Abstract
Accurate and automatic lung nodule segmentation is of prime importance for the lung cancer analysis and its fundamental step in computer-aided diagnosis (CAD) systems. However, various types of nodule and visual similarity with its surrounding chest region make it challenging to develop lung nodule segmentation algorithm. In this paper, we proposed the Deep Deconvolutional Residual Network (DDRN) based approach for the lung nodule segmentation from the CT images. Our approach is based on two key insights. Proposed deep deconvolutional residual network trained end to end and captures the diverse variety of the nodules from the 2D set of the CT images. Summation-based long skip connection from convolutional to deconvolutional part of the network preserves the spatial information lost during the pooling operation and captures the full resolution features. The proposed method is evaluated on the publicly available Lung Image Database Consortium and Image Database Resource Initiative (LIDC/IDRI) dataset. Results indicate that our proposed method can successfully segment nodules and achieve the average Dice scores of 94.97%, and Jaccard index of 88.68%.
Keywords: Computer-aided diagnosis, Lung nodule segmentation, Pulmonary nodule, Juxtapleural nodule
Introduction
Lung cancer is a leading cause of cancer-related deaths and the most commonly detected cancer worldwide both in men and women. According to the American Cancer Society, the 5-year survival rate of lung cancer is 17.8% which is lower than colon 65.4%, breast 90.35%, and prostate 99.6% [29]. The presence of a pulmonary nodule is the possible reason for lung cancer. If these nodules are detected at primary stage then the chances of survival can be increased from 10–15% to 60–80% [14]. The imaging modality like computer tomography(CT) is primarily used for the diagnosis and detection of lung cancer. According to the National Lung Screening Trial (NLST), use of CT over the other radiology techniques decreases the mortality of lung cancer by 20%. But due to advancements in scanner technology, CT scanner produces a large amount of data which makes the analysis and diagnosis of lung cancer very challenging for the radiologist. Detection of the lung nodules in such huge data is very time consuming and it increases their workload. Therefore, to speed up the diagnosis and detection process and to assist the radiologist, computer-aided diagnosis(CAD)/detection(CADx) systems are proposed. Lung nodule segmentation is a fundamental step in the CAD. Recently, many researchers have proposed the lung nodule segmentation algorithm [2, 6–8, 11, 13, 20, 25]. But achieving accurate lung nodule segmentation is very challenging because of the size, location, shape, and texture of pulmonary nodule. Recently, methods based on the intensity [2, 13], region growing [3, 12] have been proposed by many researchers. Also, the methods based on energy optimizer like a level set [10], graph cut [9], etc. are also studied. However, the accuracy and robustness of the lung nodule segmentation are still challenging, especially when pulmonary nodules are similar to the chest region like juxtapleural nodules or in case of morphological operation–based methods where size and shape of the structuring element are very difficult to set due various sizes and shapes of pulmonary nodules.
The algorithms proposed for the segmentation of pulmonary nodules in lung CT images are generally divided into four different categories: methods based on morphological operations, region growing–based method, optimization method, and machine learning–based methods. Many methods based on the morphological operation are proposed for lung nodule segmentation, Kostis et al. [21] used a morphological opening operation to remove the vessels attached to lung nodule along with the connected component. However, segmentation of nodules with a fixed size morphological operator is difficult. Kuhnigk et al. [22] have shown that radius of vessels decreases while vessels evolved along the periphery of the lung. Messay et al. [2] used a rolling ball filter with rule-based analysis for the juxtapleural nodule segmentation. Although these methods are fast and easy to implement, the size of the morphological operator is difficult to set due to the various sizes of the nodules. Small size of morphological template leads to under segmentation for larger sized nodules while the larger size of structuring element leads to over-segmentation of smaller sized nodules. According to Diciotti et al. [13], non-solid nodules are challenging for morphological operations.
In region growing–based methods, a seed point is initialized by a user to obtain the nodule segmentation. Region growing internally adds voxels to nodules set until the predefined convergence criteria is satisfied. This method gives excellent results for the isolated nodules but fails in case of juxtapleural nodule due to the similarity between juxtapleural nodules and surrounding regions. Deshmeshki et al. [12]. have proposed a contrast-based region growing method to segment juxtapleural nodules. Kubota et al. [3] used probability to determine the likelihood of each voxel belonging to nodules based on local intensity values then region growing method was used to separate the nodules from the background. The biggest challenge in region growing method is the convergence criteria which are difficult to set because of the wide variety and irregular shapes of nodules.
