Abstract
Purpose
MicroRNAs play key regulatory roles in the tumorigenesis of hepatitis B virus-related hepatocellular carcinoma (HBV-HCC). This study aimed to explore the regulatory effects of microRNA-98-5p (miR-98-5p) on the proliferation, migration, invasion, and apoptosis of HBV-HCC cells, as well as the underlying mechanisms involving nuclear factor-κB-inducing kinase (NIK).
Materials and Methods
The expressions of miR-98-5p and NIK in HBV-HCC tissues and cells, and the level of HBV DNA in HBV-HCC cells were measured by quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, migration, invasion, and apoptosis of HBV-HCC cells were analyzed by cell counting kit-8, wound healing, transwell, and flow cytometry assay, respectively. The targeting relationship between miR-98-5p and NIK was predicted by StarBase3.0 and verified by dual-luciferase reporter assay. HBV-HCC xenograft tumor model was constructed in mice to observe the tumor growth in vivo.
Results
The expression of miR-98-5p was declined in HBV-HCC tissues and cells. Overexpression of miR-98-5p markedly reduced the level of HBV DNA; inhibited the proliferation, migration, and invasion; and promoted the apoptosis of HBV-HCC cells. NIK was a target of miR-98-5p. Overexpression of miR-98-5p markedly decreased the protein expression of NIK in MHCC97H-HBV cells. NIK reversed the tumor-suppressing effect of miR-98-5p on HBV-HCC cells. Furthermore, overexpression of miR-98-5p significantly inhibited the xenograft tumor growth and decreased the expression of NIK in mice.
Conclusion
MiR-98-5p inhibits the secretion of HBV, proliferation, migration, and invasion of HBV-HCC cells by targeting NIK.
Keywords: MicroRNA-98-5p, hepatitis B virus-related hepatocellular carcinoma, proliferation, invasion, nuclear factor-κB-inducing kinase
INTRODUCTION
Hepatocellular carcinoma (HCC) now ranks the sixth most common malignancy worldwide, generating the third important reason of mortality attributed to cancer.1 There is a great difference in the incidence of HCC worldwide, with the highest rate being reported in East Asia and China accounting for approximately 50% of HCC malignancies.2 Persistent infection by hepatitis B virus (HBV) is the principal reason for HCC worldwide, which has a very poor prognosis.3 Liver resection and transplantation remain the main therapeutic strategies for HCC.4 However, the prognosis of HBV-HCC patients after surgery is not optimal due to the high HBV replication rates, viral mutations, and infection-associated inflammation.5 Therefore, it is necessary to explore new strategies for the treatment of HBV-HCC.
MicroRNAs (miRNAs) are a series of small, non-coding RNAs with a length of 18–25 nt, which negatively mediate the expression of target genes by the interactions with 3′ untranslated regions (3′ UTRs).6 MiRNAs play critical regulatory roles in the tumorigenesis of HBV-HCC. For examples, miR-152 acts as a tumor suppressor in HBV-HCC by regulating DNMT1.7 MiR-34c markedly inhibits the growth of HBV-HCC cells by targeting TGIF2.8 MiR-98 is known as a tumor suppressor in HCC. Wang, et al.9 have shown that miR-98 inhibits the proliferation, migration, and invasion of HCC cells by targeting CTHRC1. Jiang, et al.10 have proved that microRNA-98-5p (miR-98-5p) inhibits proliferation and promotes the apoptosis of HCC cells by targeting IGF2BP1. Notably, a miRNA profile in HBV-induced infection showed that miR-98 is involved in HBV infection.11 However, the specific regulatory role of miR-98-5p in HBV-HCC remains unclear.
There is constitutive activation of nuclear factor-κB (NF-κB) in most tumor cells.12 The canonical and non-canonical pathways both play an essential role in the mediation of NF-κB activation.13 In non-canonical NF-κB pathway, NF-κB-inducing kinase (NIK) is involved in initiating receptor signaling.14 A previous study showed that the activation of NF-κB by NIK obviously promotes inflammatory reaction, carcinogenic signaling, and proliferation of epithelial cells.15 Notably, NIK acted as a potential therapeutic target in HCC. For example, the silencing of NIK proved to be a valuable therapeutic strategy for reducing the constitutive NF-κB activation in HCC.16 MiR-520e attenuated the growth of HCC cells by inhibiting the expression of NIK.17 NIK-specific siRNA-1397 inhibited the proliferation of HBV-derived HCC cells by regulating NIK activation.18 Nevertheless, the potential regulatory mechanism of miR-98-5p involving NIK in HBV-HCC still remains unknown.
In the present study, we evaluated the expression of miR-98-5p in HCC-HBV tissues and cell lines, such as MHCC97H-HBV and Huh7-1.3. Then, we explored the effects of miR-98-5p on the secretion of HBV, proliferation, migration, invasion, and apoptosis of HCC-HBV cells, as well as tumorigenesis in nude mice. The association between miR-98-5p and NIK was evaluated in HBV-HCC. Our study may aid in discovering a hopeful therapeutic target for HBV-HCC, as well as the underlying mechanisms for HBV-HCC treatment.
MATERIALS AND METHODS
Tissue samples
Thirty pairs of tumor and adjacent noncancerous tissues were collected from patients with HBV-HCC at our hospital. There were 23 males and seven females. The average age of patients was 56.85±10.98 years. The patients were divided into either low group (n=12) or high group (n=18), based on the median expression of miR-98-5p (0.4656). Informed consent was obtained from all patients before the collection of samples. This study was approved by the Hospital Ethics of Human Research Committee, and performed in accordance with the Declaration of Helsinki.
Cell culture
MHCC97H and Huh7 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 (Hyclone, Novato, CA, USA), and supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, NY, USA). The cells were divided into two groups: MHCC97H-HBV cell group (MHCC97H cells were transfected with HBV genome) and Huh7-1.3 cell group (Huh7 cells were transfected with pcDNA 3.0–1.3 mer, containing the 1.3-mer fragment of HBV genome).
