean total EEG power for each state. Data shown: mean ± SEM. *p < .05; **p < .01; ***p < .001. N = 8–10 mice per group.
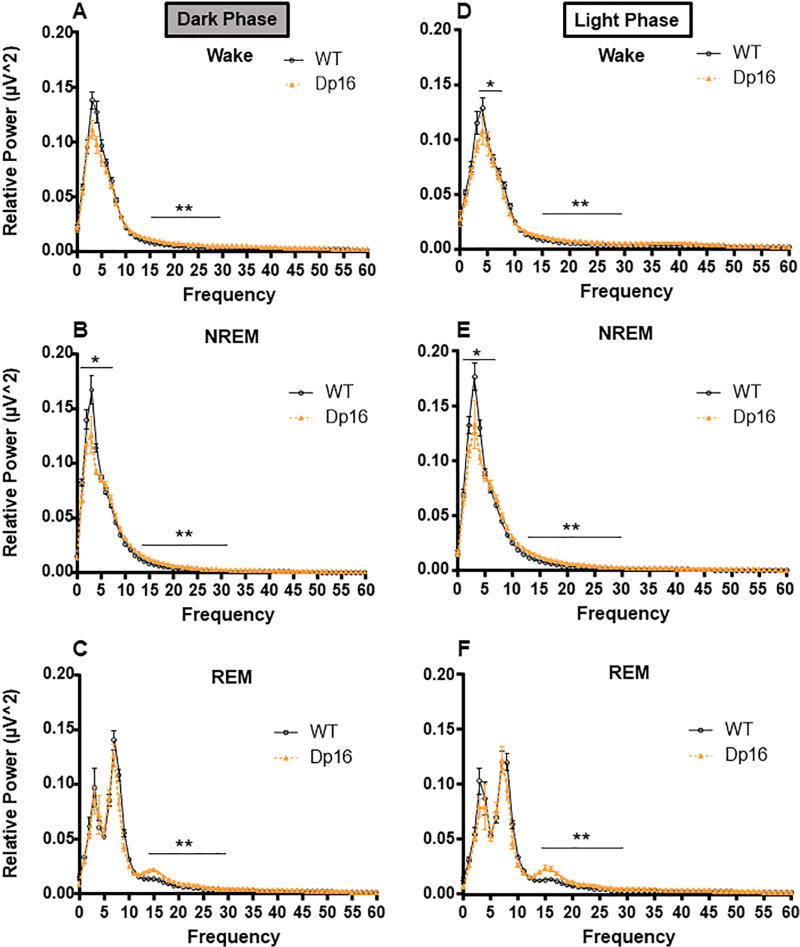
Fig. 5.
Relative spectral power across behavioral states during the dark phase (panels A–C) and light phase (panels D–F). Relative power density is displayed as the mean of total EEG power ± SEM. N = 8–10 mice per group. The frequency ranges used for power band calculations and statistical analyses were delta (0.5–4 Hz), theta (4.5–8 Hz), alpha (8.5–14 Hz), beta (15–30 Hz), and gamma (30–60 Hz). *p < .05; **p < .01; ***p < .001.
DISCUSSION
In the present study, we examined sleep behavior and EEG oscillations in the aged Dp16 mouse, a mouse model of DS that is trisomic for ~102 Hsa21 syntenic genes on mouse chromosome Mmu16. We found that aged Dp16 mice exhibit a sleep phenotype that consists of: (1) increased activity during the light phase of the light/dark cycle, (2) more time spent awake at the expense of NREM sleep and, (3) difficulty maintaining NREM sleep. Importantly, the sleep disruption we observed in our investigations mimic those reported in humans and in other genetic mouse models of DS.
We observed increased locomotor activity in aged Dp16 mice during the light phase, but not dark phase compared to WT controls, suggesting defects in sleep in these DS model mice. Using EEG to examine of percent time spent in awake, NREM, and REM states we also found that Dp16 mice spent more time awake and less time in NREM during both dark and light phases. While this observation supports the idea that Dp16 mice are indeed sleeping less than their WT counterparts, they are not expressing this loss of sleep as locomotor activity. This raises the possibility that while Dp16 mice are more awake during the dark phase, they are not engaged in increased non-sleep-related activities that are captured by locomotor activity measurements. This also may highlight the need to measure sleep in mouse models using multiple modalities as measurements based solely on locomotor activity may fail to capture some sleep-related abnormalities.