In the energy optimization, methods like level set and graph cut are used for the nodule segmentation. In the method proposed by Chan and Vase et al. [10], level set function is used to describe the image and this function is minimized when the segmented contour matches with the lung boundary. Farag et al. [16] used level set with shape prior to hypothesis. Boykov and Kolmogorov et al. [9] used a graph cut method for lung nodule segmentation by framing the problem into a maximum flow optimization task. The energy optimization methods perform well in case of isolated nodules but fail in case of juxtapleural nodules and ground glass nodules.
In machine learning–based methods, lung nodule segmentation is performed by extracting high-level features then these features are classified with the help of different machine learning classifiers. Lu et. al. classified the set of texture and shape features of voxel with the help of conditional random field classifiers. Recently, many researchers have proposed methods [6–8, 11] based on advanced machine learning methods like convolutional neuronal network(CNN). These CNN-based methods efficiently learn various discriminative features in the images. As a result of this unique feature learning ability and multiple layers, CNN has shown promising results in medical image analysis. Ciresan et al. [11] used CNN for neuronal membrane segmentation in electron microscopy. Ronneberger et al. [27] proposed the UNET-based approach for the biomedical image segmentation. Mukherjee et al. [24] proposed a deep learning–based lung nodule segmentation, where information of the location of the object is obtained using deep learning and graph cut method is used to preserve the morphological details of the object. Roy et al. [7]. proposed the synergistic combination of deep learning and level sets for the pulmonary nodule segmentation. Wang et al. [6] proposed a central focused convolutional neural network(CF-CNN) to segment the lung nodules. Although many methods are proposed by the researcher for the lung nodule segmentation, out of these methods, few are semiautomatic while few methods required user interaction. Some methods fail to segment the juxtapleural nodules due to the similarity between the nodule and surrounding chest region. In methods like a level set, automatic initialization of level set function is a challenging task.
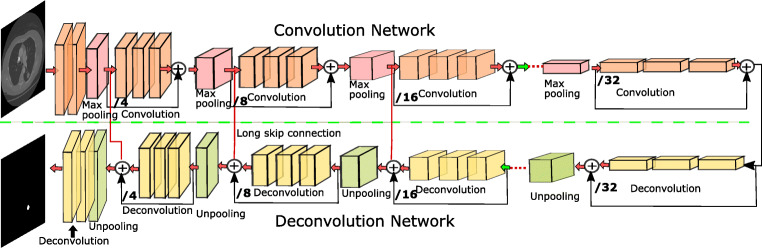
In this paper, we proposed a deep residual deconvolution network–based novel learning architecture with the combination of long and short skip connections for automatic pulmonary nodule segmentation. Our proposed architecture adopts the scheme proposed in [15, 26] and trained end to end to generate an efficient and precise prediction. Performance of our deconvolution network is enhanced by adding residual connections across the residual block. This extended residual connection exchanges the information from convolution and deconvolution part by combining long and short connection. In our proposed deconvolution, architecture information can be smoothly transferred throughout the network which improves the discriminative power of the network and increases the training accuracy. The proposed algorithm considered two types of nodules, i. e., solitary pulmonary nodules and juxtapleural nodules. Accurate segmentation of juxtapleural nodule is difficult because of its similarity with the surrounding region. To solve these challenges in nodule segmentation, we used the deep residual network [18] which segments the various type of nodules with improved accuracy. Our work based on the 2D nodule segmentation since training time required for the 2D image is less than the 3D volumetric data. The schematic diagram of our proposed approach is shown in Fig. 1. Our main contributions can be summarized as follows:
The proposed Deep Deconvolutional Residual Network (DDRN) achieves appealing segmentation accuracy for a diverse variety of nodules especially juxtapleural nodules without any post-processing.
The long and short connection in convolutional and deconvolutional part of the network preserves the spatial information and captures the full resolution features; also, it helps in faster convergence of the deep networks.
Fig. 1.
Architecture of the Deep Residual Deconvolutional Network(DRDN)
Method
Network Architecture
The detailed architecture of our proposed method is illustrated in Fig. 1. Our network is inspired by the convolutional autoencoder [26], which consists of an encoding path, i.e., convolution network and decoding path, and deconvolution network. Convolution network retrieves the features from an input image to transform the input image to a multidimensional feature vector, whereas the deconvolution part of a network acts as a shape generator that gives the object segmentation from the extracted features from the convolution network. Convolution and deconvolution parts consist of different resolution levels to retrieve the features in a different scale. The proposed architecture is constructed by using different types of building blocks. The residual block is shown in Fig. 2. consisting three convolution layers (1 × 1 filter, 3 × 3 filter, 1 × 1 filter) with each convolution layer followed by ReLu as an activation function. Batch normalization layer is added between each pair of convolutional layer and ReLu, this batch norm layer reduces the internal covariance shift and speeds up the training process. Each residual block consists of short residual skip connection. The max-pooling layer located in the convolutional part performs the downsampling of the image for the feature compression. The deconvolution layer present in the deconvolution part of the architecture consists of an unpooling layer. Unpooling operation is exactly opposite to pooling operation. Unpooling is performed to restore original activation as selected during pooling operation. The output of the deconvolution block is a probability map whose size is equal to the size of input image. It gives the probability of each pixel that belongs to the predefined label.