Transient transfection
The miR-98-5p mimics and miR-negative control of mimics (mimics-NC) were obtained by Shanghai Genepharma (Shanghai, China). The pcDNA-3.1NIK and pcDNA3.1-NC were obtained by Shanghai Sangon Biotech (Shanghai, China). MHCC97H-HBV cells grown to 60% confluency were transfected with the aforementioned agents using Lipofectamine 2000 reagent (Invitrogen).
Quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA of cells and tissues was extracted by using TRIZOL regent Trizol (Invitrogen), and then reverse-transcribed into cDNA using Takara PrimeScript RT reagent kit (Takara, Otsu, Japan). PCR reaction was performed on ABI 7500 HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) under the following conditions: 95℃ for 3 min, 40 cycles of 95℃ for 15 s, and 60℃ for 30 s. Relative expression was calculated by the 2−ΔΔCt method. U6 and β-actin were used for the normalization of miR-98-5p and NIK, respectively. Primer sequences (Shanghai Sangon Biotech) are shown in Supplementary Table 1 (only online).
Dual-luciferase reporter assay
The potential binding sites of NIK and miR-98-5p were predicted according to StarBase3.0. The NIK-Wt and NIK-Mut were cloned and combined with psiCHECK-2 vectors (Promega, Madison, WI, USA). NIK-Wt or NIK-Mut was co-transfected with miR 98a 3p mimics or mimics-NC (Shanghai Genepharma) into HEK-293T cells with Lipofectamine 3000 (Invitrogen). Luciferase activity was detected by Dual-luciferase reporter gene assay system (Promega).
Western blot
HBV-HCC cells were lysed in RIPA lysis buffer (Santa Cruz, Dallas, TX, USA) to obtain total protein. Proteins were run on sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibody [rabbit anti-human-GAPDH (1:1000, #5174, CST, Danvers, MA, USA); NIK (1:1000, #4994, CST)] overnight at 4℃. Then, the membranes were incubated with secondary antibody [anti-rabbit (1:1000, Sigma, St. Louis, MO, USA)] for 1 h. Finally, the protein bands were visualized with electrogenerated chemiluminescence reagent (Millipore, Bedford, MA, USA), and analyzed by Image LabTM Software (Bio-Rad, Hercules, CA, USA). GAPDH was introduced as the internal reference.
Cell proliferation assay
To measure the cell proliferation, 10 µL of cell counting kit-8 (CCK-8) reagents (BD Biosciences, Franklin Lakes, NJ, USA) were separately added into each well of the 96-well plate. The optical density at 450 nm (OD450) was measured using a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
Flow cytometry
Cells were seeded in 96-well plates, and cultured for 24 h. Then, 500 µL of cells were stained with 5 µL of V-FITC and 10 µL of PI for 15 min in the dark. The apoptosis rate was detected on flow cytometry (BD Biosciences).
Wound healing assay
Cells were seeded into 6-well plates (5×105 cells/well). When the cells were cultured at 90% confluence, an artificial scratch was created using 10 µL of pipette tip. Cells were incubated for 48 h, and then observed under an inverted microscope (Olympus, Tokyo, Japan). Wound healing rates were calculated by the fraction of cell coverage across the line.
Transwell assay
Transwell assay was performed to determine the cell invasion using transwell chambers (8-nm pore size) (Corning Inc., Corning, NY, USA). Cells (2×105) in serum free medium were added to upper chambers, and upper chambers were pre-coated with matrigel (BD Biosciences). RPMI-1640 medium containing 10% FBS was added to the matched lower chambers. After 48 h of incubation at 37℃, cells were removed from the upper chambers with a cotton swab, and cells in lower chambers were fixed in methanol and stained with 0.5% crystal violet for 2 min. Five random fields were photographed and counted.
Extraction and quantification of HBV DNA
After transfection for 48 h, HBV DNA in culture supernatants was collected. PCR was performed using ABI PRISM 7500 Sequence Detection System (Applied BioSystems). The concentration of HBV DNA was determined by the average threshold cycle values.
Detection of HBsAg and HBeAg
The levels of HBV surface antigen (HBsAg) and antigen (HBeAg) were detected by commercially available enzyme-linked immunosorbent assay kits (Autobio Diagnostics, Zhengzhou, China). Positive rates were measured by the following formula: positive rate (%)=(Ctested/Ccontrol)×100.
Tumor formation in mice
Male BALB/c nude mice (5 weeks old, n=24) were purchased from Shanghai Laboratory Animal Center (Shanghai, China). Mice were maintained in a controlled environment at 23–25℃, 50–60% humidity, and a 12-h light/dark cycle with free access to food and water. The MHCC97H-HBV cells transfected with mimics-NC (mimics-NC group) and miR-98-5p mimics (miR-98-5p mimics group) (5×106 cells, 150 µL each mouse) were subcutaneously injected into the left axilla of mice (n=8 in each group). Mice injected with MHCC97H-HBV cells without transfection were considered as the mock group (n=8). Tumor volume was measured every week with calipers, and it was calculated using the following formula: (A · B2)/2 (A, longest diameter; B, shortest diameter). After the last measurement at 4-week post-injection, the mice were anesthetized by intraperitoneal injection of diazepam (5 mg/kg) and ketamine (50 mg/kg), and then sacrificed by cervical dislocation. The xenografted tumors were immediately removed and weighed. All animal experiments and programs were approved by the Animal Care and Use Committee of our hospital.
Statistical analysis
All analyses of results were performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA) and GraphPad Prism software 7.0 (GraphPad, La Jolla, CA, USA). Qualitative data are presented as number, and the difference was determined by χ2 test (two groups). Quantitative data are presented as mean±SD, and the difference were analyzed by Student's t-test (two groups) or one-way ANOVA followed by multiple comparisons test (multi-groups). p<0.05 was considered to be statistically significant.
