Abstract
Quantification of neurochemical concentrations from 1H MR spectra is challenged by incomplete knowledge of contributing signals. Some experimental conditions hinder acquisition of artifact free spectra and impede acquisition of condition specific macromolecule (MM) spectra. This work studies differences caused by fitting solutions routinely employed to manage resonances from MM and lipids. High quality spectra (free of residual water and lipid artifacts and for which condition specific MM spectra are available) are used to understand the influences of spline baseline flexibility and non-condition specific MM on neurochemical quantification. Fitting with moderate spline flexibility or using non-condition specific MM led to quantification that differed from when an appropriate, fully specified model was used. This happened for all neurochemicals to an extent that varied in magnitude among and within approaches. The spline baseline was more tortuous when less constrained and when used in combination with non-condition specific MM. Increasing baseline flexibility did not reproduce concentrations quantified under appropriate conditions when spectra were fitted using a MM spectrum measured from a mismatched cohort. Using the non-condition specific MM spectrum led to quantification differences that were comparable in size to using a fitting model that had moderate freedom, and these influences were additive. Although goodness of fit was better with greater fitting flexibility, quantification differed from when fitting with a fully specified model that is appropriate for low noise data. Notable GABA and PE concentration differences occurred with lower estimates of measurement error when fitting with greater spline flexibility or non-condition specific MM. These data support the need for improved metrics of goodness of fit. Attempting to correct for artifacts or absence of a condition-specific MM spectrum via increased spline flexibility and usage of non-condition specific MM spectra cannot replace artifact free data quantified with a condition-specific MM spectrum.
Keywords: macromolecules, ultra-high field, 7 T, magnetic resonance spectroscopy, LCModel
Graphical abstract
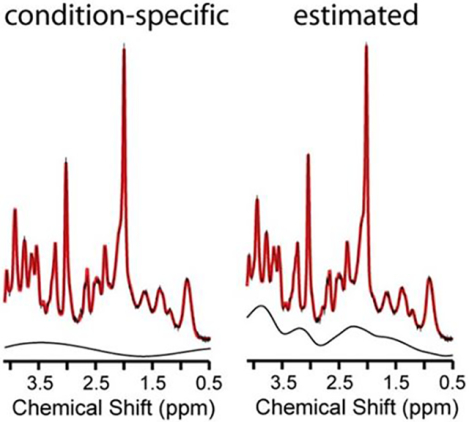
Experimental conditions often hinder acquisition of artifact free spectra or impede acquisition of condition-specific macromolecular spectra. Fitting estimates were unable to recover concentrations that were quantified from artefact free 1H MR spectra using an mismatched macromolecular basis spectrum. Estimation caused differences in quantified concentration for all of the neurochemicals that varied in magnitude among and within approaches. The spline was more tortuous when less constrained and when used in combination with estimated macromolecules and lipids.
Introduction
Experimental conditions often hinder acquisition of artifact free spectra or impede acquisition of condition specific macromolecule (MM) spectra. Fitting algorithms manage artifacts such as coherences from residual water or lipid contamination via a flexible spline baseline or added basis spectra for lipids. Broad macromolecular resonances, originating from proteins in the human brain, underlie metabolite resonances in the 2.0–4.5‐ppm range, but are readily apparent in the 0.5–2.0‐ppm range.1 Macromolecule spectra are comprised of resonances from several compounds. Therefore, the relative amplitudes of the broad resonances in the MM spectrum may vary with the macromolecular composition of the sample. Macromolecular contributions are often assumed to be the same in similar cohorts and among brain regions2,3, or managed by the fitting algorithm via added basis spectra and a flexible spline baseline. The default for the widely used LCModel fitting approach4,5 is to include several independent basis spectra to account for resonances from macromolecules and lipids, and a flexible spline baseline to fit: incompletely suppressed water, lipids, and MM resonating in the chemical shift range ≥ 3 ppm.
The goal of this project is to evaluate the influence of modeling basis spectra for MM and using a highly flexible spline baseline on quantification of neurochemical concentrations. The approach is to fit spectra that are free of residual water or lipid contamination and for which a measured MM spectrum from matched experimental conditions is available. A fully specified modelling approach, i.e., one with the condition-matched MM spectrum in the basis set and one that does not leave the spline baseline free is logically the appropriate approach to fit these spectra. LCModel is used for fitting in its default configuration, which is designed to manage artifacts,4 and in configurations that use measured MM spectra and heavily constrained spline baseline. An important aspect of this approach is to characterize consequences of using condition-mismatched MM and whether allowing high spline baseline flexibility in this circumstance can recover accurate quantification.
Experimental
Fitting approaches
Table 1 summarizes the following five fitting approaches performed in LCModel to quantify the neurochemical profile from artifact-free older adult human brain metabolite spectra. 1) The fully specified approach, i.e., the appropriate fitting approach used a MM spectrum measured in older adults and a constrained, i.e., stiff, spline baseline. No modeled basis spectra for MM were included. This fitting approach deviates most from the default LCModel configuration. Preselection of the condition-specific measured MM spectrum and stiff spline baseline as the appropriate fitting approach is in agreement with a similar recent report.6 2) The partially specified approach used the older adult MM spectrum (no modeled basis spectra for MM were included) and the default unconstrained, i.e., flexible spline baseline. 3) The condition-mismatched approach used a MM spectrum measured from young adults along with a stiff spline. Young and older adults are known to have differing MM spectra.7 A novel aspect of the current study is that the condition-matched and condition-mismatched MM spectra were different (i.e., older and young adult MM spectra were notably different in this project, while the MM spectra from two brain regions were not different in the prior study by Giapitzakis et al.6). 4) The condition-mismatched approach of using the wrong MM spectrum was used along with the default flexible spline to learn whether allowing high spline baseline flexibility can recover the quantification as the fully specified approach. 5) For comparison, the default LCModel approach of modeling basis spectra for MM and using a highly flexible spline baseline was used.
Table 1.