Fig. 2.
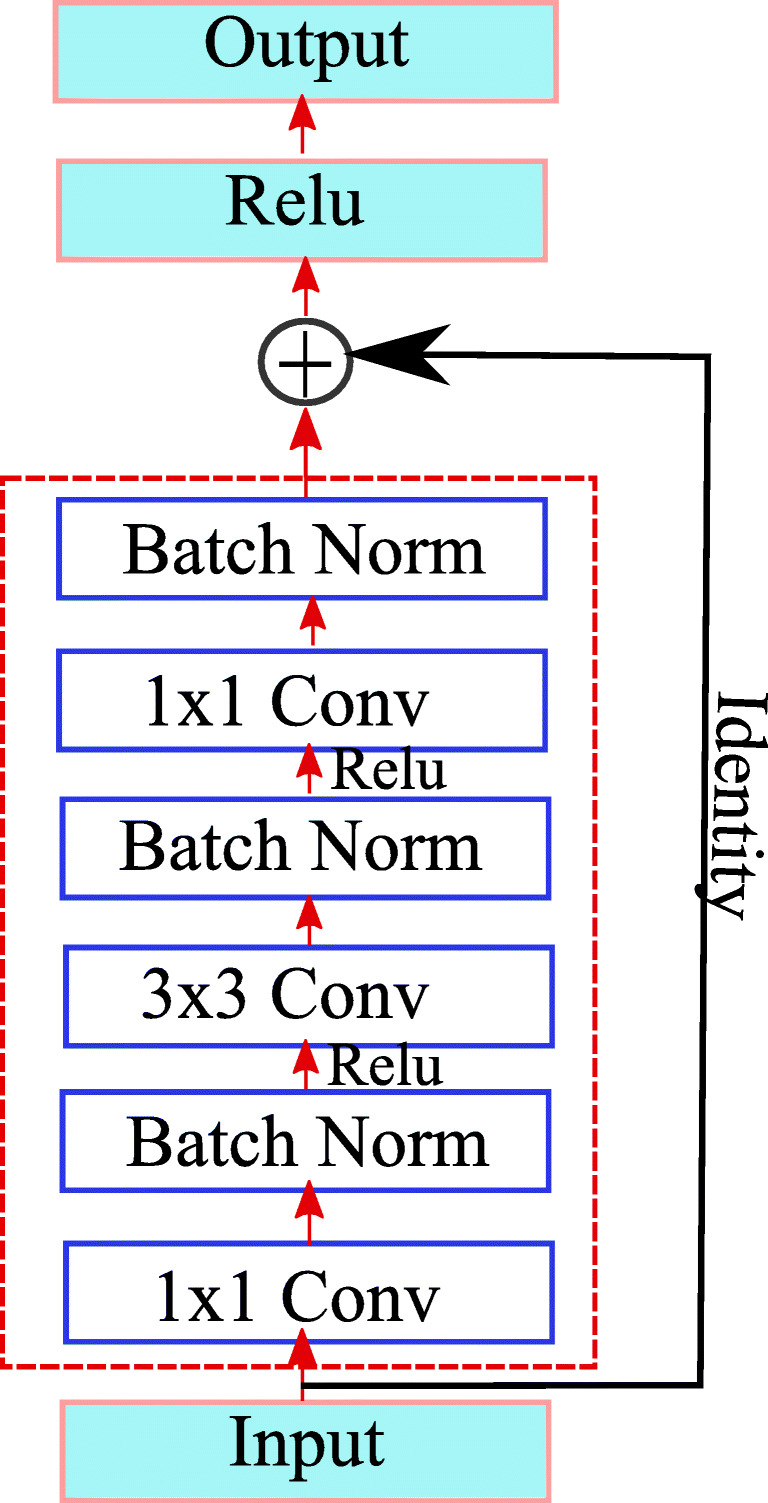
A basic residual network block
We have used Resnet-50 architecture for the lung nodule segmentation. Connection from /4, /8, and /16 scales are element wise added with the output from deconvolution from the respective scales by long skip connection as shown in Fig. 1. Our CT images are of size 512 × 512, so this long skip connection is taken from the 128 × 128, 64 × 64, and 32 × 32 output images. These connections are formed to preserve the pixel level resolution and improve the dense level prediction. Long skip connection from convolutional to deconvolutional parts of the network preserves the spatial information lost during the pooling operation and captures the full resolution features. Also, short skip connection in residual block allows direct connection from the previous layer. The difference between our proposed architecture and UNET is that in UNET-based architecture concatenation of feature maps is performed by using long skip connections only. This concatenation of the feature map is replaced by element-wise addition in our deep residual deconvolutional network. The combination of the nested long and short connections boosts the information flow among the different layers of the network. Overall, our deep residual deconvolutional network with long and short residual connections performs end to end segmentation from input CT image data to the final segmentation result.