RESULTS
MiR-98-5p expression was declined in HBV-HCC tissues and cells
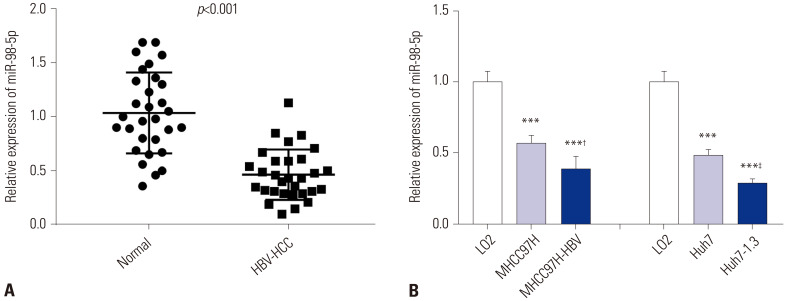
By performing qRT-PCR, we detected the expression level of miR-98-5p in HBV-HCC tissues and adjacent tissues. The expression of miR-98-5p in HBV-HCC tissues was down-regulated compared to adjacent normal tissues (p<0.001) (Fig. 1A). The expression of miR-98-5p in normal liver cells (LO2 cells), HCC cells (MHCC97H and Huh7 cells), and HBV-HCC cells (MHCC97H-HBV and Huh7-1.3 cells) was measured by qRT-PCR. The expression of miR-98-5p in MHCC97H, MHCC97H-HBV, Huh7 and Huh7-1.3 cells was lower than that in LO2 cells (p<0.001). The expression of miR-98-5p in MHCC97H-HBV and Huh7-1.3 cells was lower than that in MHCC97H and Huh7 cells, respectively (p<0.05) (Fig. 1B). We then analyzed the relationship between miR-98-5p expression in HBV-HCC tissues and clinicopathological features. As shown in Table 1, the alpha fetoprotein (AFP), HBV DNA levels, tumor node metastasis (TNM) stage, Barcelona Clinic Liver Cancer (BCLC) stage, metastasis, and time to recurrence were significantly different between high and low groups (p<0.05).
Fig. 1. The expression of microRNA-98-5p (miR-98-5p) was down-regulated in hepatocellular carcinoma-related hepatitis B virus (HBV-HCC) tissues and cells. (A) Relative expression of miR-98-5p in adjacent normal tissues and HBV-HCC tissues was detected by qRT-PCR (p<0.001). (B) Relative expression of miR-98-5p in LO2, MHCC97H, MHCC97H-HBV, Huh7, and Huh7-1.3 cells was detected by qRT-PCR. ***p<0.001 vs. LO2; †p<0.05 vs. MHCC97H; ‡p<0.05 vs. Huh7. qRT-PCR, quantitative real-time polymerase chain reaction.
Table 1. Correlations between MicroRNA-98-5p Expression and Clinicopathological Features of Patients with HBV-Related Hepatocellular Carcinoma.
| Clinicopathological features | No. of cases | miR-98-5p | p value | |
|---|---|---|---|---|
| High (n=12) | Low (n=18) | |||
| Sex | 0.840 | |||
| Male | 23 | 9 | 14 | |
| Female | 7 | 3 | 4 | |
| Age (yr) | 0.660 | |||
| <60 | 11 | 3 | 8 | |
| ≥60 | 19 | 9 | 10 | |
| AFP (µg/mL) | 0.025* | |||
| <400 | 16 | 8 | 8 | |
| ≥400 | 14 | 4 | 10 | |
| Cirrhosis | 0.270 | |||
| Positive | 20 | 7 | 13 | |
| Negative | 10 | 5 | 5 | |
| Use of antivirals | 0.526 | |||
| Entecavir | 23 | 10 | 13 | |
| Tenofovir disoproxil | 7 | 4 | 3 | |
| HBV DNA level | 0.001** | |||
| <1.0×10e3 | 13 | 11 | 2 | |
| ≥1.0×10e3 | 17 | 2 | 15 | |
| Tumor size (cm) | 0.800 | |||
| <3 | 12 | 5 | 13 | |
| ≥3 | 18 | 12 | 10 | |
| TNM stage | 0.010* | |||
| I/II | 22 | 12 | 10 | |
| III/IV | 8 | 0 | 8 | |
| BCLC stage | 0.026* | |||
| 0 | 4 | 3 | 1 | |
| A | 17 | 9 | 8 | |
| B | 7 | 0 | 7 | |
| C | 2 | 0 | 2 | |
| Metastasis | 0.001** | |||
| No | 10 | 8 | 2 | |
| Yes | 20 | 4 | 16 | |
| Time to recurrence (month) | 0.004** | |||
| <12 | 9 | 6 | 3 | |
| ≥12 | 21 | 6 | 15 | |
AFP, alpha fetoprotein; HBV, hepatitis B virus; TNM, tumor node metastasis; BCLC, Barcelona Clinic Liver Cancer.
Present significant difference at *p<0.05 and **p<0.01, respectively.
MiR-98-5p inhibits HBV secretion
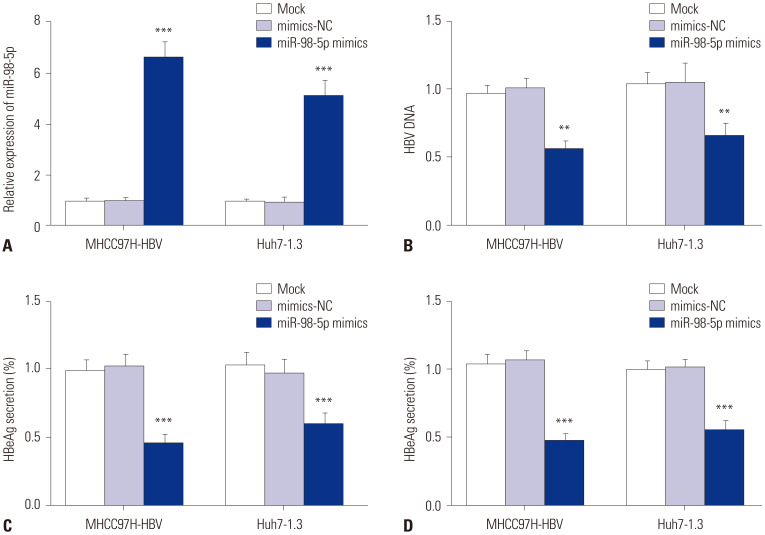
The miR-98-5p transfection efficiency was measured by qRT-PCR. In MHCC97H-HBV and Huh7-1.3 cells, the transfection of miR-98-5p mimics significantly promoted miR-98-5p expression (p<0.001) (Fig. 2A). The level of HBV DNA was measured by qRT-PCR. The level of HBV DNA in miR-98-5p mimics group was lower than that in mimics-NC and mock group (p<0.01) (Fig. 2B). The ratio of HBsAg and HBeAg secretion in miR-98-5p mimics group was lower than that in mimics-NC and mock group (p<0.001) (Fig. 2C and D).