Summary of different fitting approaches used for fitting older adult spectra from the OCC using LCModel.
| Approach | Abbreviation | MM used for fitting | Spline baseline constraint (DKNTMN) |
|---|---|---|---|
| Older MM, stiff | O-MM Stiff | Measured Older Adult | 5 |
| Older MM, flexible | O-MM Flex | Measured Older Adult | default (0.15) |
| Young MM, stiff | Y-MM Stiff | Measured Young Adult | 5 |
| Young MM, flexible | Y-MM Flex | Measured Young Adult | default (0.15) |
| LCModel, flexible | LCModel | LCModel NSIMUL = 11 | default (0.15) |
Subjects
Sixteen older adults (12 males, 4 females; age: 75 ± 6 years; age range 67 to 88 years) participated in the study after providing informed consent according to procedures approved by the Human Subjects’ Protection Committee at the University of Minnesota, Institutional Review Board and passing the clinical and neurological examination. All participants underwent an MRI exam at 7 T. Exclusion criteria for older adults were Montreal Cognitive Assessment (MoCA8) score < 24, memory or cognitive complaints, history of brain tumor or brain cancer, stroke, Parkinson’s disease, multiple sclerosis, cerebral palsy, Huntington’s disease, encephalitis, meningitis, medical condition (including kidney dialysis or liver disease) or neurological diagnosis (including vascular dementia or having undergone brain surgery to clear arteries in the brain) that potentially could affect cognition, or poor creatinine clearance (GFR < 45 mL/min9). All sixteen spectra were used in prior work7 which focused on quantification of macromolecular contribution in the older human brain.
MR acquisition
In vivo data were obtained on a 7-T, 90-cm horizontal bore magnet (Magnex Scientific Inc., Oxford, UK) interfaced with a Siemens console running syngo VB17 (Siemens, Erlangen, Germany). The magnet was equipped with a body gradient coil (maximum amplitude: 70 mT/m, slew rate: 200 mT/m/ms). A 16-channel transmission line head array radiofrequency (RF) coil10 was used to transmit and receive the signal. The transmit phase of each coil channel was controlled with an independent 1-kW RF amplifier (CPC, Brentwood, NY, USA). An RF power monitoring system measured the forward and reflected power for each channel to ensure that the local specific absorption rate did not exceed 3 W/kg.
Magnetization-prepared rapid gradient-echo (MPRAGE) images11 (TR = 3 s; TE = 3.27 ms; TI = 1.5 s; flip angle = 5°; field of view: 256 × 256 mm2; matrix: 256 × 256; slice thickness = 1 mm, 176 slices, acceleration factor = 3, acquisition time: 4 min 17 s) were acquired to position 8 mL volume-of-interest (VOI) in the occipital cortex (OCC) and to obtain gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) content. The VOI was positioned symmetrically across hemispheres and rotated to align with the boundaries of the occipital lobe and placed in the center of the occipital lobe vertically while the back corner was placed 4 mm anterior to the back of the occipital lobe. Proton-density (PD) images (TR = 1.41 s; TE = 3.27 ms; field of view: 256 × 256 mm2; matrix: 256 × 256; slice thickness = 1 mm, 176 slices, acceleration factor = 3, acquisition time: 2 min) were also acquired to correct for intensity field bias in the T1-weighted MPRAGE images12. To maximize the transmit B1 in the VOI, fast local shimming of the phase of B1+ for each coil channel was employed13,14. B0 shimming of first- and second-order terms was achieved using the fast automatic shimming technique by mapping along projections with echo planar imaging readout (FAST(EST)MAP,15,16). MR spectra were measured using an ultra-short echo time stimulated echo acquisition mode (STEAM) sequence (TR = 5 s; TE = 8 ms; mixing time (TM) = 32 ms, 64 averages) and with 3D outer volume suppression (OVS) interleaved with variable power and optimized relaxation delay (VAPOR) water suppression.17 The B1 field and the water suppression pulses were calibrated in the VOI for each individual. OVS was optimized for the coil used and for the morphometry of older adult brains. The chemical shift displacement of the VOI was 4% of the voxel dimension per ppm. Each free induction decay was acquired with 2048 complex data points using a spectral width of 6 kHz. A non-suppressed water spectrum was acquired for eddy current correction and quantification (TR = 5 s; TE = 8 ms; TM = 32 ms; 1 average).
For half of the participants, MPRAGE (TR = 2.4 s; TE = 2.24 ms; TI = 1.06 s; flip angle = 8°; field of view: 256 × 256 mm2; matrix: 320 × 320; slice thickness = 0.8 mm, 208 slices, acceleration factor = 2, acquisition time: 6 min 38 s) images were acquired at 3 T to obtain GM, WM, and CSF content.
Image processing and analysis
7 T MPRAGE images were pre-flattened by dividing by the PD images12 using FSL, a comprehensive library of analysis tools for fMRI, MRI, and DTI brain imaging data18. Images were further flattened using statistical parametric mapping (SPM)19 and segmented into GM, WM, and CSF content using Freesurfer20. The tissue composition was extracted for each VOI using analysis of functional neuro images (AFNI)21. 3 T MPRAGE images were segmented using Freesurfer20 via the HCP protocol22. In-session 7 T MPRAGE images were aligned to the 3 T MPRAGE images, and this transformation matrix was applied to each VOI using AFNI21 to obtain tissue composition for each VOI using Freesurfer20.
The VOIs contained 39 ± 5% of GM, 38 ± 6% of WM, and 24 ± 5% of CSF in older adults.
Spectral processing and quantification
The acquired spectra were processed in Matlab (MathWorks Inc., Natick, MA). Eddy-current effects were corrected using the non-suppressed water spectrum. Single-shot metabolite spectra were frequency and phase aligned using a cross-correlation algorithm.