Deep Residual Network
For the lung nodule segmentation, we have used the transfer learning with the deep residual network [17]. Multiple stack layers improve the discrimination capabilities of the network but these networks are difficult to train as compared with shallow network. If we go on increasing the number of layers, then it not only affects the performance of the network but also leads to bad convergence of the network. This problem is known as the degradation problem. Deep residual network with the residual block has effectively solved this degradation problem. It has shown the improved accuracy and convergence on tasks like ImageNet [28] and MSCOCO [23].
The deep residual network proposed by He et al. [17] effectively solves the degradation problem because of the residual learning mechanism. It makes the optimization of the network easy using skip connection and activation after addition. Our proposed approach is based on the Resnet-50 architecture which consists of a number of stacked residual units. The residual unit shown in Fig. 2 is used to construct the deep residual network. These residual blocks or units are the building blocks of the Resnet-50 architecture. In the residual unit shown in Fig. 2, signal from one block to another block is directly transferred through skip connection. Each residual unit consists of three convolution layers (1 ×1 filter, 3 × 3 filter, 1 × 1 filter) and each convolution layer is followed by a batch normalization layer. The residual block is shown in Fig. 2 will learn the following function.
| 1 |
where F denotes the residual function, i.e., a stack of two convolutional layers with batch normalization layer (BN) is shown in Fig. 2. xl and xl+ 1 are input and output features of the l th Resnet unit. Wl is a set of weights and biases associated with the I th Resnet unit.
Experimental Results on the Clinical Data
In this section, we describe the results of our proposed algorithm on the clinical dataset.
Datasets
We have used publicly available data from the Lung Image Database Consortium (LIDC) [1]. The LIDC database contains 1018 helical thoracic CT scans of the patients, and these scans are collected from seven academic institutes and eight medical imaging companies in the world. The CT images in this database contain 2610 pulmonary lung nodules, whose diameter is ranging from 2.03 to 38.12 mm. The distance between two successive slices is ranging from 0.45mm to 5.0m. A team of four different radiologists has annotated the nodule as < 3mm and nodule > 3mm. In total, 893 pulmonary nodules are annotated by radiologists. After the series of blinded and unblinded reviews, ground truths of 893 pulmonary nodules are marked and stored in .XML format. Please refer to the LIDC paper [1] for more details. In our proposed method, we have randomly divided the dataset into training, validation, and testing set. Our training set consists of 272, validation set consists of 54, and testing set consists of 89. Our proposed algorithm is trained on the training set and test set is used for determining the accuracy of the algorithm.
Evaluation Parameters
The accuracy of the proposed approach is quantitatively evaluated by using performance measures like the Dice similarity coefficient (DSC) and Jaccard index (JI). These parameters are computed by finding the difference between our automatic segmentation result and manual segmented reference standard. DSC is defined as the ratio between twice the intersection of two binary masks by sum of total number of elements in each set, i.e.,
| 2 |
Let Aauto is binary mask generated by proposed method. Bref is the binary mask of the ground truth or reference standard. DSC always in the range of [0, 1], where 1 shows the 100 % overlap and 0 indicates no overlap between output and reference standard. Jaccard index (JI) is given by the expression
| 3 |
Implementation Details
Our proposed DDRN for pulmonary nodule segmentation is implemented by using publicly available C++ Caffe library [19]. Proposed method train and tested on the system equipped with NVIDIA TITAN X GPU. We train our DRDN using back propagation with a stochastic gradient descent method. The learning rate was set as 0.000001 and models were trained for up to 500000 iterations. We employed a weight decay of 0.0005 and a momentum of 0.9. It took about 8 h to train the network. In the test phase, we used a 2D CT image of size 512 × 512 for testing and obtained probability maps give a prediction for whole CT image.