Fig. 2. MiR-98-5p inhibited the secretion of HBV. (A) Relative expression of miR-98-5p in MHCC97H-HBV and Huh7-1.3 cells was detected by qRT-PCR. (B) The level of HBV DNA in MHCC97H-HBV and Huh7-1.3 cells was detected by qRT-PCR. (C) The ratio of HBeAg secretion in MHCC97H-HBV and Huh7-1.3 cells. (D) The ratio of HBsAg secretion in MHCC97H-HBV and Huh7-1.3 cells. **p<0.01, ***p<0.001 vs. Mock and mimics-NC. MiR-98-5p, microRNA-98-5p; HBV, hepatitis B virus; qRT-PCR, quantitative real-time polymerase chain reaction.
MiR-98-5p inhibits the proliferation, migration, and invasion of HBV-HCC cells
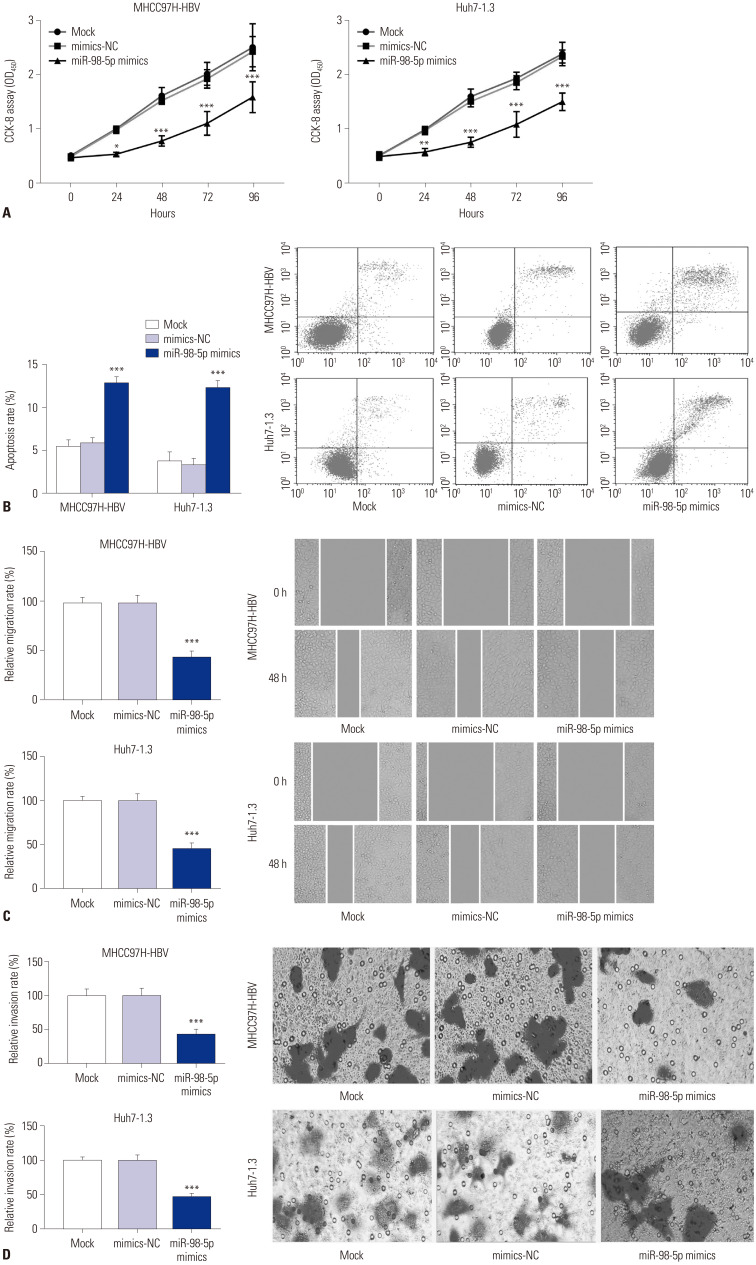
The biological effect of miR-98-5p on cell proliferation was measured by CCK-8 assay. The cell proliferation in miR-98-5p mimics group was down-regulated compared to that in mimics-NC and mock group (p<0.05) (Fig. 3A). The apoptosis rate of cell was measured by flow cytometry. Compared to the mimics-NC and mock groups, the apoptosis rate of cell was markedly increased in miR-98-5p mimics group (p<0.001) (Fig. 3B). The migration of cell was detected by wound healing assay. The cell migration in miR-98-5p mimics group was declined than that in mimics-NC and mock group (p<0.001) (Fig. 3C). The invasion of cell was detected by transwell assay. Compared to the mimics-NC and mock groups, the cell invasion in miR-98-5p mimics group was markedly increased (p<0.001) (Fig. 3D).