Spectra were analyzed using LCModel4,5 6.3–1J (Stephen Provencher, Inc., Oakville, ON, Canada) with the basis set simulated in Matlab using the density matrix formalism23 with ideal pulses and actual echo time and mixing time and previously published chemical shifts and J-couplings24–26. The pertinent fitting parameters are provided in the appendix. The following metabolites were included in the basis set: ascorbate (Asc), aspartate (Asp), creatine (Cr), γ-aminobutyric acid (GABA), glucose (Glc), glutamine (Gln), glutamate (Glu), glutathione (GSH), glycerophosphorylcholine (GPC), lactate (Lac), myo-inositol (mIns), N-acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphocreatine (PCr), phosphorylcholine (PCho), phosphorylethanolamine (PE), scyllo-inositol (sIns), and taurine (Tau). No baseline correction, zero-filling, or line broadening were applied to the in vivo data before the analysis. Spectra were fitted between 0.5 and 4.1 ppm. The average age-specific MM spectra published previously7 were also included in the basis set. They were experimentally measured using the inversion-recovery technique1 (TR = 2 s, TE = 8 ms, inversion time (TI) = 0.68 s) from the OCC of 4 young (total of 1532 averages) and 3 older (total of 960 averages) individuals and processed as previously described7. They have no apparent extraneous coherences.
A subset of spectra measured from older adults from prior work7 with no evidence of extraneous coherences were fitted using the five approaches summarized in Table 1. Macromolecular contribution was fitted with three different approaches: 1. condition-specific (older adults) MM spectrum; 2. condition-mismatched young-adults MM spectrum; and 3. LCModel simulated macromolecules (NSIMUL = 11). Stiff and flexible approaches for the spline baseline fit were accomplished by using the LCModel parameter that controls knot spacing (DKNTMN)4 of 527 and default of 0.15, respectively. A DKNTMN of 5 imposed the greatest possible stiffness, since DKNTMN is the minimum spacing of the knots in ppm, except that the spacing cannot exceed one third of the fitted range. There are also spline knots at both ends of the fitted region.
Quantification was performed using the unsuppressed water signal obtained from the same VOI and corrected for GM, WM, and CSF content as previously described28. The water content for CSF and WM were assumed to be 0.9729 and 0.7130–32, respectively. For GM, the water content was assumed to be 0.79 for older women30 and 0.76 for older men30. T1 and T2 relaxation time constants of water used in the calculation of the attenuation factors were taken from published reports [T1(GM) = 2130 ms, T1(WM) = 1220 ms, T1(CSF) = 442533; T2(GM) = 50 ms, T2(WM) = 55 ms, T2(CSF) = 141 ms34] and included a 10% reduction in T2 of tissue water in older adults35 [T2(GM) = 45 ms, T2(WM) = 49.5 ms]. The T1 and T2 relaxation time constants of neurochemicals were not taken into consideration as attenuation factors since their T1 are shorter and their T2 are longer than water, thus the effect would be very small given the use of repetition time of 5 s and echo time of 8 ms.
Neurochemical concentrations with Cramér-Rao lower bounds (CRLB, an estimate of the lower limit of the variance) lower than 50% were retained for statistical analysis. This CRLB cutoff was selected to avoid over-filtering of the data. CRLB were retained in the database to allow further consideration of neurochemical concentrations with CRLB in the range between 20 and 50%. If the spectral overlap caused the covariance between two neurochemicals to be high (characterized by strong negative correlation, i.e., correlation coefficient < −0.7) for all of the participants, the sum of the neurochemical concentrations was reported, specifically total creatine (tCr) = Cr + PCr, and total choline (tCho) = PCho + GPC. Glc and Lac were not quantified because they are present in CSF at concentrations of 2–3 mM, i.e., comparable to tissue36.
Statistical analysis
Differences between neurochemical concentrations quantified using the Older-MM, stiff approach (Table 1) and other fitting approaches were compared using paired two-tailed Student’s t-tests in Excel (Microsoft Corporation, Redmond, WA). To control for multiple testing (14 neurochemicals and 4 estimated fitting approaches), the threshold of significance was 0.00089 (0.05/56).
Results
High-quality spectra were consistently measured with narrow resonances and flat baseline without evidence of contamination from unsuppressed water or lipids from outside of the VOI. The tCr linewidths measured as the full width at half maximum of the resonance at 3.03 ppm were on average (± standard deviation) 13.2 ± 0.8 Hz. The signal-to-noise ratios (SNR) of the highest signal in the spectra divided by the root mean square of the noise were 94 ± 15. Figure 1 illustrates the LCModel fits quantified from the spectrum measured from a representative older adult’s occipital cortex (tCr linewidth = 13.9, SNR = 88) using the fitting approaches of Table 1. The average standard deviations of the residuals (± standard deviations) for all spectra were 0.044 ± 0.010 for O-MM Stiff, 0.037 ± 0.010 for O-MM Flex, 0.049 ± 0.009 for Y-MM Stiff, 0.036 ± 0.010 for Y-MM Flex, and 0.032 ± 0.008 for LCModel. The spline baseline is increasingly tortuous progressing from O-MM Stiff to O-MM Flex and then to LCModel (Figures 1 and 2). The fit of the spline baseline with the LCModel approach has a large rise on the 4 ppm side (Figures 1 and 2), which is expected given that there are no basis spectra to model MM in this region.
Figure 1.
