Figure 1.
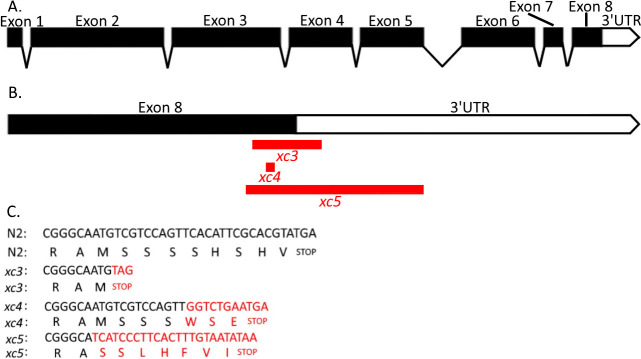
Figure 1. A. Map of exons, introns and the 3’UTR of cls-2 (R107.6). B. The eighth exon and 3’UTR of cls-2 (R107.6) with the position of the xc3, xc4, and xc5 mutations indicated in red. C. Alignment of DNA and amino acid sequences in mutant and wildtype worms with mutations in red.
Description
We have generated novel mutant alleles, named xc3, xc4, and xc5, of the gene cls-2 (R107.6) that encode one of the three predicted orthologs of mammalian CLASPs and of Drosophila ORBIT/MAST, microtuble-binding proteins (Akhmanova et al., 2001; Maiato et al., 2002). In C. elegans CLS-2 is required for meiosis and mitosis (Cheeseman et al., 2005; Dumont et al., 2010; Espiritu et al., 2012; Maton et al., 2015; Nahaboo et al., 2015). The alleles were isolated from gene mutations generated by Non-Homologous End Joining (NHEJ) mediated repair of Cas9-generated breaks (Dickinson et al., 2013; Ran et al., 2013). The alleles were detected by PCR using the following primers, 5’- CGATACGTCGGAGCAGAGC -3’ and 5’- CGGGGGTCGAAAATCATAAGG -3’. Next Generation Sequencing allowed us to identify 30 bp flanking sequences of the alleles xc3, xc4, and xc5 as TTGTCCAAGTCTACGTCAATCGGGCAATGT – [42 bp deletion] – AGCCCATAATTCCCCCGTATTCGTATCCCA, TCTACGTCAATCGGGCAATGTCGTCCAGTT – [3 bp deletion, 41 bp insertion (GGTCTGAATGACTTTCGCACTATTCCCCTATTCGCACGCCT)] – ATTCGCACGTATGATTCGTCGTTGCAATGT, and AACCTTGTCCAAGTCTACGTCAATCGGGCA – [111 bp deletion ] – TCATCCCTTCACTTTGTAATATAATTTTAT, respectively.
Based on information about cls-2 (R107.6) (WormBase, http://www.wormbase.org, WS261), the xc3, xc4, and xc5 mutant alleles effect the eighth exon and the 3’-UTR in the same way in each splicing isoform (Fig.1). In the xc3 mutant, 16 bp of the 3’UTR is deleted and a new stop codon was introduced after an 8 amino acid deletion (SSSSHSHV) of the C-terminus of the protein. In xc4 due to an insertion causing a frameshift mutation, 5 wildtype amino acids (SHSHV) from the C-terminus will be replaced by 3 amino acids (WSE). In xc5 the endogenous stop codon is deleted as well as 81 bp of the 3’UTR, while a new stop codon is introduced 21 bp after the mutation. Because of the deletion and new stop codon, in the xc5 mutant 9 amino acids (MSSSSHSHV) in the C-terminus of the protein will be replaced by 7 new amino acids (SSLHFVI). Previous researchers replaced serine residues with non-phosphorylatable alanine residues to study the effect of human CLASP2 phosphorylation (Kumar et al., 2017). The mutations we have generated have multiple serine residues deleted which presents a unique opportunity to study the effect of cls-2 (R107.6) phosphorylation. Since more of the 3’UTR is deleted in xc5 than xc3, the 3’UTR’s function could also be studied using these mutants.
Reagents
Alt-R® CRISPR-Cas9 crRNA Alt-R® CRISPR-Cas9 tracrRNA Alt-R® S.p. Cas9 Nuclease
Strains:XC125 cls-2 (xc3) unc-119 (ed3) III; ieSi38 (IV)XC126 cls-2 (xc4) unc-119 (ed3) III; ieSi38 (IV)XC127 cls-2 (xc5) unc-119 (ed3) III; ieSi38 (IV)
Acknowledgments
Funding
NSF RUI 1244517, NIH R15 HD068996
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001 Mar 23;104(6):923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, MacLeod I, Yates JR 3rd, Oegema K, Desai A. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005 Apr 26;15(8):771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013 Sep 01;10(10):1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol. 2010 Aug 22;12(9):894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu EB, Krueger LE, Ye A, Rose LS. CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev Biol. 2012 May 19;368(2):242–254. doi: 10.1016/j.ydbio.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009 Mar 16;184(6):895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol. 2002 May 28;157(5):749–760. doi: 10.1083/jcb.200201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maton G, Edwards F, Lacroix B, Stefanutti M, Laband K, Lieury T, Kim T, Espeut J, Canman JC, Dumont J. Kinetochore components are required for central spindle assembly. Nat Cell Biol. 2015 Apr 13;17(5):697–705. doi: 10.1038/ncb3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahaboo W, Zouak M, Askjaer P, Delattre M. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner. Mol Biol Cell. 2015 Apr 01;26(11):2020–2029. doi: 10.1091/mbc.E14-12-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]