In the clinical DS population, parental questionnaires and patient EEG recordings consistently show sleep architecture to be altered in children (Hamaguchi et al., 1989; Levanon et al., 1999; Miano et al., 2008; Churchill et al., 2012). These alterations typically consist of a reduction in time spent in NREM, increased sleep latency, and increased night waking. One of the key difficulties in characterizing the genetic influence of Hsa21 trisomy on sleep disturbances in the DS population is the high comorbidity of OSA, which is present in upwards of 60% of individuals with DS (Alexander et al., 2016). However, it is important to note that sleep disturbances persist even when treatment for OSA is obtained. In support of this notion, children with DS maintain an increased arousal index following adeno-tonsillectomy, the most common treatment for OSA in children (Dyken et al., 2003). Further, Shete et al. and colleagues found sleep distribution and sleep efficiency remain impaired despite improvements in breathing when measured using a post adeno-tonsillectomy apnea hypopnea index (Shete et al., 2010). Taken together, the findings of the current study support the hypothesis that Hsa21 trisomy produces a unique sleep phenotype.
The development of mouse strains trisomic for specific genetic regions of Hsa21, including the Down Syndrome Critical Region (DSCR), allows for a more direct examination of the effects of these genes on sleep behavior. Whether sleep and locomotor activity are altered in mouse models of DS appears to be contingent on the strain used. Ts65Dn mice are more active during the light phase as compared to controls and we observed a similar pattern in Dp16 mice, while Tc1 mice show increased activity during the dark phase (Colas et al., 2008; Heise et al., 2015). In support, the Ts65Dn strain, which contains ~90 genes from Zbtb21 to Mrpl139, exhibited increased waking activity at the expense of NREM (Colas et al., 2008). The Tc1 strain, which contains ~125 genes spanning Hsa21, has been shown to exhibit fragmented sleep during the light phase, in addition to hyperactivity similar to that of the DS clinical population (Heise et al., 2015). However, these commonly used mouse strains are not without weaknesses. For example, as stated earlier, Ts65Dn mice are trisomic for ~60 additional genes located on Mmu17 not homologous to any region on Hsa21 (Franken et al., 2001; Yu et al., 2010). Tc1 mice present varying levels of genetic expression of Hsa21 across tissues, in addition to genetic deletion resulting in only ~83% of the Hsa21 genes being triplicated (Rueda et al., 2012). Conversely, Ts1Cje mice do not exhibit abnormal activity and sleep disturbances (Colas et al., 2008). Since Ts1Cje mice are only syntenic for ~71 genes located on Hsa21 and not the entire region, it is possible that these mice lack genes involved in the disturbed sleep phenotype in DS. Thus, each DS model mouse may present advantages and disadvantages depending on the specific experimental question being asked.
Our findings are important due to the role of sleep on cognitive performance. Poor sleep is associated with a reduction in cognitive performance and recent findings suggest that poor sleep patterns may serve as a biomarker in the development of AD phenotypes, such as Aβ overexpression in individuals with DS (Fernandez and Edgin, 2013). While we did not determine the presence of these AD characteristics in our aged mice, previous studies have shown that increased wakefulness significantly enhances Aβ basal levels in the hippocampi of mice and in the CSF of adults with presenilin mutations (Kang et al., 2009; Roh et al., 2012). Paired with the genetic predisposition in DS for increased Aβ production, it is plausible that sleep disturbances in DS may be involved in the development of AD-like phenotypes observed in DS. However, the effect of disturbed sleep on the development of AD-like phenotypes in DS warrants further investigation. In sum, these data suggest that the genetic regions triplicated in DS are differentially involved in sleep disturbances, and these sleep disturbances may play a key role in the development of the cognitive and EEG abnormalities observed in DS.
A critical finding in our study is the presence of EEG abnormalities in aged Dp16 mice. Changes in EEG rhythms reflect changes in brain function and cognitive activity (Fingelkurts and Fingelkurts, 2010). Abnormalities in spectral EEG power, such as increased theta, beta, and alpha power have been reported in both child and adult individuals with DS (Politoff et al., 1996; Śmigielska-Kuzia et al., 2005; Velikova et al., 2011). We found that alpha power was consistently increased in Dp16 mice during NREM sleep. Only a single study has examined EEG changes during sleep in DS and that study showed a decrease in alpha power during REM (Śmigielska-Kuzia et al., 2005). It is important to note that the participants in that study were children between the ages of 1 and 8, whereas the mice (12–14 months) in the current study are aged.
