Abstract
There have been many studies suggesting that probiotics are effective in patients with diarrhea-predominant irritable bowel syndrome (IBS-D). However, its mechanism of action as well as prediction of response is still to be elucidated. In the present study, to find out metabolomic characteristics of probiotic effect in IBS-D, we compared IBS symptom changes and metabolomic characteristics in the subjects’ urine samples between multi-strain probiotics (one strain of Lactobacillus sp. and four strains of Bifidobacterium sp.) group (n = 32) and placebo group (n = 31). After 8 weeks’ administration (3 times/day), dissatisfaction in bowel habits and stool frequencies were significantly improved. Also, probiotics group had significantly changed seven metabolites including palmitic acid methyl ester (PAME) and 4,6-dihydroxyquinoline, 4-(2-aminophenyl)-2,4-dioxobutanoic acid (DOBA). According to IBS-SSS and IBS-QoL questionnaires, IBS-SSS responders showed higher PAME levels and IBS-QoL responders showed lower DOBA levels. This suggests potential role of these metabolites as a biomarker to predict probiotics effect in IBS-D patients.
Electronic supplementary material
The online version of this article (10.1007/s10068-019-00717-2) contains supplementary material, which is available to authorized users.
Keywords: Irritable bowel syndrome, Probiotics, Metabolomics
Introduction
Irritable bowel syndrome (IBS) is common (Gwee et al., 2009), but the precise pathophysiology is still need to be elucidated (Barbara et al., 2009; Camilleri, 2013; Ghoshal and Ranjan, 2011; Hammerle and Surawicz, 2008; Jeffery et al., 2012; Keszthelyi et al., 2012; Malagelada and Malagelada, 2016; Spiller, 2004). Pharmacologic, psychologic, and complementary approaches are considered as therapeutic options in IBS patients (Quigley, 2012) and probiotics are one of those options.
Probiotics alter the composition of the gut flora by exerting antibacterial, antiviral and anti-inflammatory effects at mucosal surface. One of the most active research areas is about the mechanisms how the efficacy of probiotics shows in some IBS patients (Harper et al., 2018; O’Hara and Shanahan, 2007). One of possible action is anti-inflammatory effects of probiotics (Dunne et al., 1999). For example, in an experimental animal with interleukin (IL)-10 knockout model of colitis, both a Lactobacillus and a Bifidobacterium showed a marked and parallel reduction in inflammation in the colon and the pro-inflammatory cytokines, such as interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and IL-12, while levels of the anti-inflammatory cytokine, such as transforming growth factor (TGF)-β, were maintained (McCarthy et al., 2003). We also have shown that B. bifidum BGN4, B. lactis AD011 and Lb. acidophilus AD031 were effective to improve symptoms in IBD patients who excrete normal or loose stools (Hong et al., 2009).
Recently, metabolomic approaches have studied systematic metabolism related to several diseases, such as inflammatory bowel diseases (IBD) and celiac disease (Bertini et al., 2009; Marchesi et al., 2007; William et al., 2009; Yu et al., 2018). Some metabolic changes in serum, urine and feces are known to be associated with the gut microflora, intestinal absorption, and host energy metabolism. However, there are only few studies with IBS patients, so far.
The aim of present stud was to examine the effect of a multi-species probiotics mixture on IBS symptoms and metabolomics characteristics. First, we compared IBS symptom changes between probiotics and placebo groups. Secondly, we tested effects of a multi-species probiotics mixture on metabolomic characteristics in the subjects’ urine samples and if there are any metabolomics differences between probiotics and placebo group responders and non-responders.
Materials and methods
Patients
Patients were enrolled prospectively at the Department of Gastroenterology of Seoul National University Hospital, between March 2013 and May 2013. The inclusion criteria were: (a) age between 18 and 75 years, (b) diagnosed with diarrhea-dominant IBS according to Rome II criteria (c) without any organic abnormalities by physical and laboratory examination during the screening period. Exclusion criteria were: (a) intolerance to probiotics or lactose, (b) pregnancy or lactation, (c) severe systemic illness (liver cirrhosis, congestive heart failure, chronic renal failure, angina, uncontrolled hypertension, endocrine disorder, metabolic disorder, or malignant tumors), (d) history of inflammatory bowel disease or psychiatric disorder, (e) alcohol or drug addiction, (f) previous abdominal surgery other than appendectomy, (g) being judged ineligible for participation in clinical trials by clinicians.