Fig. 3. MiR-98-5p inhibited the proliferation, migration, and invasion of HBV-HCC cells. (A) Proliferation of MHCC97H-HBV and Huh7-1.3 cells was assessed by cell counting kit-8 (CCK-8) assay. (B) Apoptosis of MHCC97H-HBV and Huh7-1.3 cells was assessed by flow cytometry. (C) Migration of MHCC97H-HBV and Huh7-1.3 cells was assessed by wound healing assay. (D) Invasion of MHCC97H-HBV and Huh7-1.3 cells was assessed by transwell assay. *p<0.05, **p<0.01, ***p<0.001 vs. Mock and mimics-NC. MiR-98-5p, microRNA-98-5p; HBV-HCC, hepatitis B virus-related hepatocellular carcinoma.
NIK is a direct target of miR-98-5p
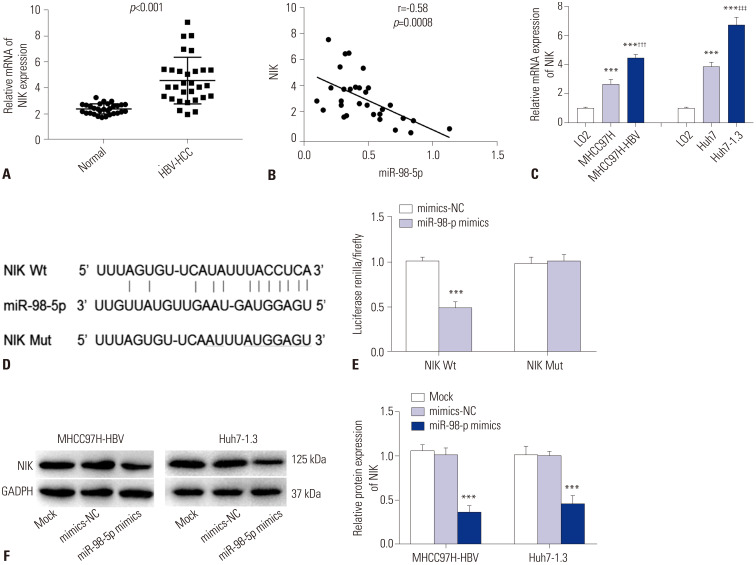
The expression of NIK mRNA in adjacent normal tissues and HBV-HCC tissues was measured by qRT-PCR. The expression of NIK mRNA in HBV-HCC tissues was higher than that in adjacent normal tissues (p<0.001) (Fig. 4A). The expression of NIK mRNA was negatively correlated with miR-98-5p (r=−0.58, p=0.0008) (Fig. 4B). The expression of NIK mRNA in normal liver cells (LO2 cells), HCC cells (MHCC97H and Huh7 cells), and HBV-HCC cells (MHCC97H-HBV and Huh7-1.3 cells) was measured by qRT-PCR. The results showed that the expression of NIK mRNA in MHCC97H, MHCC97H-HBV, Huh7 and Huh7-1.3 cells was significantly higher than that in LO2 cells. The expression of NIK mRNA in MHCC97H-HBV and Huh7-1.3 cells was higher than that in MHCC97H and Huh7 cells, respectively (p<0.001) (Fig. 4C). A computational Star-Base3.0 search predicted one binding site for miR-98-5p in NIK 3′ UTR (Fig. 4D). Luciferase renilla/firefly activity was markedly decreased in the HEK-293T cells transfected with NIK Wt and miR-98-5p mimics (p<0.001) (Fig. 4E). Relative protein levels of NIK in MHCC97H-HBV and Huh7-1.3 cells were detected by western blot. The relative protein expression of NIK in MHCC97H-HBV and Huh7-1.3 cells in miR-98-5p mimics group was lower than that in mock and mimics-NC group, respectively (p<0.001) (Fig. 4F).
Fig. 4. NIK was a direct target of miR-98-5p. (A) Relative mRNA expression of NIK in normal liver tissues and HBV-HCC tissues was detected by qRT-PCR (p<0.001). (B) The mRNA expression of NIK was negatively correlated with that of miR-98-5p (p=0.0008). (C) Relative mRNA expression of NIK in LO2, MHCC97H, MHCC97H-HBV, Huh7, and Huh7-1.3 cells was detected by qRT-PCR (***p<0.001 vs. LO2; †††p<0.001 vs. HepG2; ‡‡‡p<0.001 vs. Huh7). (D) Schematic diagram of the target site between miR-98-5p and NIK. (E) Luciferase renilla/firefly activity of HEK-293T cells transfected with NIK Wt/Mut and miR-98-5p mimics/mimics-NC (***p<0.001 vs. mimics-NC). (F) Relative protein expression of NIK in MHCC97H-HBV and Huh7-1.3 cells was detected by western blot (***p<0.001 vs. Mock and mimics-NC). NIK, nuclear factor-κB-inducing kinase; MiR-98-5p, microRNA-98-5p; HBV-HCC, hepatitis B virus-related hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