We also found a significant increase in beta frequency power in aged Dp16 mice. Prior to the current study, beta power had not been examined in any mouse models of DS, but increases in beta frequency power have been previously reported in the DS population, and beta rhythm dysregulation has been associated with sleep dysregulation (Velikova et al., 2011; Zielinski et al., 2016). Furthermore, alterations in beta power have been associated with neuropathology in AD (Karageorgiou and Vossel, 2017). Abnormalities in beta power are also associated with impairments in perception, attention, and motor action (Shin et al., 2017). Beta oscillations are generally considered a subharmonic of gamma activity and are thought to be generated by GABAergic interneurons that contain parvalbumin (PV) (Brown et al., 2012). Previous studies have shown that PV cells are involved in both beta and gamma oscillations in vivo and an alteration in PV cells may impact beta oscillations (Roopun et al., 2006; Kuki et al., 2015). In Ts65Dn mice, the number of PV-expressing interneurons are increased in the brain, and this overexpression has been linked to oligodendrocyte transcription factors (e.g. OLIG2) (Chakrabarti et al., 2010). In contrast, a previous study found OLIG2 to be decreased in fetal Dp16 brains (Goodliffe et al., 2016). Whether this reduction in OLIG2 expression persists in aged Dp16 mice and its effect on beta and gamma EEG activity have yet to be determined.
Finally, the decrease in delta power during NREM sleep may be a critical finding, especially when coupled with the simultaneous increase in alpha power. Delta waves are characterized by a slow negative pattern that occurs synchronously over the entire neocortical surface during NREM sleep. First-order neurons of the thalamus receive excitatory input from layer 6 and significant inhibitory input from GABAergic interneurons of the thalamic reticular nucleus and send spatially limited projections to the cortex (Buzsaki, 2006). A decrease in power in the delta bandwidth during NREM sleep suggests that the synchrony of the deeper neurons of the thalamus that results in the slow negative pattern across the cortical surface may be disrupted, and this disruption may involve GABAergic signaling. It is also plausible to suggest that alterations in GABAergic signaling may contribute to increases in alpha power. Alpha rhythms are thought to result from both thalamic and neocortical circuity, and GABAergic interneuron excitation has been proposed to be involved in the generation of alpha rhythms (Buzsaki, 2006). However, the mechanisms underlying the observed increase in alpha power during NREM sleep in aged Dp16 mice is currently unknown.
In summary, the present study examined sleep architecture in aged Dp16 mouse. Aged Dp16 mice exhibit a sleep phenotype that mirrors the both the clinical population of DS and more commonly used genetic mouse models of DS. In addition, the EEG abnormalities in these aged Dp16 mice mirror EEG findings in the AD population, which may serve as a biomarker for the development of AD phenotypes. Delineating the mechanisms underlying the sleep disturbances, EEG abnormalities, and the role GABAergic neuronal populations in DS phenotypes will lay the groundwork for the development of interventions designed to improve the quality of life in the DS population.
Acknowledgments
Supported by: NIH R01, NS086933-01; DA036673-01; Linda Crnic Institute, Linda Crnic Seed Grant; Simon’s Foundation, Sie postdoctoral award.
Abbreviations
- Dp16
Dp(16)1Yey/+ mice
- DS
Down syndrome
- DSCR
Down Syndrome Critical Region
- EEG
electroencephalography
- FFT
Fast Fourier transform
- Hsa21
human chromosome 21
- Mmu16
mouse chromosome 16
- OSA
obstructive sleep apnea
- PV
parvalbumin
- WT
wild type.