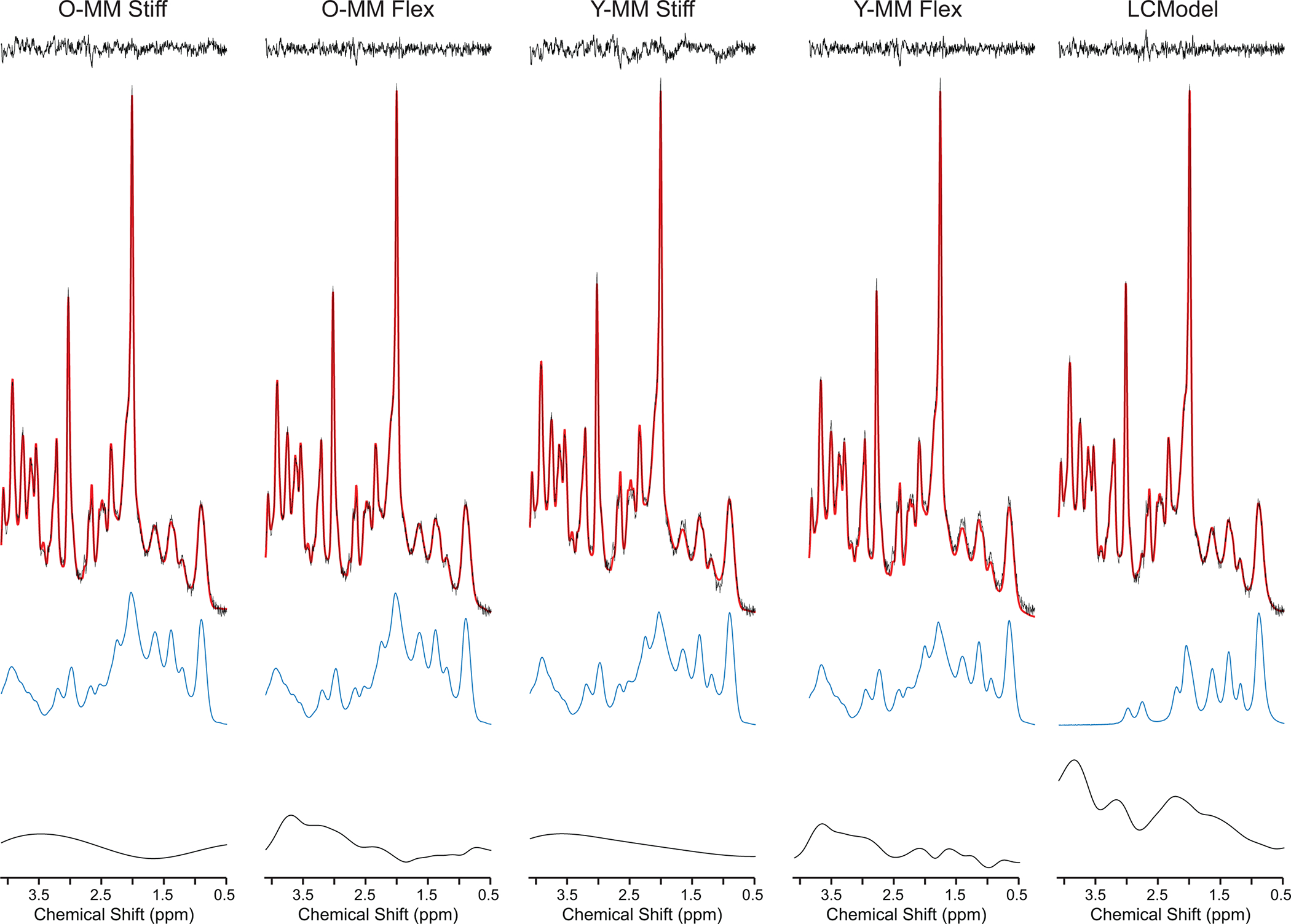
Data and fitting quality. Representative 1H MR spectrum from one older adult acquired from 8-mL VOI placed in the OCC (STEAM, 7 T, TR = 5 s, TE = 8 ms, number of averages = 64) fitted with five approaches summarized in Table 1 using LCModel. In vivo spectrum is shown together with LCModel fits (red lines), residuals (above), spline baseline contributions (below), and MM/lipid contributions (blue lines). Spectra are shown with no line-broadening applied.
Figure 2.
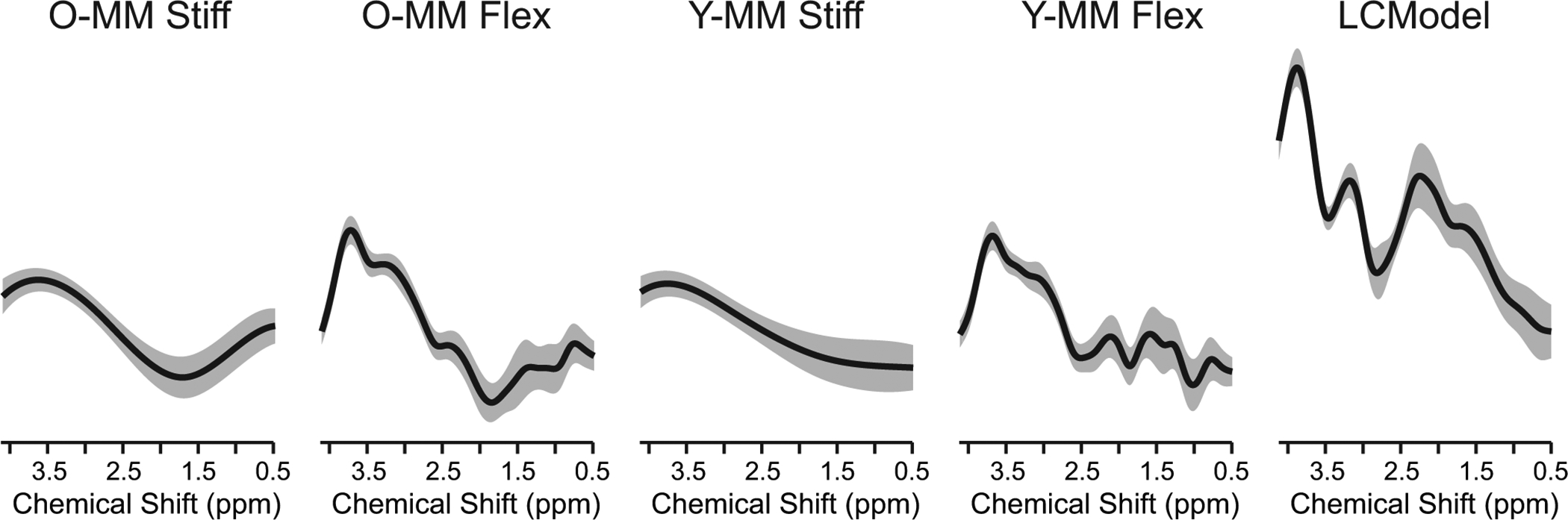
Spline baselines obtained with LCModel fitting. Average (black line) and standard deviation (shaded area) of individual spline baselines are shown for all 16 spectra fitted using five approaches summarized in Table 1. Spline baselines are shown with no line broadening.