Signed informed consent was obtained from each patient prior to the enrolment. The study was conducted according to the Declaration of Helsinki and was approved by the Institutional Review Boards of Seoul National University Hospital (ClinicalTrials.gov Identifier: NCT01637714).
Study design
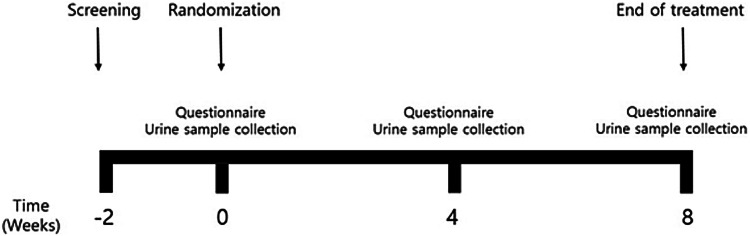
Figure 1 is a flow chart of the study design. Patients fulfilling the inclusion criteria were checked with the baseline (week 0) stool form, stool frequency, and visual analogue scale (VAS). After urine samples were collected, patients were randomized to receive either placebo or a multi-species probiotic mixture through a computer-generated table. All patients and investigators, except for the study coordinator, were kept blinded to allocation until study completion.
Fig. 1.
A flow diagram illustrating the study design
Participants visited the clinic to get their investigational products and were examined for IBS symptoms at 4 weeks and 8 weeks after the first administration. At these visits, patients submitted self-administered questionnaires on stool form/frequency and VAS score for each IBS symptom using IBS-SSS (severity scoring system) and IBS-QoL (quality of life) questionnaires. According to IBS-SSS, responders were defined as subjects who showed more than 50 points decrease after the probiotics treatment. And according to IBS-QoL, responders were defined as subjects who showed more than 10 points decrease after the probiotics treatment. At 4 and 8 weeks, we also collected a follow-up urine samples.
Study medication
Combined probiotics, which included 5 strains of probiotics (Bifidobacterium longum BORI, Bifidobacterium bifidum BGN4, Bifidobacterium lactis AD011, Bifidobacterium infantis IBS007, and Lactobacillus acidophilus AD031) were used in this study. Three strains BGN4, AD011 and AD031 were reported to be safe and effective, especially in IBD patients who excrete normal or loose stools by Kim et al. (Hong et al., 2009), and BORI and IBS007 were screened through preliminary animal test (data not shown). A capsule was composed of a total of 5 × 109 viable cells in a lyophilized powder form with the other ingredients including maltodextrin, corn starch, and silicon dioxide. The placebo capsule had almost the same contents as the active medication, even though the bacteria were replaced with maltodextrin. Probiotics (n = 32) or placebo (n = 31) was administered 3 times daily.
Assessment of urine metabolites
Urine samples 100 μL with 100% cold H2O 400 μL were vortexed for 10 min and centrifuged at 18,341×g for 20 min at 4 °C to remove particulates, 5 μL of which was injected into HPLC for the chromatographic separation. Chromatographic separations of metabolites in urine were performed with a Zorbax SB-C18, 50 × 2.1 mm, 1.8 μm (Agilent Technologies, Santa Clara, CA) analytical column using an Agilent 1200 series HPLC system (Agilent Technologies, Santa Clara, CA): 2 mM ammonium formate in H2O (0.1% formic acid) as a mobile phase A, 100% methanol (0.1% formic acid) as a mobile phase B, and flow rate was 0.4 ml/min. Q-TOF MS was done with Agilent Q-TOF 6530 mass spectrometer and the overall quality of the analysis procedure was monitored using repetition of a pooled urine sample (data not shown). The multivariate data matrix was analyzed by Mass Profiler Professional (MPP) software B.12.01 (Agilent Technologies) package which was used to principal component analysis (PCA). The identities of the specific metabolites were confirmed by using the human metabolome database (HMDB) and METLIN, and comparing chromatographic retention times and their mass spectra (MS/MS fragmentation patterns) to those obtained using commercially available reference standards. Levels of metabolites discriminating placebo and probiotics groups were normalized to creatinine levels in urine.
Statistical analysis
All statistical analyses were performed usingSPSS for Windows (ver. 18.0; SPSS, Chicago, IL). Student’s t-tests was used for continuous variables and Chi square tests or Fisher’s exact tests were used for categorical variables. To compare the quantitative changes of urine metabolites before and after the study period in both groups, paired t-tests were used. Spearman rank correlation coefficient were used to check the correlation. The results with a p value less than 0.05 were considered statistically significant.