References
- Akeson EC, Lambert JP, Narayanswami S, Gardiner K, Bechtel LJ, Davisson MT. Ts65Dn – localization of the translocation breakpoint and trisomic gene content in a mouse model for Down syndrome. Cytogenet Cell Genet. 2001;93:270–276. doi: 10.1159/000056997. [DOI] [PubMed] [Google Scholar]
- Alexander M, Petri H, Ding Y, Wandel C, Khwaja O, Foskett N. Morbidity and medication in a large population of individuals with Down syndrome compared to the general population. Dev Med Child Neurol. 2016;58:246–254. doi: 10.1111/dmcn.12868. [DOI] [PubMed] [Google Scholar]
- Andreou G, Galanopoulou C, Gourgoulianis K, Karapetsas A, Molyvdas P. Cognitive status in Down syndrome individuals with sleep disordered breathing deficits (SDB) Brain Cogn. 2002;50:145–149. doi: 10.1016/s0278-2626(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Bliwise DL. Sleep Disorders Sleep disorders in Alzheimer’s disease and other dementias. Clin Cornerstone. 2004a;6:S16–S28. doi: 10.1016/s1098-3597(04)90014-2. [DOI] [PubMed] [Google Scholar]
- Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of Sleep and Wakefulness. Physiol Rev. 2012;92:1087–1187. doi: 10.1152/physrev.00032.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. Rhythms of the brain. Oxford University Press; 2006. [Google Scholar]
- Chakrabarti L, Best TK, Cramer NP, Carney RSE, Isaac JTR, Galdzicki Z, Haydar TF. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill SS, Kieckhefer GM, Landis CA, Ward TM. Sleep measurement and monitoring in children with Down syndrome: A review of the literature, 1960–2010. Sleep Med Rev. 2012;16:477–488. doi: 10.1016/j.smrv.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas D, Valletta JS, Takimoto-Kimura R, Nishino S, Fujiki N, Mobley WC, Mignot E. Sleep and EEG features in genetic models of Down syndrome. Neurobiol Dis. 2008;30:1–7. doi: 10.1016/j.nbd.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SS. Down syndrome: a review of the literature. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1997;84:279–285. doi: 10.1016/s1079-2104(97)90343-7. [DOI] [PubMed] [Google Scholar]
- Dyken ME, Lin-Dyken DC, Poulton S, Zimmerman MB, Sedars E. Prospective polysomnographic analysis of obstructive sleep apnea in down syndrome. Arch Pediatr Adolesc Med. 2003;157:655–660. doi: 10.1001/archpedi.157.7.655. [DOI] [PubMed] [Google Scholar]
- Fan Z, Ahn M, Roth HL, Li L, Vaughn BV. Sleep apnea and hypoventilation in patients with down syndrome: analysis of 144 polysomnogram studies. Children. 2017;4:55. doi: 10.3390/children4070055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, Edgin JO. Poor sleep as a precursor to cognitive decline in down syndrome: a hypothesis. J Alzheimers Dis Park. 2013;3:124. doi: 10.4172/2161-0460.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingelkurts AA, Fingelkurts AA. Short-term EEG spectral pattern as a single event in EEG phenomenology. Open Neuroimaging J. 2010;4:130–156. doi: 10.2174/1874440001004010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodliffe JW, Olmos-Serrano JL, Aziz NM, Pennings JLA, Guedj F, Bianchi DW, Haydar TF. Absence of Prenatal Forebrain Defects in the Dp(16)1Yey/+ Mouse Model of Down Syndrome. J Neurosci. 2016;36:2926–2944. doi: 10.1523/JNEUROSCI.2513-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi H, Hashimoto T, Mori K, Tayama M. Sleep in the down syndrome. Brain Dev. 1989;11:399–406. doi: 10.1016/s0387-7604(89)80024-5. [DOI] [PubMed] [Google Scholar]
- Heise I, Fisher SP, Banks GT, Wells S, Peirson SN, Foster RG, Nolan PM. Sleep-like behavior and 24-h rhythm disruption in the Tc1 mouse model of Down syndrome: Sleep and rhythm disruption in Tc1 mice. Genes Brain Behav. 2015;14:209–216. doi: 10.1111/gbb.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-E, Lim MM, Bateman RJ, Lee JJ, Smyth LP, Cirrito JR, Fujiki N, Nishino S, Holtzman DM. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science. 2009;326:1005–1007. doi: 10.1126/science.1180962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgiou E, Vossel KA. Brain rhythm attractor breakdown in Alzheimer’s disease: Functional and pathologic implications. Alzheimers Dement. 2017;13:1054–1067. doi: 10.1016/j.jalz.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuki T, Fujihara K, Miwa H, Tamamaki N, Yanagawa Y, Mushiake H. Contribution of parvalbumin and somatostatin-expressing GABAergic neurons to slow oscillations and the balance in beta-gamma oscillations across cortical layers. Front Neural Circuits. 2015;9 doi: 10.3389/fncir.2015.00006. Available at: https://www.frontiersin.org/articles/10.3389/fncir.2015.00006/full#h4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon A, Tarasiuk A, Tal A. Sleep characteristics in children with Down syndrome. J Pediatr. 1999;134:755–760. doi: 10.1016/s0022-3476(99)70293-3. [DOI] [PubMed] [Google Scholar]