NIK eliminates the effect of miR-98-5p on HBV-HCC cells
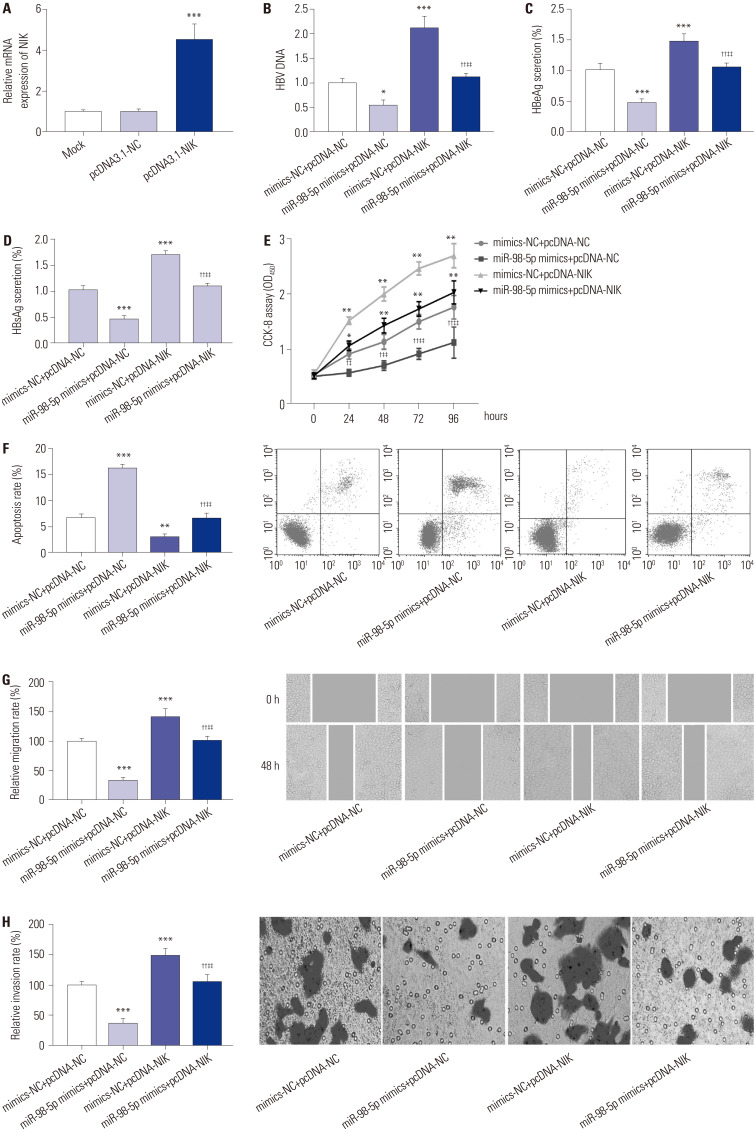
The transfection of pcDNA3.1-NIK significantly promoted mRNA expression of NIK in MHCC97H-HBV cells (p<0.05) (Fig. 5A). Compared to mimics-NC+pcDNA-NC group, the level of HBV DNA, secretion ratio of HBsAg and HBeAg, CCK-8 assay (OD450), migration, and invasion of MHCC97H-HBV cells were markedly promoted in mimics-NC+pcDNA-NIK group (p<0.01). Compared to mimics-NC+pcDNA-NC group, the abovementioned indexes of MHCC97H-HBV cells were markedly reduced in miR-98-5p mimics+pcDNA-NC group (p<0.05). The transfection of pcDNA3.1-NIK markedly rescued the inhibiting effect of miR-98-5p mimics on the above indexes in MHCC97H-HBV cells (p<0.01) (Fig. 5B–E, G, and H). The apoptosis of MHCC97H-HBV cells was contrary to CCK-8 assay (OD450) (p<0.01) (Fig. 5F).
Fig. 5. NIK eliminated the effect of miR-98-5p on HBV-HCC cells. (A) Relative mRNA expression of NIK in MHCC97H-HBV cells was detected by qRT-PCR (***p<0.001 vs. Mock and pcDNA3.1-NC). (B) The level of HBV DNA in MHCC97H-HBV cells was detected by qRT-PCR. (C) Secretion ratio of HBeAg in MHCC97H-HBV cells. (D) Secretion ratio of HBsAg in MHCC97H-HBV cells. (E) Proliferation of MHCC97H-HBV cells was assessed by cell counting kit-8 (CCK-8) assay. (F) Apoptosis of MHCC97H-HBV was assessed by flow cytometry. (G) Migration of MHCC97H-HBV cells was assessed by wound healing assay. (H) Invasion of MHCC97H-HBV cells was assessed by transwell assay. *p<0.05, **p<0.01, ***p<0.001 vs. mimics-NC+pcDNA-NC; †p<0.05, ††p<0.01 vs. miR-98-5p mimics+pcDNA-NC; ‡p<0.05, ‡‡p<0.01 vs. mimics-NC+pcDNA-NIK. NIK, nuclear factor-κB-inducing kinase; MiR-98-5p, microRNA-98-5p; HBV-HCC, hepatitis B virus-related hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
MiR-98-5p prevents subcutaneous tumor growth in mice
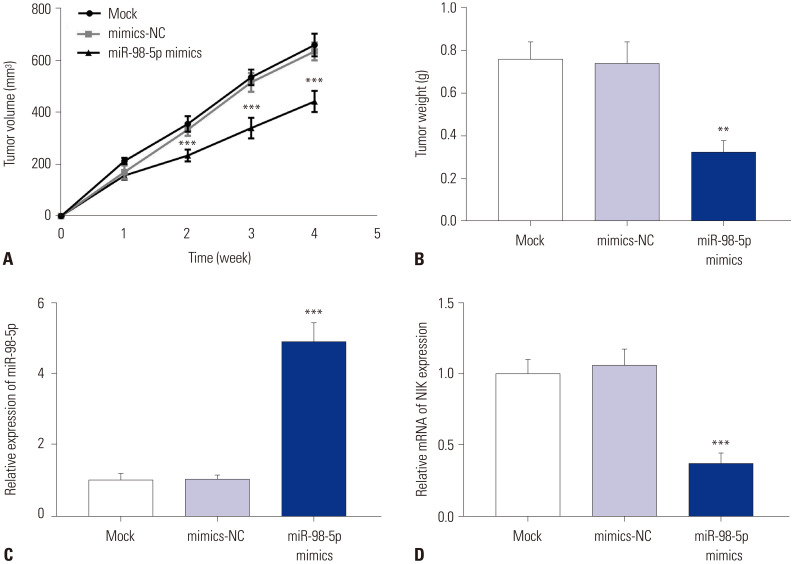
To reveal the anti-tumor effect of miR-98-5p in vivo, MHCC97H-HBV cells were injected into mice. Compared to mimics-NC group, the tumor volume and tumor weight were markedly declined in miR-98-5p mimics group (p<0.01) (Fig. 6A and B). Relative expression of miR-98-5p in mice tumor in mimics-NC group was lower than that in miR-98-5p mimics group (p<0.001) (Fig. 6C). Relative mRNA of NIK expression in mice tumor in miR-98-5p mimics group was lower than that in mimics-NC group (p<0.001) (Fig. 6D).