Figure 3 illustrates neurochemical concentrations quantified from metabolite spectra measured from the occipital cortex of the older adults using the fitting approaches of Table 1. Applying approaches other than the fully specified, i.e., moderate baseline flexibility or non-condition specific MM, leads to differences in quantified concentration for all of the neurochemicals. Which neurochemicals are impacted is different for each approach. Asc, GSH, PE, sIns, and tCho concentrations are impacted by all approaches. They are not necessarily the smallest signals. In fact, the concentration of NAA is substantially influenced, i.e., 14% by the LCModel approach. On the other hand, Gln is minimally influenced (≤ 7%) by all approaches other than LCModel.
Figure 3.
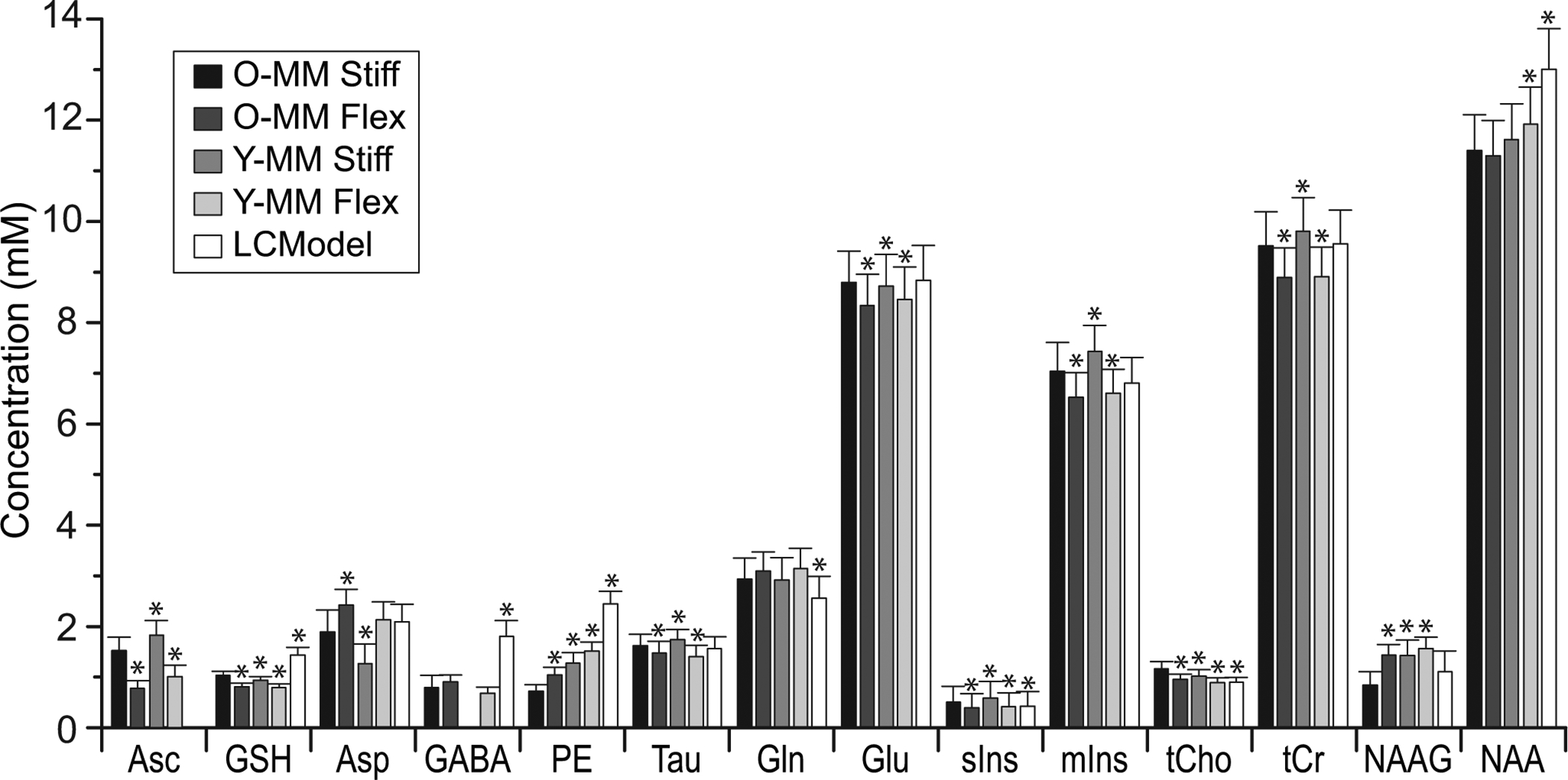
Concentrations of metabolites quantified using LCModel. Older adult spectra were fitted using five approaches summarized in Table 1. * - significant difference (p < 0.00089) between O-MM Stiff and other fitting approaches.
Going from a stiff to a flexible spline baseline leads to concentration differences that consistently increase or decrease for each neurochemical except for NAA, and the size of the difference is of similar size (i.e., within 30%) for all neurochemicals except NAAG and Asp whether O-MM or Y-MM is used. While using O-MM in the basis set, going from a stiff to a flexible spline baseline leads to concentrations that are significantly lower more often than higher and the biggest differences are for Asc (49%), NAAG (70%), and PE (45%). The concentration of Asc is substantially lower (49%) when fitting with flexible spline baselines.
Going from young to older MM only (i.e., going from O-MM Stiff to Y-MM Stiff, Table 1) causes concentrations to be significantly higher more often than lower and the greatest differences are for GABA (not fitted), Asp (33%), PE (78%), and NAAG (69%). Using young MM but allowing flexible spline baseline recovers the same Asp and GABA levels as the fully specified approach, but this does not occur for PE and NAAG.
The LCModel default approach finds an equal number of higher concentrations and lower concentrations with the greatest difference for Asc (not fitted), GABA (126%), and PE (241%), and noteworthy for NAA (14%). GSH concentration is noticeably higher (39%) when fitted without a measured MM spectrum. The LCModel approach leads to the largest total percent (519%) and absolute concentration difference from the fully specified approach, when absolute values of all neurochemical concentration differences are summed. This overall concentration difference is impacted to a similar extent (258% and 301%, respectively) by use of the condition-mismatched MM spectrum (stiff spline) and a flexible spline (right MM). The overall concentration difference is impacted by going from O-MM Stiff to LCModel approximately twice as much as use of the condition-mismatched MM spectrum or the default DKNTMN individually.