Results and discussion
Baseline clinical characteristics and IBS symptom changes
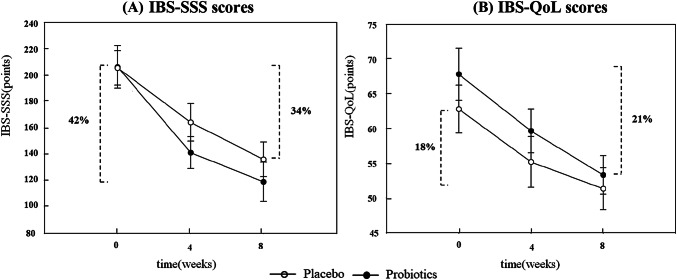
A total of 63 patients were enrolled and randomized to probiotics (n = 32) or placebo (n = 31) group. There were no significant differences in the baseline clinical and laboratory characteristics between the probiotic a placebo groups (Supplementary Table 1). When comparing the effects of probiotics and placebo on IBS symptoms, probiotics significantly changed defecation discomfort (− 29.1 in the probiotics group vs. − 21.4 in the placebo group; p = 0.0423) and stool frequency (− 1.3 in the probiotics group vs. + 0.7 in the placebo group; p = 0.0431), however, in terms of other symptoms, there was no significant difference (Table 1). It might be due to the nature of IBS symptoms. It usually takes a long time to prove a symptomatic improvement to any treatment. According to ‘FDA Guidance for Industry Irritable Bowel Syndrome—Clinical Evaluation of Drugs for Treatment’(US Department of Health and Human Services, 2012), a treatment period of at least 8 weeks duration was recommended at the section of trial design. In addition to the minimum duration of treatment, low numbers of subjects in the present study might be the cause of negative response in the IBS-SSS and IBS-QoL scores. The probiotics treatment resulted in better clinical relief in IBS patients by showing more IBS-SSS and IBS-QoL score decreases compared to those of placebo group; − 87.3 and − 14.5 in the probiotics group (42% and 21% improvements), and − 69.5 and − 11.5 in the placebo group (34% and 18% improvements), respectively (Fig. 2). However, the numbers of clinical responders, according to IBS-SSS or IBS-QoL scores, were not significantly different between those two groups (Table 2). Even though this association could not guarantee any definite causal relationships between them, this might be the valuable first step to elucidate the pathophysiology how probiotics alleviate IBS symptoms in some IBS-D patients. Many hypotheses have been advanced to explain the pathophysiology of IBS, including dysmotility, visceral hypersensitivity, aberrant cerebral representation of visceral events, and abnormal stress responses. However, none of them has provided definite explanations for all the IBS patients with various phenotypes (Quigley and Craig, 2012). More recently, the focus in IBS research has shifted to more local factors such as impaired gut barrier function, low-grade inflammation/immune activation, and an altered microbiome. Given its heterogeneous nature, it is likely that IBS involves more than one pathophysiological entity and may ultimately prove to encompass a number of distinct entities that cannot currently be separated on the basis of symptom clustering systems. Evidences have accumulated to suggest the presence of a low grade inflammatory state or immune activation in some IBS patients. Some findings in the colonic mucosa, such as increased mast cell numbers, enhanced mast cell degranulation, more intense lymphocyte infiltration, and an up-regulation of both pro-inflammatory cytokines and Toll-like receptors, supports this theory (Barbara and Cremon, 2008; Barbara et al., 2004; Brint et al., 2011; Chadwick et al., 2002; Crowell et al., 2004; Dinan et al., 2006; O’Mahony et al., 2005; O’Sullivan et al., 2000; Piche et al., 2008; Scully et al., 2010; Spiller et al., 2000; Talley and Butterfield, 1996; Weston et al., 1993).
Table 1.