Fig. 6. MiR-98-5p inhibited xenograft tumor growth in mice. (A) Tumor volume. (B) Tumor weight. (C) Relative expression of miR-98-5p in xenograft tumor was detected by qRT-PCR. (D) Relative mRNA expression of NIK in xenograft tumor was detected by qRT-PCR. **p<0.01, ***p<0.001 vs. Mock and mimics-NC. NIK, nuclear factor-κB-inducing kinase; MiR-98-5p, microRNA-98-5p; qRT-PCR, quantitative real-time polymerase chain reaction.
DISCUSSION
Numerous studies have reported that abnormal expression of miRNAs is related to the tumorigenesis of cancer, such as HCC.19,20 Here, the expression of miR-98-5p in HBV-HCC tissues was markedly up-regulated by qRT-PCR compared to adjacent non-cancerous tissues. Our research is similar to previous studies on HBV-HCC, such as miR-27a,21 miR-12222 and miR 34c.8 We suspect that miR-98-5p plays an essential role in inhibiting tumorigenesis in HBV-HCC. The clinicopathological features demonstrated that the AFP, HBV DNA levels, TNM stage, BCLC stage, metastasis, and time to recurrence were significantly different between high and low groups. Expression of miR-630 was significantly increased at the advanced TNM stage, and it revealed a positive correlation with the levels of serum AFP.23 Moreover, miR-187-3p inhibited metastasis of HCC by targeting S100A4.24 The inhibitory effect of miR-214 was much more obvious in HCC patients with early-recurrence than in HCC patients with no recurrence.25 In our study, the function of miR-98-5p was similar to the abovementioned miRNAs on clinicopathological features, suggesting that miR-98-5p may be a valuable diagnostic and prognostic factor for HBV-HCC.
HBV is part of the Hepadna virus family, which mainly infects hepatocytes and causes acute and chronic hepatopathy, and persistent HBV infection is the primary etiological factor in HCC.26 Overexpression of miR-199a-3p results in downregulation of HBV replication in HBV-HCC cells.27 MiR-29c has been found to reduce HBV DNA replication and the secretion ratio of HBsAg and HBeAg by targeting TNFAIP3,28 and miR-125a-5p inhibits the secretion of HBV proteins by targeting ErbB3.29 Similar with previous studies, the results of our study also showed that miR-98-5p markedly reduced the level of HBV DNA in HBV-HCC cells. Taken together, our findings suggest that the expression of miR-98-5p is negatively associated with the secretion of HBV in HBV-HCC cells.
MiRNAs plays an essential regulatory function in the proliferation, migration, invasion, and apoptosis of HCC cells.11 Our research showed that miR-98-5p significantly decreased the proliferation, migration, and invasion of HBV-HCC cells, while increasing the apoptosis of HBV-HCC cells. Huang, et al.30 have shown that miR-128-3p significantly inhibited proliferation of HCC cells by regulating G1 phase cell arrest. MiR-325-3p suppressed HBV-HCC cells proliferation and induced apoptosis by the inhibition of aquaporin 5.31 Lang, et al.32 reported that the overexpression of miR-124 markedly inhibited tumorigenesis in HCC mice. Here, we also found that miR-98-5p markedly reduced the HBV-HCC xenograft tumor volume and weight in nude mice, and suppressed proliferation, migration, and invasion of HBV-HCC cells. Taken together, our results in vivo and in vitro suggest that miR-98-5p suppresses tumor growth by inhibiting cells proliferation and inducing apoptosis.
MiRNAs can down-regulate the expression of important cancer-related genes by base pairing with 3′-UTR, and it might be involve in the diagnosis and therapy of HCC.33 NIK effectively participates in the activation of NF-κB, thereby indirectly mediating cell growth and transformation.15 The expression of NIK is usually up-regulated in tumor cells, and NIK represents a valuable therapeutic target for cancers.34,35 Similar with previous studies, our research found that the expression of NIK was markedly up-regulated in HBV-HCC tissues and negatively associated with miR-98-5p. Zhang, et al.17 also reported that miR-520e suppresses the tumorigenesis of HCC by targeting NIK participating in the NIK/p-ERK1/2/NF-κB signaling pathway. Here, NIK reversed the effect of miR-98-5p on the levels of HBV DNA, proliferation, migration, and invasion of HBV-HCC cells. Therefore, our results suggest that miR-98-5p may attenuate the progress of HBV-HCC by targeting NIK.
In summary, our findings demonstrated that miR-98-5p was decreased in HBV-HCC tissues and cells. Also, MiR-98-5p inhibited secretion of HBV, proliferation, migration, and invasion of HBV-HCC cells, while inducing apoptosis. MiR-98-5p prevented the tumorigenesis of HBV-HCC by targeting NIK. MiR-98-5p may act as a potential therapeutic target for HBV-HCC.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Yuanyuan Liu and Xiukun Fei.
- Data curation: Xiukun Fei, Peipei Zhang, and Yu Pan.
- Formal analysis: Yuanyuan Liu and Xiukun Fei.
- Funding acquisition: Yuanyuan Liu and Xiukun Fei.
- Investigation: all anthors.
- Methodology: all anthors.
- Project administration: Xiukun Fei.
- Resources: all authors.
- Software: Yuanyuan Liu and Xiukun Fei.
- Supervision: Xiukun Fei.
- Validation: Yuanyuan Liu and Xiukun Fei.
- Visualization: all authors.
- Writing—original draft: Yuanyuan Liu.
- Writing—review & editing: all authors.
- Approval of final manuscript: all authors.