No difference in GABA concentration is found when a free spline baseline is allowed with the condition-specific MM spectrum. Using the fully specified approach, GABA is fit with CRLB < 50% in all cases except 2 (remainder CRLB average 19%, range 10 – 38%). When the Y-MM spectrum (stiff spline baseline) is used, GABA is only fit in 1 case (with CRLB 32%). When using LCModel to fit MM, GABA appears to fit in all cases (CRLB ≤ 31%). The quantification difference that is caused by going to the young MM spectrum with a stiff spline baseline is reversed by the flexible spline baseline, but overshoots equally far when using the LCModel approach. The large difference encountered when using the LCModel estimation (126%) is predictable given the known age-associated differences in the MM spectrum near 1.9 ppm (i.e., the chemical shift of the 3CH2 of GABA).
Quantification of PE is particularly sensitive to the fitting approach. PE is fitted in more cases with lower CRLB using all estimated fitting approaches (CRLB for: O-MM Stiff ≥ 50% for 5 cases, range: 22–999%; O-MM Flex one at 64%, remainder in range: 13–26%; LCModel no values ≥ 50%, range: 7–11%).
NAAG and Asp have resonances in chemical shift ranges where the older and younger adult MM spectra are notably different7 (i.e., 3.75 ppm region for Asp and 1.2–2.2 range for NAAG), and they lack other substantial resonances that, if present, would aid with fitting. The 33% difference in measured Asp concentration between Y-MM and O-MM (stiff) is consistent with the fitting model having compensated for the difference between these basis spectra in the 3.75 ppm chemical shift region. This phenomenon is comparably (i.e., 31%) observed for NAAG in the 1.2–2.2 chemical shift range. As such, the greatest impact is logically predicted by a compound-associated resonance in the spectrum. Using free spline baselines, quantification differences occur for NAAG and Asp even though there are no spectral contributions that would be comparably expected to explain them. This is anticipated, given the unspecific nature of the spline baseline.
Discussion
This project used high quality spectra (free of residual water or lipid contamination and for which a measured MM spectrum from matched experimental conditions was available), that did not warrant high spline baseline flexibility or modeling MM spectra in the basis set for fitting, to understand the influences of spline baseline flexibility and modeled MM, which is of interest when fitting conventional spectra. Forcing a smoother baseline leaves the portion of the data that would otherwise have been accounted for unfit. However, a flexible baseline takes up a mixture of signal and noise. In this study, spectra with low noise fit well with only basis spectra (i.e., without spline flexibility). This allows us to address the question whether allowing a free spline baseline leads to a different outcome than when the spectra are well modelled without this unassigned component.
Fitting older adult metabolite spectra with a young MM spectrum had a notable impact on quantification that was comparable in magnitude to allowing a flexible spline baseline. Using the young MM spectrum also had a readily apparent impact on the shape of the spline baseline. The default LCModel approach (with flexible spline baseline) led to approximately twice as much quantification difference as using the condition-mismatched MM spectrum or a flexible spline baseline. The previous work in which condition-matched and condition-mismatched measured MM spectra were not different found that using the wrong MM spectrum did not notably affect the spline baseline or quantification.6 The current and previous6 studies agree that the impact of using the LCModel, flexible spline baseline approach has more influence on quantification than using the condition-mismatched MM spectrum. However, there was no overlap between the two studies with regard to which neurochemicals were influenced by using the condition-mismatched MM spectrum or a flexible spline baseline. Other approaches to manage MM contributions have been proposed37,38, but are not widely used, thus they were not tested in this study.
GABA and PE concentration differences were notable consequences of estimated fitting approaches, yet the fits that were not fully specified were accompanied by lower CRLB estimates of measurement error. This exemplifies the inadequacy of CRLB provided by LCModel as a reliability criterion. Similarly, more freedom in the fit via flexible spline baseline or using LCModel to estimate the MM contribution leads to an apparent better fit (smaller CRLB and fit residual) at the consequence of greater quantification differences from the fully specified approach. Also, allowing a flexible spline baseline did not consistently reverse the quantification differences that arose from using the condition-mismatched MM spectrum. When fitting older adult spectra with Y-MM, the impact on quantification is logically linked to molecules in the sample that contribute resonances to the spectrum, whereas when fitting older adult spectra with a flexible spline baseline, the impact is not linked to a tangible entity. In the former case, although wrongly identified, presence of a compound is accurately detected (which would be useful as a biomarker). On the other hand, using a moderately flexible spline baseline is more likely to cause neurochemical differences that are not present in the sample. For noisy spectra or when a condition-specific MM spectrum is not accessible, spline flexibility and use of non-condition specific MM may be necessary to achieve a good fit. However, a better fit performance does not necessarily indicate more reliable quantification.
Relevance of the quantification differences reported here can be cast in light of meaningful concentration differences detected using the fully specified approach, i.e., spectra measured from young and older adults using the same protocol39. In the aging work39, the following neurochemical concentration differences were measured in older relative to young adults: NAA −10%, Glu −11%, Asc −14%, tCho 19%, NAAG −20%, and PE −51%. For NAA and Glu, none of the differences between the fully specified and other fitting approaches (−1% to 14%, −5% to 0%, respectively) were as large as the meaningful difference measured in the study on aging. For Asc and NAAG, the differences in the fitting approaches (−49% - 20% for fitted Asc cases, 31% - 85% for NAAG) were always larger than the meaningful difference. This circumstance was intermediate for tCho and PE (differences among fitting approaches −24% to −13% for tCho, 45% - 241% for PE). From this perspective, fitting approaches have a mild influence on quantification of the strongly represented NAA and Glu resonances, a moderate influence on tCho and PE, and a marked influence on weakly represented Asc and NAAG resonances. The nuances of fitting are more relevant when knowledge on the more challenging neurochemicals to quantify is sought.