- Li Z, Yu T, Morishima M, Pao A, LaDuca J, Conroy J, Nowak N, Matsui S-I, Shiraishi I, Yu YE. Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum Mol Genet. 2007;16:1359–1366. doi: 10.1093/hmg/ddm086. [DOI] [PubMed] [Google Scholar]
- López-Loeza E, Rangel-Argueta AR, López-Vázquez MÁ, Cervantes M, Olvera-Cortés ME. Differences in EEG power in young and mature healthy adults during an incidental/spatial learning task are related to age and execution efficiency. Age. 2016;38 doi: 10.1007/s11357-016-9896-z. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5005903/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano S, Bruni O, Elia M, Scifo L, Smerieri A, Trovato A, Verrillo E, Terzano MG, Ferri R. Sleep phenotypes of intellectual disability: A polysomnographic evaluation in subjects with Down syndrome and Fragile-X syndrome. Clin Neurophysiol. 2008;119:1242–1247. doi: 10.1016/j.clinph.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Nisbet LC, Phillips NN, Hoban TF, O’Brien LM. Characterization of a sleep architectural phenotype in children with Down syndrome. Sleep Breath. 2015;19:1065–1071. doi: 10.1007/s11325-014-1094-6. [DOI] [PubMed] [Google Scholar]
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, Anderson P, Mason CA, Collins JS, Kirby RS, Correa A National Birth Defects Prevention Network. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birt Defects Res A Clin Mol Teratol. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- Politoff AL, Stadter RP, Monson N, Hass P. Cognition-related EEG abnormalities in nondemented Down syndrome subjects. Dement Basel Switz. 1996;7:69–75. doi: 10.1159/000106856. [DOI] [PubMed] [Google Scholar]
- Roh JH, Huang Y, Bero AW, Kasten T, Stewart FR, Bateman RJ, Holtzman DM. Disruption of the sleep-wake cycle and diurnal fluctuation of β-amyloid in mice with Alzheimer’s disease pathology. Sci Transl Med. 2012;4:150ra122. doi: 10.1126/scitranslmed.3004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopun AK, Middleton SJ, Cunningham MO, LeBeau FE, Bibbig A, Whittington MA, Traub RD. A beta2-frequency (20–30 Hz) oscillation in nonsynaptic networks of somatosensory cortex. Proc Natl Acad Sci U S A. 2006;103:15646–15650. doi: 10.1073/pnas.0607443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda N, Flórez J, Martínez-Cué C. Mouse models of down syndrome as a tool to unravel the causes of mental disabilities. Neural Plast. 2012 doi: 10.1155/2012/584071. Available at: https://www.hindawi.com/journals/np/2012/584071/ [DOI] [PMC free article] [PubMed]
- Salem LC, Sabers A, Kjaer TW, Musaeus C, Nielsen MN, Nielsen A-G, Waldemar G. Quantitative electroencephalography as a diagnostic tool for Alzheimer’s dementia in adults with down syndrome. Dement Geriatr Cogn Disord Extra. 2015;5:404–413. doi: 10.1159/000438857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shete MM, Stocks RMS, Sebelik ME, Schoumacher RA. Effects of adeno-tonsillectomy on polysomnography patterns in Down syndrome children with obstructive sleep apnea: A comparative study with children without Down syndrome. Int J Pediatr Otorhinolaryngol. 2010;74:241–244. doi: 10.1016/j.ijporl.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Shin H, Law R, Tsutsui S, Moore CI, Jones SR. The rate of transient beta frequency events predicts behavior across tasks and species. eLife. 2017;6 doi: 10.7554/eLife.29086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmigielska-Kuzia J, Sobaniec W, Kuak W, Boćkowski L, Soowiej E. Quantitative EEG analysis of REM sleep in children with Down syndrome. Adv Med Sci. 2005;50:20–22. [PubMed] [Google Scholar]
- Velikova S, Magnani G, Arcari C, Falautano M, Franceschi M, Comi G, Leocani L. Cognitive impairment and EEG background activity in adults with Down’s syndrome: A topographic study. Hum Brain Mapp. 2011;32:716–729. doi: 10.1002/hbm.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, et al. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19:2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski MR, McKenna JT, McCarley RW. Functions and mechanisms of sleep. AIMS Neurosci. 2016;3:67–104. doi: 10.3934/Neuroscience.2016.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]