Comparison with Different Methods
In this paper, experimentation is performed on the LIDC-IDRI dataset [1] which includes various types of nodules with different sizes, shapes, and textures, located at a different locations in the lungs. The proposed method is compared with the state-of-the-art methods published on pulmonary nodule segmentation with their results evaluated on the LIDC database. We observe that proposed methods in literature include level set [10], graph cut [9], and UNET [27], and Mukharji et al. [24], Roy et al. [7], and Wang et al. [6] used DSC as a key performance indicator. For a comprehensive performance analysis, we have measured the DSC and Jaccard index (JI). Our method achieves a DSC of 94.68% and JI of 88.37 ± 12.00% on the LIDC database which is a significantly higher value than the other state-of-the-art methods; also, this method successfully segments all challenging nodule cases including pleural nodules. The qualitative results of nodule segmentation can be seen in Figs. 3 and 4. Also, results of the proposed algorithm for the juxtapleural nodules are shown in Fig. 3. and magnified version is shown in Fig. 5.
Fig. 3.
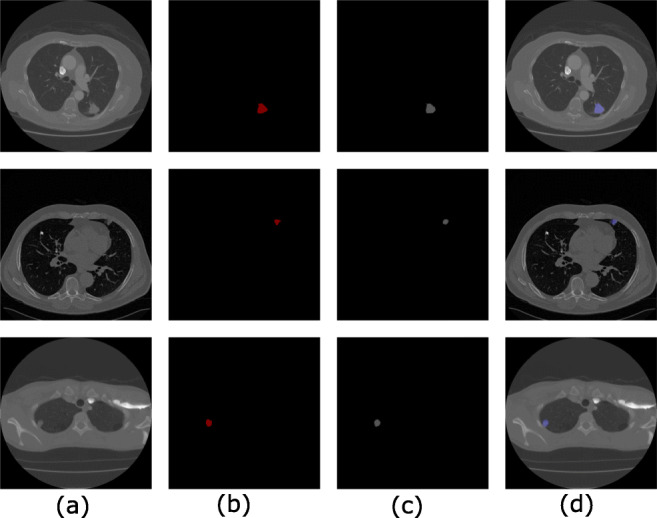
Juxtapleural nodule segmentation results at three different locations. a Original CT image. b. Ground truth. c Output. d Final output
Fig. 4.
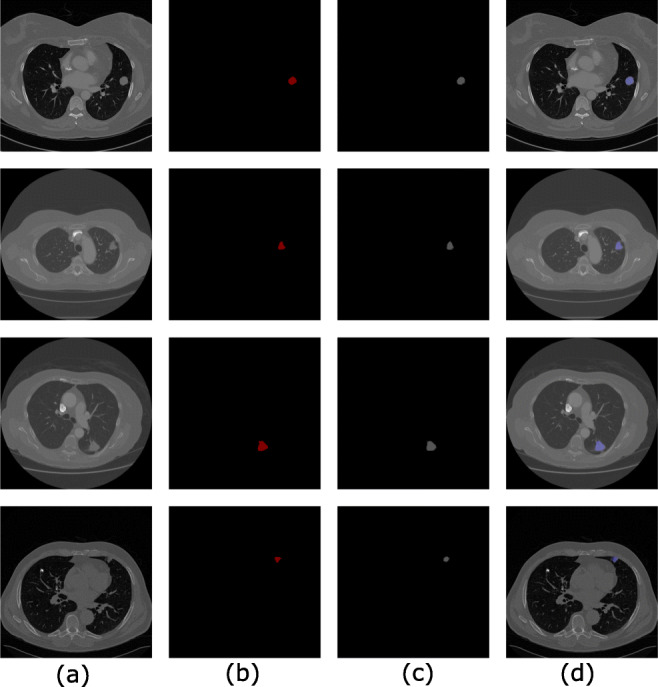
Segmentation results. a Original CT image. b Ground truth. c Output. d Final output
Fig. 5.