The large impact on quantification when the condition-mismatched MM spectrum is used on fitting GABA is reminiscent of the impact that inadequate consideration of MM signal contributions has on quantifying GABA concentration from edited spectra.40 Regardless of the acquisition or fitting approach, awareness of MM signal contributions is particularly important when attempting to measure GABA concentration. Given widespread interest in GABA41, improved knowledge of whether MM spectra differ with experimental conditions is warranted.
Conclusion
Attempting to correct for artifacts or absence of an accurate MM spectrum via flexible spline baseline and addition of modeled MM spectra cannot replace artefact free data quantified with an accurate MM spectrum. Influences on quantification of fitting with condition-mismatched MM spectra and/or a flexible spline are highly variable with regard to both fitting approach and which particular neurochemicals are influenced. Using the condition-mismatched MM spectrum leads to quantification errors that are comparable in size to using a fitting model that has moderate freedom. Using both the condition-mismatched MM and moderate freedom has an overall additive influence on quantification. In this study of artefact free spectra, the appearance of a tortuous spline when flexibility was allowed warns that signal from neurochemicals (i.e., not artefacts) may be over-fitted by the spline baseline, causing quantification errors, including those that propagate to neurochemicals with overlapping chemical shifts42. Such errors lack logical linkage to physical entities, i.e., resonances from compounds that are present in the sample. The influence of fitting approach on GABA quantification and lack of linkage to CRLB as a quality control criterion warrants increased characterization of the MM signal contributions. More broadly, these data support the need for improved quality control metrics.
Acknowledgements
The authors thank: Stephen Provencher for providing the definition of and clarification on the usage of the LCModel control parameter, DKNTMN; Laura Hemmy, Ph.D. and J. Riley McCarten, M.D. for screening of older adults; James S. Hodges, Ph.D. for discussion on statistical analysis; Dinesh K. Deelchand for help with figures; Edward J. Auerbach, Ph.D. for implementing the FAST(EST)MAP and STEAM sequences on the Siemens platform; Pierre-Francois Van de Moortele, M.D., Ph.D., and Julien Sein, Ph.D. for the T1-flattening script; and Emily Kittelson for image segmentation. We also acknowledge the University of Minnesota Retiree’s Volunteer Center for assistance with recruiting older adult candidates.
Sponsors: This work was supported by the National Institutes of Health [grant numbers R01AG055591, R01AG039396, P41 EB015894, P30 NS076408] and the W.M. Keck Foundation.
List of abbreviations:
- CSF
cerebrospinal fluid
- GM
gray matter
- MM
macromolecules
- MPRAGE
magnetization-prepared rapid gradient echo
- O-MM Flex
macromolecules measured in older adult, DKNTMN = default (0.15)
- O-MM Stiff
macromolecules measured in older adult, DKNTMN = 5
- OCC
occipital cortex
- OVS
outer volume suppression
- PD
proton density
- RF
radiofrequency
- STEAM
stimulated echo acquisition mode
- VOI
volume of interest
- WM
white matter
- Y-MM Flex
macromolecules measured in young adult, DKNTMN = default (0.15)
- Y-MM Stiff
macromolecules measured in young adult, DKNTMN = 5
Appendix. Pertinent fitting parameters, that deviated from default, used in LCModel to fit the data. The correction for GM, WM, and CSF content was performed outside of the LCModel.
DOWS=T
WSMET='NAA'
WSPPM=2.01
N1HMET=3
NUSE1=5
CHUSE1(1)='NAA'
CHUSE1(2)='Cr'
CHUSE1(3)='PCr'
CHUSE1(4)='Ins'
CHUSE1(5)='Glu'
NCALIB=0
DESDSH=0.004
NSDSH=3
CHSDSH(1)='Scyllo'
CHSDSH(2)='GPC'
CHSDSH(3)='Gly'
ALSDSH(1)=0.004
ALSDSH(2)=0.004
LSDSH(3)=0.004
DEGZER= 0.00
SDDEGZ=20.00
DEGPPM=0.00
SDDEGP=6.00
SHIFMN=-0.2,−0.1
SHIFMX=0.3,0.3
NRATIO=0
FWHMBA=0.00673
RFWHM=1.8
DKNTMN=5
PPMST=4.1
PPMEND=0.5
NSIMUL=0
CHKEEP=0
HZPPPM=297.2175
NUNFIL=2048
DELTAT=0.0001666
References
- 1.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. September 1994;32(3):294–302. [DOI] [PubMed] [Google Scholar]
- 2.Schaller B, Xin L, Gruetter R. Is the macromolecule signal tissue-specific in healthy human brain? A 1H MRS study at 7 Tesla in the occipital lobe. Magn Reson Med. October 2014;72(4):934–940. [DOI] [PubMed] [Google Scholar]
- 3.Snoussi K, Gillen JS, Horska A, et al. Comparison of brain gray and white matter macromolecule resonances at 3 and 7 Tesla. Magn Reson Med. September 22 2015;74(3):607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. December 1993;30(6):672–679. [DOI] [PubMed] [Google Scholar]
- 5.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. June 2001;14(4):260–264. [DOI] [PubMed] [Google Scholar]
- 6.Giapitzakis IA, Borbath T, Murali-Manohar S, Avdievich N, Henning A. Investigation of the influence of macromolecules and spline baseline in the fitting model of human brain spectra at 9.4T. Magn Reson Med. February 2019;81(2):746–758. [DOI] [PubMed] [Google Scholar]
- 7.Marjanska M, Deelchand DK, Hodges JS, et al. Altered macromolecular pattern and content in the aging human brain. NMR Biomed. February 2018;31(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasreddine ZS, Phillips NA, Bedirian V, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc April 2005;53(4):695–699. [DOI] [PubMed] [Google Scholar]
- 9.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16(1):31–41. [DOI] [PubMed] [Google Scholar]
- 10.Adriany G, De Moortele PFV, Ritter J, et al. A geometrically adjustable 16-channel transmit/receive transmission line array for improved RF efficiency and parallel imaging performance at 7 Tesla. Magn Reson Med. March 2008;59(3):590–597. [DOI] [PubMed] [Google Scholar]
- 11.Brant-Zawadzki M, Gillan GD, Nitz WR. MP RAGE: a three-dimensional, T1-weighted, gradient-echo sequence--initial experience in the brain. Radiology March 1992;182(3):769–775. [DOI] [PubMed] [Google Scholar]