Changes in symptom scores
| Placebo (n = 31) | Probiotics (n = 32) | |
|---|---|---|
| Abdominal pain | ||
| Baseline (± SD) | 35.5 (± 8.9) | 42.5 (± 11.4) |
| Δ 4 week | − 15.1 | − 20.0 |
| Δ 8 week | − 18.2 | − 23.3 |
| Bloating | ||
| Baseline (± SD) | 37.4 (± 15.1) | 40.2 (± 18.9) |
| Δ 4 week | − 15.3 | − 25.1 |
| Δ 8 week | − 16.2 | − 27.2 |
| Defecation discomfort | ||
| Baseline (± SD) | 51.1 (± 22.8) | 50.0 (± 20.5) |
| Δ 4 week | − 20.7 | − 26.3 |
| Δ 8 week | − 21.4 | − 29.1* |
| Stool frequency | ||
| Baseline (± SD) | 8.1 (± 2.1) | 7.5 (± 3.5) |
| Δ 8 week | 0.7 | − 1.3* |
| Stool consistency | ||
| Baseline (± SD) | 5.1 (± 2.2) | 5.3 (± 1.2) |
| Δ 8 week | 0.35 | − 0.1 |
Abdominal pain, bloating, defecation discomfort, stool frequency, and stool consistency after probiotics and placebo administrations for 4 and 8 weeks
*p < 0.05 versus placebo group
Fig. 2.
Comparison of (A) IBS-SSS and (B) IBS-QoL scores between placebo and probiotics groups
Table 2.
Comparison of responders and non-responders by IBS-SSS or IBS-QoL scores differences between 0- and 8-week in probiotics group
| Probiotics (n = 32) | ||
|---|---|---|
| ΔIBS-SSS | ||
| Responders | ≥ 50 points | 21 (66%) |
| Non-responders | < 50 points | 11 (34%) |
| ΔIBS-QoL | ||
| Responders | ≥ 10 points | 19 (59%) |
| Non-responders | < 10 points | 13 (41%) |
Changes of urine metabolites after probiotics treatment
We performed untargeted metabolome analyses and generated principal component analysis (PCA) score plots to find metabolites characterizing different endogenous metabolome of probiotics administered IBS patients. Four and eight weeks’ treatment of probiotics changed metabolome of IBS patients (Supplementary Fig. 1). There are 7 metabolites distinguishing metabolomic characteristics between probiotics and placebo groups: 2-ketobutyric acid, l-kynurenine, 4,6-dihydroxyquinoline, 4-(2-aminophenyl)-2,4-dioxobutanoic acid (DOBA), palmitic acid methyl ester (PAME), cholic acid, and palmitoleoyl ethanolamide (PEA) (Table 3). Among them, urinary levels of PAME and PEA were increased, and those of others were decreased with high significances (p < 0.001) except cholic acid after probiotics treatment for 8 weeks (Supplementary Fig. 2).
Table 3.
Endogenous metabolites discriminating the probiotics treatment group from the placebo group
| Retention time (min) | m/z | Formula | Fold change (probiotics/placebo) | Identity |
|---|---|---|---|---|
| 0.5 | [101.0244]− | C4H6O3 | 0.72 | 2-Ketobutyric acid |
| 1.7 | [207.0775]− | C10H12N2O3 | 0.36 | l-Kynurenine |
| 3.3 | [162.0550]+ | C9H7NO2 | 0.59 | 4,6-Dihydroxyquinoline |
| 3.6 | [206.0459]− | C10H9NO4 | 0.31 | 4-(2-Aminophenyl)-2,4-dioxobutanoic acid (DOBA) |
| 10.2 | [288.2982]+ | C17H34O2 | 1.77 | Palmitic acid methyl ester (PAME) |
| 11.2 | [407.2803]− | C24H40O5 | 0.48 | Cholic acid |
| 12.1 | [320.2647]+ | C18H35NO2 | 2.26 | Palmitoleoyl ethanolamide (PEA) |
Comparison of urine metabolites between responders and non-responders