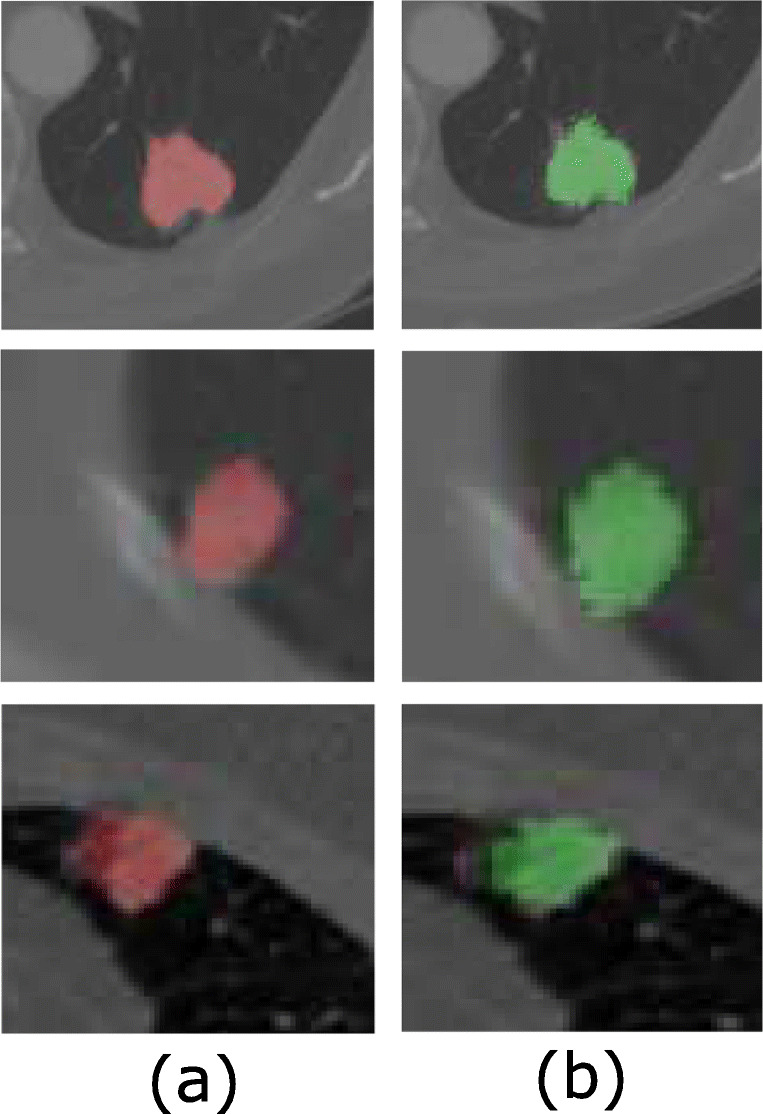
Segmentation results. a Ground truth. b Output
Table 1 shows the comparison of our proposed method with several state-of-the-art methods. Comparison in Table 1 shows that deep learning–based method like UNET [27], Mukharji et al. [24], Roy et al. [7], Wang et al. [6], and our proposed method achieves better performance as compared with traditional methods like level set [10] and graph cut [9]. Level set, graph cut, and UNET-based methods falsely segment the cavity region as a background. Also, it gives the under segmentation since these methods are unable to distinguish background chest region and nodules. Mukharji et al. [24] proposed lung nodule segmentation using graph cut and deeply learned probability map, and achieved good results. Roy et al. [7] applied the combination of deep learning and shape driven level set and achieve compelling performance. Wang et al. [6] proposed a central focused convolutional neural network and achieves good performance on both 2D and 3D CT images.
Table 1.
Comparison of the proposed method with other state-of-the-art methods
Results in Table 2. show that our method performs well in terms of Jaccard index (JI) as compared with other methods. Shakibapur et al. [8] proposed an unsupervised pulmonary nodule segmentation method and it does not need any manual intervention. Methods by Kubota et al. [3], Lassen et al. [4], Messay et al. [5], and Tan et al. [30] require manual interaction whereas our method is a fully automatic pulmonary nodule segmentation method which does not require any manual interaction. From Tables 1 and 2,we can see that proposed method outperforms other methods due to the fact that our residual learning–based approach overcome the degradation problem when training deeper network. So performance is improved by increasing the network depth. Our method is built with 50 layers; hence, large variations of pulmonary nodules are covered by generating more representative features as compared with its deep learning–based methods. Our result analysis confirmed that proposed deeper architecture with residual learning achieves improved accuracy as compared with other deep learning–based method without any post-processing
Table 2.