SUPPLEMENTARY MATERIAL
Primer Sequences Used in Quantitative Real-Time Polymerase Chain Reaction
References
- 1.Bravi F, Bosetti C, Tavani A, Gallus S, La Vecchia C. Coffee reduces risk for hepatocellular carcinoma: an updated meta-analysis. Clin Gastroenterol Hepatol. 2013;11:1413–1421.e1. doi: 10.1016/j.cgh.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: a comprehensive review. World J Hepatol. 2015;7:2648–2663. doi: 10.4254/wjh.v7.i26.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang A, Ju W, Yuan X, Han M, Wang X, Guo Z, et al. Comparison between liver resection and liver transplantation on outcomes in patients with solitary hepatocellular carcinoma meeting UNOS criteria: a population-based study of the SEER database. Oncotarget. 2017;8:97428–97438. doi: 10.18632/oncotarget.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Y, Su T, Ding Y, Cao G. Effects of antiviral therapy on the recurrence of hepatocellular carcinoma after curative resection or liver transplantation. Hepat Mon. 2012;12:e6031. doi: 10.5812/hepatmon.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 8.Liang WL, Cao J, Xu B, Yang P, Shen F, Sun Z, et al. miR-892a regulated PPP2R2A expression and promoted cell proliferation of human colorectal cancer cells. Biomed Pharmacother. 2015;72:119–124. doi: 10.1016/j.biopha.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang CY, Zhang JJ, Hua L, Yao KH, Chen JT, Ren XQ. MicroRNA-98 suppresses cell proliferation, migration and invasion by targeting collagen triple helix repeat containing 1 in hepatocellular carcinoma. Mol Med Rep. 2016;13:2639–2644. doi: 10.3892/mmr.2016.4833. [DOI] [PubMed] [Google Scholar]
- 10.Jiang T, Li M, Li Q, Guo Z, Sun X, Zhang X, et al. MicroRNA-98-5p inhibits cell proliferation and induces cell apoptosis in hepatocellular carcinoma via targeting IGF2BP1. Oncol Res. 2017;25:1117–1127. doi: 10.3727/096504016X14821952695683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer. 2017;17:805. doi: 10.1186/s12885-017-3816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hironaka N, Mochida K, Mori N, Maeda M, Yamamoto N, Yamaoka S. Tax-independent constitutive IkappaB kinase activation in adult T-cell leukemia cells. Neoplasia. 2004;6:266–278. doi: 10.1593/neo.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Qi H, Ren J, Cui J, Li Z, Waldum HL, et al. Involvement of NF-κB/IL-6 pathway in the processing of colorectal carcinogenesis in colitis mice. Int J Inflam. 2014;2014:130981. doi: 10.1155/2014/130981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thu YM, Su Y, Yang J, Splittgerber R, Na S, Boyd A, et al. NF-κB inducing kinase (NIK) modulates melanoma tumorigenesis by regulating expression of pro-survival factors through the β-catenin pathway. Oncogene. 2012;31:2580–2592. doi: 10.1038/onc.2011.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noort AR, van Zoest KP, Weijers EM, Koolwijk P, Maracle CX, Novack DV, et al. NF-κB-inducing kinase is a key regulator of inflammation-induced and tumour-associated angiogenesis. J Pathol. 2014;234:375–385. doi: 10.1002/path.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T, et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-κB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology. 2014;60:1659–1673. doi: 10.1002/hep.27312. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-κB-inducing kinase (NIK) Oncogene. 2012;31:3607–3620. doi: 10.1038/onc.2011.523. [DOI] [PubMed] [Google Scholar]
- 18.Cho HA, Park IS, Kim TW, Oh YK, Yang KS, Kim JS. Suppression of hepatitis B virus-derived human hepatocellular carcinoma by NF-kappaB-inducing kinase-specific siRNA using liver-targeting liposomes. Arch Pharm Res. 2009;32:1077–1086. doi: 10.1007/s12272-009-1714-z. [DOI] [PubMed] [Google Scholar]
- 19.Wei W, Wanjun L, Hui S, Dongyue C, Xinjun Y, Jisheng Z. miR-203 inhibits proliferation of HCC cells by targeting survivin. Cell Biochem Funct. 2013;31:82–85. doi: 10.1002/cbf.2863. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Wang G, Zhou X, Song X, Gao H, Ma C, et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792–797. doi: 10.1016/j.biopha.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 21.Wu XJ, Li Y, Liu D, Zhao LD, Bai B, Xue MH. miR-27a as an oncogenic microRNA of hepatitis B virus-related hepatocellular carcinoma. Asian Pac J Cancer Prev. 2013;14:885–889. doi: 10.7314/apjcp.2013.14.2.885. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193–2205. doi: 10.1128/JVI.02831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JW, Li Y, Zeng XC, Zhang T, Fu BS, Yi HM, et al. miR-630 overexpression in hepatocellular carcinoma tissues is positively correlated with alpha-fetoprotein. Med Sci Monit. 2015;21:667–673. doi: 10.12659/MSM.892515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q, et al. miR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380–390. doi: 10.1016/j.canlet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Xia H, Ooi LL, Hui KM. MiR-214 targets β-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7:e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 27.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, et al. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Zhang W, Gong L, Huang X. MicroRNA 125a-5p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by downregulation of ErbB3. Oncol Res. 2019;27:449–458. doi: 10.3727/096504017X15016337254623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CY, Huang XP, Zhu JY, Chen ZG, Li XJ, Zhang XH, et al. miR-128-3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients. Oncol Rep. 2015;33:2889–2898. doi: 10.3892/or.2015.3936. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Han Y, Sun G, Liu X, Jia X, Yu X. MicroRNA-325-3p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by down-regulation of aquaporin 5. Cell Mol Biol Lett. 2019;24:13. doi: 10.1186/s11658-019-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426:247–252. doi: 10.1016/j.bbrc.2012.08.075. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in hepatocellular carcinoma. Hepat Mon. 2014;14:e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitoh Y, Yamamoto N, Dewan MZ, Sugimoto H, Martinez Bruyn VJ, Iwasaki Y, et al. Overexpressed NF-kappaB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood. 2008;111:5118–5129. doi: 10.1182/blood-2007-09-110635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer Sequences Used in Quantitative Real-Time Polymerase Chain Reaction