Patients treated with probiotics were divided into responders and non-responders by IBS-SSS or IBS-QoL scores differences from 0- to 8-week (∆IBS-SSS and ∆IBS-QoL, respectively) (Table 2). According to responders criteria based on IBS-SSS and IBS-QoL (decrease more than 50 points and 10 points, respectively), we compared relative intensity differences of urine metabolites listed in Table 3. In the IBS-SSS responders group, urinary levels of PEA and PAME were increased and, DOBA, l-kynurenine and cholic acid were decreased after probiotics treatment for 8 weeks (Supplementary Fig. 3). In the IBS-QoL responders group, 4,6-dihydroxyquinoline, 2-ketobutyric acid and cholic acid were decreased after probiotics treatment for 8 weeks (Supplementary Fig. 4). Notably, l-kynurenine, 4,6-dihydroxyquinoline, and DOBA are associated with tryptophan metabolism. This metabolism is known for important in inflammatory bowel disease (Knights et al., 2013), and kynurenine pathway plays a key role in the tryptophan metabolism. Indeed, previous researches stated that 4,6-dihydroxyquinoline and DOBA were increased in IBD animal models (Zhang et al., 2012), and in our study, those metabolites were decreased after probiotics treatment in IBS patients. Saturated free fatty acids are known for inducing the expression of cyclooxygenase-2 (Lee et al., 2001), thus palmitic acid could increase inflammation as a member of them. However, this inflammatory effect could be ameliorated by adduction methyl ester to palmitic acid via microbial organisms (Cavigelli et al., 1995), and many studies elucidated that PAME decreases inflammation related interleukins, cytokines and prostaglandins (Cai et al., 2005), and PEA, a kind of N-acylethanolamines, also has been reported to have anti-inflammatory effects (Syed et al., 2012). Therefore, increased PAME and PEA might indicate anti-inflammatory effects of probiotics in IBS patients. Also, decreased cholic acid represents bile acid homeostasis stabilization since there are many studies demonstrated elevated cholic acid in IBD animal model and Crohn’s disease patients (Tagesson et al., 1985; Zhang et al., 2012). And 2-ketobutyric acid, a well-known product of homocysteine degradation, was reduced after probiotics treatment. Given that homocysteine induces cell injuries as a results of inflammation (Schlüssel et al., 1995), reduced 2-ketobutyric acid might attributable to suppressed inflammation through homocysteine and 2-ketobutyric acid pathway.
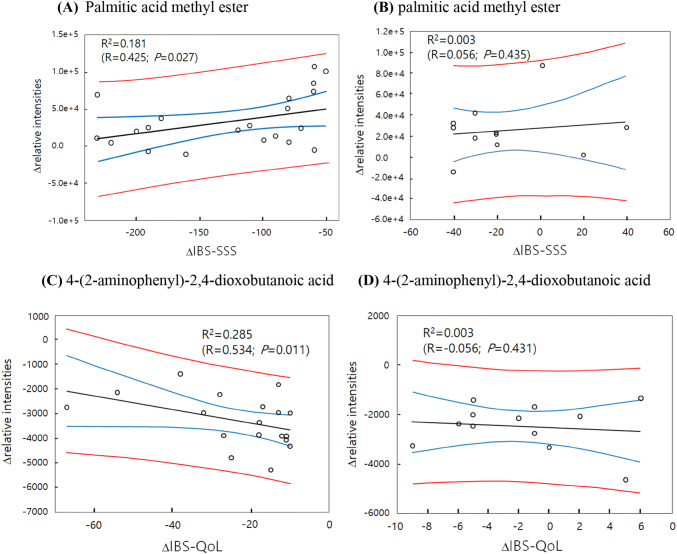
Also, we performed correlation analysis to find metabolites associated with IBS improvements following probiotics treatment in patients. Among distinctive 7 metabolites between probiotics and placebo group, urinary levels of PAME showed significant correlations with IBS-SSS improvement in responders (Fig. 3A, R = 0.425; p = 0.027). And DOBA with IBS-QoL showed improvements in responders (Fig. 3C, R = − 0.534; p = 0.011).
Fig. 3.
Correlation analysis of metabolites and IBS-SSS or IBS-QoL in the probiotics treatment group. Palmitic acid methyl ester and IBS-SSS in responders (A) and in non-responders (B). 4-(2-Aminophenyl)-2,4-dioxobutanoic acid and IBS-QoL in responders (C) and in non-responders (D) excluding outliers. Black line: linear trend; blue line: 95% confidence band; red line: 95% prediction band
However, in non-responders, there was no significnat correlation: R = 0.056; p = 0.435 for PAME, and R = − 0.056; p = 0.431 for DOBA (Fig. 3B, D). Compared with non-responders, IBS-SSS responders showed initially higher PAME levels and IBS-QoL responders showed initially lower DOBA levels (Fig. 3A, C).