Average JI for different nodule segmentation proposals on the LIDC-IDRI dataset
| Method | Number of Nodules | Performance(JI) | |
|---|---|---|---|
| Training | Testing | ||
| Messay et al. [2] | – | 68 | 63.00 ± 16.00% |
| Kubota et al. [3] | – | 23 | 69.00 ± 18.00% |
| – | 82 | 59.00 ± 19.00% | |
| Tan et al. [30] | – | 23 | 65.00 |
| Messay et al. [5] | 300 | 66 | 71.70 ± 19.89% |
| – | 77 | 69.23 ± 13.82% | |
| Wang et al. [6] | 350 | 493 | 71.16 ± 12.22% |
| Shakibapour et al. [8] | – | 705 | 70.37 ± 08.00% |
| Proposed method | 272 | 89 | 88.37 ± 12.00% |
Conclusion
In this paper, we developed a novel deep residual deconvolutional network and analyzed its ability in automatically segmenting pulmonary nodules in lung CT images. Our method uses a deeper network like deep residual network for solving the challenging problem of pulmonary nodule segmentation as compared with earlier methods proposed on the lung nodule segmentation. Our end to end deep residual deconvoluional network considered multi-level contextual information and automatically learned nodule sensitive features from the 2D CT images to improve the segmentation performance. With extensive experiments on a well-known LIDC dataset and by comparing with other nodule segmentation methods, it validates the efficacy of our method for pulmonary nodule segmentation. The proposed method can support the research on automated pulmonary nodule segmentation as well as offer a powerful and effective tool for computer-aided diagnosis of lung cancer, where accurate segmentation of pulmonary nodule is essential. Given the accuracy of our algorithm, our automated nodule detection tool has the potential to be a powerful clinical tool which can be implemented in the clinical practice for accurate detection of nodules in lung cancer–screening projects and also for patients undergoing metastatic work-up for pulmonary metastasis. This tool will not only enhance the manual identification of pulmonary nodules but will also reduce the reading time when using assistive tool. In the future, we plan to improve the accuracy of pulmonary nodule segmentation for all other types of nodules.
Acknowledgments
We would like to acknowledge the assistance and support provided by Center of Excellence in Signal and Image Processing, SGGS Institute of Engineering and Technology, Nanded, India.
Funding
This work is funded by Rajiv Gandhi Science and Technology Commission (RGSTC), Government of Maharashtra, India, grant number RGSTC/File-2015/DPP-153/CR-58.
Compliance with Ethical Standards
Conflict of interests
The authors declare that they have no conflict of interest.
Footnotes
Ethical approval
This article does not contain any studies with human participants or animals performed by of the authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ganesh Singadkar, Email: singadkarganesh@sggs.ac.in.
Sanjay Talbar, Email: sntalbar@sggs.ac.in.
References
- 1.Lung Image Database Consortium - Imaging Database Resources Initiative LIDC-IDRI. http://imaging.nci.nih.gov/ncia/login.jsf
- 2.Messay T, Hardie RC, Rogers SK. A new computationally efficient cad system for pulmonary nodule detection in ct imagery. Med Image Anal. 2010;14(3):390–406. doi: 10.1016/j.media.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Kubota T, Jerebko AK, Dewan M, Salganicoff M, Krishnan A. Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med Image Anal. 2011;15(1):133–154. doi: 10.1016/j.media.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Lassen BC, Jacobs C, Kuhnigk JM, van Ginneken B, van Rikxoort EM. Robust semi-automatic segmentation of pulmonary subsolid nodules in chest computed tomography scans. Phys Med Biol. 2015;60(3):1307–1323. doi: 10.1088/0031-9155/60/3/1307. [DOI] [PubMed] [Google Scholar]
- 5.Messay T, Hardie RC, Tuinstra TR. Segmentation of pulmonary nodules in computed tomography using a regression neural network approach and its application to the lung image database consortium and image database resource initiative dataset. Med Image Anal. 2015;22(1):48–62. doi: 10.1016/j.media.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y, Dong D, Gevaert O, Tian J. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy R, Chakraborti T, Chowdhurya AS. A deep learning-shape driven level set synergism for pulmonary nodule segmentation. Pattern Recogn Lett. 2019;123:31–38. [Google Scholar]
- 8.Shakibapour E, Cunha A, Aresta G, Mendonça AM, Campilho A. An unsupervised metaheuristic search approach for segmentation and volume measurement of pulmonary nodules in lung ct scans. Expert Syst Appl. 2019;119:415–428. [Google Scholar]