- 12.Van de Moortele PF, Auerbach EJ, Olman C, Yacoub E, Ugurbil K, Moeller S. T1 weighted brain images at 7 Tesla unbiased for Proton Density, T2* contrast and RF coil receive B1 sensitivity with simultaneous vessel visualization. Neuroimage June 2009;46(2):432–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger GJ, Snyder C, Akgun C, Vaughan T, Ugurbil K, Van de Moortele PF. Local B1+ shimming for prostate imaging with transceiver arrays at 7T based on subject-dependent transmit phase measurements. Magn Reson Med. February 2008;59(2):396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Moortele PF, Akgun C, Adriany G, et al. B1 destructive interferences and spatial phase patterns at 7 T with a head transceiver array coil. Magn Reson Med. December 2005;54(6):1503–1518. [DOI] [PubMed] [Google Scholar]
- 15.Automatic Gruetter R., localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. June 1993;29(6):804–811. [DOI] [PubMed] [Google Scholar]
- 16.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. February 2000;43(2):319–323. [DOI] [PubMed] [Google Scholar]
- 17.Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at 7 T. Magn Reson Med. September 2001;46(3):451–456. [DOI] [PubMed] [Google Scholar]
- 18.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. August 15 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 19.Friston KJ, Holmes AP, Worsley KJ, Poline J-P, Frith CD, Frackowiak RSJ. Statistical parameteric maps in functional imaging: A general linear approach. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- 20.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. January 31 2002;33(3):341–355. [DOI] [PubMed] [Google Scholar]
- 21.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. June 1996;29(3):162–173. [DOI] [PubMed] [Google Scholar]
- 22.Glasser MF, Sotiropoulos SN, Wilson JA, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. October 15 2013;80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry PG, Marjanska M, Walls JD, Valette J, Gruetter R, Ugurbil K. Proton-observed carbon-edited NMR spectroscopy in strongly coupled second-order spin systems. Magn Reson Med. February 2006;55(2):250–257. [DOI] [PubMed] [Google Scholar]
- 24.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. May 2000;13(3):129–153. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser LG, Marjanska M, Matson GB, et al. 1H MRS detection of glycine residue of reduced glutathione in vivo. J Magn Reson. February 2010;202(2):259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govind V, Young K, Maudsley AA. Corrigendum: proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153. NMR Biomed. July 2015;28(7):923–924. [DOI] [PubMed] [Google Scholar]
- 27.Deelchand DK, Marjanska M, Hodges JS, Terpstra M. Sensitivity and specificity of human brain glutathione concentrations measured using short-TE 1H MRS at 7 T. NMR Biomed May 2016;29(5):600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gussew A, Erdtel M, Hiepe P, Rzanny R, Reichenbach JR. Absolute quantitation of brain metabolites with respect to heterogeneous tissue compositions in 1H-MR spectroscopic volumes. Magn Reson Mater Phy October 2012;25(5):321–333. [DOI] [PubMed] [Google Scholar]
- 29.Ernst T, Kreis R, Ross BD. Absolute Quantitation of Water and Metabolites in the Human Brain .1. Compartments and Water. J Magn Reson B. August 1993;102(1):1–8. [Google Scholar]
- 30.Neeb H, Zilles K, Shah NJ. Fully-automated detection of cerebral water content changes: study of age- and gender-related H2O patterns with quantitative MRI. Neuroimage. February 1 2006;29(3):910–922. [DOI] [PubMed] [Google Scholar]
- 31.Abbas Z, Gras V, Mollenhoff K, Keil F, Oros-Peusquens AM, Shah NJ. Analysis of proton-density bias corrections based on T1 measurement for robust quantification of water content in the brain at 3 Tesla. Magn Reson Med. December 2014;72(6):1735–1745. [DOI] [PubMed] [Google Scholar]
- 32.Reetz K, Abbas Z, Costa AS, et al. Increased cerebral water content in hemodialysis patients. PLoS One. 2015;10(3):e0122188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney WD, Johnson G, Li X, et al. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med. February 2007;57(2):308–318. [DOI] [PubMed] [Google Scholar]
- 34.Bartha R, Michaeli S, Merkle H, et al. In vivo 1H2O T2(dagger) measurement in the human occipital lobe at 4T and 7T by Carr-Purcell MRI: Detection of microscopic susceptibility contrast. Magn Reson Med. April 2002;47(4):742–750. [DOI] [PubMed] [Google Scholar]
- 35.Marjanska M, Emir UE, Deelchand DK, Terpstra M. Faster metabolite 1H transverse relaxation in the elder human brain. PLoS One. 2013;8(10):e77572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oz G, Hutter D, Tkac I, et al. Neurochemical alterations in spinocerebellar ataxia type 1 and their correlations with clinical status. Mov Disord July 15 2010;25(9):1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Povazan M, Strasser B, Hangel G, et al. Simultaneous mapping of metabolites and individual macromolecular components via ultra-short acquisition delay 1H MRSI in the brain at 7T. Magn Reson Med. March 2018;79(3):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottschalk M, Lamalle L, Segebarth C. Short-TE localised 1H MRS of the human brain at 3 T: quantification of the metabolite signals using two approaches to account for macromolecular signal contributions. NMR Biomed June 2008;21(5):507–517. [DOI] [PubMed] [Google Scholar]
- 39.Marjanska M, McCarten JR, Hodges J, et al. Region-specific aging of the human brain as evidenced by neurochemical profiles measured noninvasively in the posterior cingulate cortex and the occipital lobe using 1H magnetic resonance spectroscopy at 7 T. Neuroscience. June 23 2017;354:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. June 15 1993;90(12):5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikkelsen M, Barker PB, Bhattacharyya PK, et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. October 1 2017;159:32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terpstra M, Tkac I, Rao R, Gruetter R. Quantification of vitamin C in the rat brain in vivo using short echo-time 1H MRS. Magn Reson Med. May 2006;55(5):979–983. [DOI] [PubMed] [Google Scholar]