As a conclusion, this study demonstrated that probiotics could improve IBS symptoms in IBS-D patients and this might be associated with alteration of metabolomes. Probiotics decreased more IBS-SSS and IBS-QoL than placebo (8%p and 3%p, respectively) and changed levels of urinary metabolites mainly relating to inflammation, therefore we could come up with an idea of probiotics’ anti-inflammation effects (Fig. 2). However, not all patients were responsive to the probiotics treatment. This might be attributed to the heterogeneity of IBS patients and it would be clinically useful if we can predict who would be responsive to probiotics. In this aspect, this study suggest that urinary levels of metabolites can be the candidate biomarkers for this. Especially urinary PAME and DOBA levels showed significant correlation with clinical responsiveness. Moreover, urinary samples are much easier to get than stool samples. Several previous studies have already shown that not only stool samples but also urinary samples can reveal characteristics of intestinal environment quite precisely (Dabur et al., 2017; Marcobal et al., 2013).
However, this study had some limitations. First, even though probiotics group showed higher proportion of patients with adequate symptom relief than the placebo group, there were no significant differences between those of IBS-QoL and IBS-SSS. This can be explained as a placebo effect because the placebo group in this study showed high rate of symptom relief. This remarkably high placebo effect would be one of major obstacles in assessing the efficacy of probiotics in IBS patients. Secondly, the diagnosis of IBS-D relies mostly on the clinical features and there is no objective diagnostic test or validated biomarker for IBS-D diagnosis. Thus, patients with heterogeneous disease entities could be included simply as IBS-D patients. Lastly, even though urine metabolites discriminating probiotics and placebo groups were found, we cannot be certain that it is the cause of IBS symptoms or the results of probiotics treatment. These changes could not guarantee any causal relationships. Therefore, more careful interpretation on these data would be mandatory and further well-designed studies with mechanism experiments in vitro or in vivo are needed to fully characterize metabolic characteristics and the effect of probiotic supplementation on metabolic modulation of IBS. Also, it is necessary to do microbiome related study in the future, because the probiotics treatment can cause major changes in the microbiome, and as a result, in the metabolome.
Nevertheless, the data presented in this study are valuable suggesting that in some IBS patients, probiotics can spur the anti-inflammatory process and this might lead to the symptom relief and these metabolites might be useful to predict who would respond to the probiotics. Furthermore, this study highlights the potential of untargeted metabolic profiling as a possible monitoring method for nutritional or pharmacological interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by research fund from the Clinical Research Institute of the Seoul National University Hospital. This work was also carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01123002)”, Rural Development Administration, Republic of Korea.
Compliance with ethical standards
Conflict of interest
The authors have no financial conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinjoo Kim, Email: jjkim0727@gmail.com.
Kumsun Cho, Email: kscho615@snu.ac.kr.
Bumsik Kim, Email: bumik@yeonsung.ac.kr.
Myeong Soo Park, Email: bifidopark@bifido.com.
Joo-Youn Cho, Email: joocho@snu.ac.kr.
Kyoung Sup Hong, Email: kshong1@empas.com.
References
- Barbara G, Cremon C, Pallotti F, De Giorgio R, Stanghellini V, Corinaldesi R. Postinfectious irritable bowel syndrome. J. Pediatr. Gastroenterol. Nutr. 2009;48:S95–S97. doi: 10.1097/MPG.0b013e3181a15e2e. [DOI] [PubMed] [Google Scholar]
- Barbara G, Cremon C. Serine proteases: new players in diarrhea-predominant irritable bowel syndrome. Gut. 2008;57:1035–1037. doi: 10.1136/gut.2008.150821. [DOI] [PubMed] [Google Scholar]
- Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, Collins SM, Corinaldesi R. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Bertini I, Calalbro A, De Carli V, Luchinat C, Nepi S, Porfirio B, Renzi D, Saccenti E, Tenori L. The metabonomic signature of celiac disease. J. Proteome. Res. 2009;8:170–177. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2011;106:329–336. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- Cai P, Kaphalia BS, Ansari GA. Methyl palmitate: inhibitor of phagocytosis in primary rat Kupffer cells. Toxicology. 2005;210:197–204. doi: 10.1016/j.tox.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Camilleri M. Current and future pharmacological treatments for diarrhea-predominant irritable bowel syndrome. Expert Opin. Pharmacother. 2013;14:1151–1160. doi: 10.1517/14656566.2013.794223. [DOI] [PubMed] [Google Scholar]
- Cavigelli MA, Robertson GP, Klug MJ. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant Soil. 1995;170:99–113. [Google Scholar]
- Chadwick VS, Chen W, Shu D, Paulus B, Bethwaite P, Tie A, Wilson I. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–1783. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- Crowell MD, Shetzline MA, Moses PL, Mawe GM, Talley NJ. Enterochromaffin cells and 5-HT signaling in the pathophysiology of disorders of gastrointestinal function. Curr. Opin. Investig. Drugs. 2004;5:55–60. [PubMed] [Google Scholar]
- Dabur R, Shirolkar A, Mishra V, Yadav BS. Non-invasive qualitative urinary metabolomic profiling discriminates gut microbiota derived metabolites in the moderate and chronic alcoholic cohort. Curr. Pharm. Biotechnol. 2017;18:1175–1189. doi: 10.2174/1389201019666180308093207. [DOI] [PubMed] [Google Scholar]
- Dinan TG, Quigley EM, Ahmed SM, Scully P, O’Brien S, O’Mahony L, O’Mahony S, Shanahan F, Keeling PW. Hypothalamicpituitary-gut axis dysregulation in irritable bowel syndrome: plasma cytokines as a potential biomarker? Gastroenterology. 2006;130:304–311. doi: 10.1053/j.gastro.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Dunne C, Murphy L, Flynn S, Mahony L, O’Halloran S, Feeney M, Morrissey D, Thornton G, Fitzgerald G, Daly C, Kiely B, Quigley EM, O’Sullivan GC, Shanahan F, Collins JK. Probiotics: from myth to reality, Demonstration of functionality in animal models of disease and in human clinical trials. Antonie. Van. Leeuwenhoek. 1999;76:279–292. [PubMed] [Google Scholar]
- Ghoshal UC, Ranjan P. Post-infectious irritable bowel syndrome: the past, the present and the future. J. Gastroenterol. Hepatol. 2011;26:94–101. doi: 10.1111/j.1440-1746.2011.06643.x. [DOI] [PubMed] [Google Scholar]
- Gwee KA, Lu CL, Ghoshal UC. Epidemiology of irritable bowel syndrome in Asia: something old, something new, something borrowed. J. Gastroenterol. Hepatol. 2009;24:1601–1607. doi: 10.1111/j.1440-1746.2009.05984.x. [DOI] [PubMed] [Google Scholar]
- Hammerle CW, Surawicz CM. Updates on treatment of irritable bowel syndrome. World J. Gastroenterol. 2008;14:2639–2649. doi: 10.3748/wjg.14.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A, Naghibi MM, Garcha D. The role of bacteria, probiotics and diet in irritable bowel syndrome. Foods. 2018;7:E13. doi: 10.3390/foods7020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KS, Kang HW, Im JP, Ji GE, Kim SG, Jung HC, Song IS, Kim JS. Effect of probiotics on symptoms in korean adults with irritable bowel syndrome. Gut Liver. 2009;3:101–1077. doi: 10.5009/gnl.2009.3.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Quigley EM, Öhman L, Simrén M, O’Toole PW. The microbiota link to irritable bowel syndrome: an emerging story. Gut Microbes. 2012;3:572–576. doi: 10.4161/gmic.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi D, Troost FJ, Masclee AA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Methods to assess visceral hypersensitivity in irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G141–G154. doi: 10.1152/ajpgi.00060.2012. [DOI] [PubMed] [Google Scholar]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Sohn KH, Rhee SH, Hwang D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J. Biol. Chem. 2001;276:16683–16689. doi: 10.1074/jbc.M011695200. [DOI] [PubMed] [Google Scholar]
- Malagelada JR, Malagelada C. Mechanism-oriented therapy of irritable bowel syndrome. Adv. Ther. 2016;33:877–893. doi: 10.1007/s12325-016-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Protrome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J, O’Mahony L, O’Callaghan L, Sheil B, Vaughan EE, Fitzsimons N, Fitzgibbon J, O’Sullivan GC, Kiely B, Collins JK, Shanahan F. Double-blind, placebo-controlled trial of two probiotics strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–980. doi: 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, Shanahan F. Gut microbiota: mining for therapeutic potential. Clin. Gastroenterol. Hepatol. 2007;5:274–284. doi: 10.1016/j.cgh.2006.12.009. [DOI] [PubMed] [Google Scholar]