- 9.Boykov Y, Kolmogorov V. An experimental comparison of min-cut/max- flow algorithms for energy minimization in vision. IEEE Trans Pattern Anal Mach Intell. 2004;26(9):1124–1137. doi: 10.1109/TPAMI.2004.60. [DOI] [PubMed] [Google Scholar]
- 10.Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 11.Ciresan D, Giusti A, Gambardella LM, Schmidhuber J, Bottou L: Deep neural networks segment neuronal membranes in electron microscopy images. In: (Pereira F, Burges CJC, Weinberger KQ, Eds.) Advances in neural information processing systems, vol 25. Curran Associates, Inc., 2012, pp 2843–2851
- 12.Dehmeshki J, Amin H, Valdivieso M, Ye X. Segmentation of pulmonary nodules in thoracic ct scans: a region growing approach. IEEE Trans Med Imaging. 2008;27(4):467–480. doi: 10.1109/TMI.2007.907555. [DOI] [PubMed] [Google Scholar]
- 13.Diciotti S, Picozzi G, Falchini M, Mascalchi M, Villari N, Valli G. 3dd segmentation algorithm of small lung nodules in spiral ct images. Trans Info Tech Biomed. 2008;12:1. doi: 10.1109/TITB.2007.899504. [DOI] [PubMed] [Google Scholar]
- 14.Diederich S, Wormanns D, Semik M, Thomas M, Lenzen H, Roos N, Heindel W. Screening for early lung cancer with low-dose spiral ct: prevalence in 817 asymptomatic smokers 1. Radiology. 2002;222(3):773–781. doi: 10.1148/radiol.2223010490. [DOI] [PubMed] [Google Scholar]
- 15.Fakhry A, Zeng T, Ji S. Residual deconvolutional networks for brain electron microscopy image segmentation. IEEE Trans Med Imaging. 2017;36(2):447–456. doi: 10.1109/TMI.2016.2613019. [DOI] [PubMed] [Google Scholar]
- 16.Farag AA, Munim HEAE, Graham JH, Farag AA. A novel approach for lung nodules segmentation in chest ct using level sets. IEEE Trans Image Process. 2013;22(12):5202–5213. doi: 10.1109/TIP.2013.2282899. [DOI] [PubMed] [Google Scholar]
- 17.He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. arXiv:1512:03385
- 18.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: The IEEE Conference on computer vision and pattern recognition (CVPR)
- 19.Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T: Caffe: convolutional architecture for fast feature embedding.. In: Proceedings of the 22Nd ACM international conference on multimedia, MM ’14. ACM, New York, 2014, pp 675–678
- 20.Kalpathy-Cramer J, Zhao Bg, Goldgof D, Gu Y, Wang X, Yang H, Tan Y, Gillies R, Napel S. A comparison of lung nodule segmentation algorithms: methods and results from a multi-institutional study. J Digit Imaging. 2016;29(4):476–487. doi: 10.1007/s10278-016-9859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostis WJ, Reeves AP, Yankelevitz DF, Henschke CI. Three-dimensional segmentation and growth-rate estimation of small pulmonary nodules in helical ct images. IEEE Trans Med Imaging. 2003;22(10):1259–1274. doi: 10.1109/TMI.2003.817785. [DOI] [PubMed] [Google Scholar]
- 22.Kuhnigk J, Dicken V, Bornemann L, Bakai A, Wormanns D, Krass S, Peitgen H. Morphological segmentation and partial volume analysis for volumetry of solid pulmonary lesions in thoracic ct scans. IEEE Trans Med Imaging. 2006;25(4):417–434. doi: 10.1109/TMI.2006.871547. [DOI] [PubMed] [Google Scholar]
- 23.Lin Tsung-Yi, Maire Michael, Belongie Serge, Hays James, Perona Pietro, Ramanan Deva, Dollár Piotr, Zitnick C. Lawrence. Computer Vision – ECCV 2014. Cham: Springer International Publishing; 2014. Microsoft COCO: Common Objects in Context; pp. 740–755. [Google Scholar]
- 24.Mukherjee S, Huang X, Bhagalia RR: Lung nodule segmentation using deep learned prior based graph cut.. In: 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017), 2017, pp 1205–1208
- 25.Mukhopadhyay Sudipta. A Segmentation Framework of Pulmonary Nodules in Lung CT Images. Journal of Digital Imaging. 2015;29(1):86–103. doi: 10.1007/s10278-015-9801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noh H, Hong S, Han B (2015) Learning deconvolution network for semantic segmentation. In: The IEEE International conference on computer vision (ICCV)
- 27.Ronneberger Olaf, Fischer Philipp, Brox Thomas. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation; pp. 234–241. [Google Scholar]
- 28.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Li F-F. ImageNet large scale visual recognition challenge. Int J Comput Vis (IJCV) 2015;115(3):211–252. [Google Scholar]
- 29.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer J Clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 30.Tan Y, Schwartz LH, Zhao B. Segmentation of lung lesions on ct scans using watershed, active contours, and Markov random field. Med Phys. 2013;40(4):043502. doi: 10.1118/1.4793409. [DOI] [PMC free article] [PubMed] [Google Scholar]