- O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, Quigley EM. Lactobacillus and Bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- O’Sullivan M, Clayton N, Breslin NP, Harman I, Bountra C, McLaren A, O’Morain CA. Increased mast cells in the irritable bowel syndrome. Neurogastroenterol. Motil. 2000;12:449–457. doi: 10.1046/j.1365-2982.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- Piche T, Saint-Paul MC, Dainese R, Marine-Barjoan E, Iannelli A, Montoya ML, Peyron JF, Czerucka D, Cherikh F, Filippi J, Tran A, Hébuterne X. Mast cells and cellularity of the colonic mucosa correlated with fatigue and depression in irritable bowel syndrome. Gut. 2008;57:468–473. doi: 10.1136/gut.2007.127068. [DOI] [PubMed] [Google Scholar]
- Quigley EM. Bugs on the brain; brain in the gut-seeking explanations for common gastrointestinal symptoms. Ir. J. Med. Sci. 2012;182:1–6. doi: 10.1007/s11845-012-0865-y. [DOI] [PubMed] [Google Scholar]
- Quigley EM, Craig OF. Irritable bowel syndrome; update on pathophysiology and management. Turk. J. Gastroenterol. 2012;23:313–322. doi: 10.4318/tjg.2012.0551. [DOI] [PubMed] [Google Scholar]
- Schlüssel E, Preibisch G, Pütter S, Elstner EF. Homocysteine-induced oxidative damage: mechanisms and possible roles in neurodegenerative and atherogenic processes. Z. Naturforsch. C. 1995;50:699–707. doi: 10.1515/znc-1995-9-1017. [DOI] [PubMed] [Google Scholar]
- Scully P, McKernan DP, Keohane J, Groeger D, Shanahan F, Dinan TG, Quigley EM. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am. J. Gastroenterol. 2010;105:2235–2243. doi: 10.1038/ajg.2010.159. [DOI] [PubMed] [Google Scholar]
- Spiller RC. Irritable bowel syndrome. Br. Med. Bull. 2004;72:15–29. doi: 10.1093/bmb/ldh039. [DOI] [PubMed] [Google Scholar]
- Spiller RC, Jenkins D, Thornley JP, Hebden JM, Wright T, Skinner M, Neal KR. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in postdysenteric irritable bowel syndrome. Gut. 2000;47:804–811. doi: 10.1136/gut.47.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed SK, Bui HH, Beavers LS, Farb TB, Ficorilli J, Chesterfield AK, Kuo MS, Bokvist K, Barrett DG, Efanov AM. Regulation of GPR119 receptor activity with endocannabinoid-like lipids. Am. J. Physiol. Endocrinol. Metab. 2012;303:E1469–E1478. doi: 10.1152/ajpendo.00269.2012. [DOI] [PubMed] [Google Scholar]
- Tagesson C, Franzén L, Dahl G, Weström B. Lysophosphatidylcholine increases rat ileal permeability to macromolecules. Gut. 1985;26:369–377. doi: 10.1136/gut.26.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley NJ, Butterfield J. Mast cell infiltration and degranulation in colonic mucosa in the irritable bowel syndrome. Am. J. Gastroenterol. 1996;91:1675–1676. [PubMed] [Google Scholar]
- US Department of Health and Human Services (2012), Food and Drug Administration, Center for Drug Evaluation and Research. Guidance for industry irritable bowel syndrome—clinical evaluation of drugs for treatment. 2012.
- Weston AP, Biddle WL, Bhatia PS, Miner PB., Jr Terminal ileal mucosal mast cells in irritable bowel syndrome. Dig. Dis. Sci. 1993;38:1590–1595. doi: 10.1007/BF01303164. [DOI] [PubMed] [Google Scholar]
- William HRT, Holmes E, Khan F, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJ, Teare JP, Jakobovits S, Zeki S, Welsh KI, Taylor-Robinson SD, Orchard TR. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am. J. Gastroenterol. 2009;104:1435–1444. doi: 10.1038/ajg.2009.175. [DOI] [PubMed] [Google Scholar]
- Yu LM, Zhao KJ, Wang SS, Wang X, Lu B. Gas chromatography/mass spectrometry based metabolomic study in a murine model of irritable bowel syndrome. World J. Gastroenterol. 2018;24:894–904. doi: 10.3748/wjg.v24.i8.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi FF, Zhou Y, Leung FP, Tan S, Lin S, Xu H, Jia W, Sung JJ, Cai Z, Bian Z. Metabolite profiling of plasma and urine from rats with TNBS-induced acute colitis using UPLC-ESI-QTOF-MS-based metabonomics—a pilot study. FEBS J. 2012;279:2322–2338. doi: 10.1111/j.1742-4658.2012.08612.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.