Abstract
Automatic multi-classification of breast cancer histopathological images has remained one of the top-priority research areas in the field of biomedical informatics, due to the great clinical significance of multi-classification in providing diagnosis and prognosis of breast cancer. In this work, two machine learning approaches are thoroughly explored and compared for the task of automatic magnification-dependent multi-classification on a balanced BreakHis dataset for the detection of breast cancer. The first approach is based on handcrafted features which are extracted using Hu moment, color histogram, and Haralick textures. The extracted features are then utilized to train the conventional classifiers, while the second approach is based on transfer learning where the pre-existing networks (VGG16, VGG19, and ResNet50) are utilized as feature extractor and as a baseline model. The results reveal that the use of pre-trained networks as feature extractor exhibited superior performance in contrast to baseline approach and handcrafted approach for all the magnifications. Moreover, it has been observed that the augmentation plays a pivotal role in further enhancing the classification accuracy. In this context, the VGG16 network with linear SVM provides the highest accuracy that is computed in two forms, (a) patch-based accuracies (93.97% for 40×, 92.92% for 100×, 91.23% for 200×, and 91.79% for 400×); (b) patient-based accuracies (93.25% for 40×, 91.87% for 100×, 91.5% for 200×, and 92.31% for 400×) for the classification of magnification-dependent histopathological images. Additionally, “Fibro-adenoma” (benign) and “Mucous Carcinoma” (malignant) classes have been found to be the most complex classes for the entire magnification factors.
Electronic supplementary material
The online version of this article (10.1007/s10278-019-00307-y) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Histopathological images, Multi-classification, Handcrafted features, Transfer learning
Introduction
Breast cancer (BC) has been recognized as one of the most frequent cancers among females. The reports from the World Health Organization (WHO) and the American Cancer Society (ACS) revealed that BC is the second leading cause of death after lung cancer [1, 2]. BC is affecting about 2.1 million women every year and accounted for 627,000 deaths as per the latest report from WHO [1]. Among advanced medical imaging modalities (thermography, magnetic resonance imaging, computed tomography scan, ultrasound, mammography), histopathological modality of imaging is still acknowledged as a paragon of excellence in the diagnosis of cancer [3]. The paucity of the pathologist is a serious barrier in the analysis of histopathological images. There is a single pathologist for every 100,000 and 130,000 people in sub-Saharan Africa and China respectively [4]. A similar scenario has been found in India and the USA. In India, the availability of pathologist is one over 65,000 people, while in the USA, it is 5.7 over 100,000 people [5]. Thus, the dearth of pathologists in the developed and developing countries gives rise to the intense burden on the available pathologist.
Digital pathology is a technology that allows digitization of tissue samples into digital images and tries to imitate the pathologist by introducing computational algorithms for analysis [6, 7]. The computational algorithms in digital pathology are employed to detect fine details and information that cannot easily be determined by a human eye. Despite the emergence of such new technologies, precise diagnosis and treatment is still a challenge. Since the selection of treatment procedure for BC largely depends upon the accurate classification of cancer from histopathological images, but lacking of a skilled and experienced pathologist as well as over-weariness of the pathologist sometimes leads to misclassification which results in misdiagnosis. After the establishment of “precision medicine initiatives” by Barak Obama in 2015, the automated classification of BC from histopathological images has become one of the most active research areas in medicine [8, 9]. The demand to develop an automated classification system for BC to offer a reliable diagnosis motivates us to work in this direction.
In the classification of BC histopathological data, multi-classification is a big challenge. Some well-known factors such as similarity in clinical manifestations, coherency in cancerous cells of different classes, irregularity in color distribution during staining, and variations in appearance of images at different resolutions belonging to the same classes make the multi-classification task very complicated [10, 11]. In the medical domain, in-depth knowledge of disease is always required to provide an accurate diagnosis. Multi-classification assists in providing detailed information about the location of cancer which ultimately helps in making an accurate decision regarding diagnosis. Instead of great clinical significance, very few contributions have been made towards multi-classification by the research community. However, plenty of research studies have been conducted for binary classification. So, in the present work, we have focused on the multi-classification of the BC histopathological data. The transfer learning is employed for automated multi-classification of the histopathological images which utilizes two aspects: (1) as a feature extractor and (2) as a baseline model. A handcrafted approach has also been proposed which includes Hu moment, Haralick texture, and colored histogram for the abovementioned application. In this context, a comparative study has been conducted in which the performance metrics obtained from different classifiers for handcrafted features at various magnification levels (40×, 100×, 200×, and 400×) are compared. In a similar manner, the performances of transfer learning-based systems is compared with their corresponding counterparts for an in-depth analysis. The uniqueness of this study lies in the fact that it provides a single platform to the variety of researchers and readers with the applicability of various machine learning techniques in order to resolve the breast cancer diagnosis problem through a widely available health informatics data in a comparative and conclusive manner.
Relevant Studies
An automated classification system is mainly composed of data acquisition, data pre-processing, feature extraction, classification, and decision-making stage. Feature extraction is the most crucial stage in every kind of classification system because the extracted features significantly influence the system performance. In relevant studies, most of the proposed classification systems utilize “texture” of the images as a feature that are based on various feature descriptors like gray-level co-occurrence matrix (GLCM), local binary pattern (LBP) [12], graph run length matrix (GRLM) [13], histogram of gradient (HOG) [12], local phase quantization (LPQ), scale invariant feature transform (SIFT), and speeded-up robust features (SURF). The SIFT and SURF features were later overperformed by the oriented fast and rotated brief (ORB) method due to its comparable performance, robustness to noise, and less requirement of computational power [14]. The morphological operations were then followed by a wavelet-based covariance descriptor [15], wavelet neural network [16], spoke LBP, and ring LBP [17]. In [18], one class kernel principle component analysis (KPCA) model was proposed in which different features were extracted from each image in the class and one KPCA model was trained for each extracted feature separately. The same operation was repeated for the entire images present in other classes, and the trained KPCA models were then ensemble to make the decision. The KPCA method obtained 92% accuracy on BC histopathological images and provided a new turning point in the research.
The systems proposed in above-discussed studies utilize handcrafted features to make a classification that requires a fairly large domain expertise. The handcrafted feature descriptors extract only inadequate features which make these systems non-adaptable to the new data and incapable to provide discriminate analysis. Eventually, a classifier is always required to make a classification decision that has the ability to handle the acquired feature space. However, the selection of an appropriate classifier is a very complicated task. Logistic regression (LR), QDA [19, 20], support vector machine (SVM) [21], artificial neural network (ANN) [12], naïve bays (NB), k-nearest neighbor (k-NN), linear discriminate analysis (LDA) [13], random forests (RF) [22] are some different classifiers that were used in earlier studies.
In the past few years, many efforts have been made to apply convolutional neural networks (CNNs) on the histopathological imaging modality because CNNs have already shown impressive performance on the classification of natural images [23–28]. CNNs are famous for their ability to learn directly from the data in a hierarchical manner [29, 30]. In [31], a pre-trained CNN (AlexNet) was used for binary classification of BC tissue images (BreakHis dataset [32]) and trained from scratch. To handle the increasing complexity of the pre-trained model, the authors used two different methods for patches generation: (1) random extraction, and (2) sliding window (with 50% overlapping). Further, the results obtained from patch generation methods were combined using three fusion rules (sum, max, and average) and a comparative study was given with [20]. After successful implementation of CNN on medical images, transfer of knowledge from natural images to medical images became a big concern in research due to a substantial difference between the natural and medical images. In [33], a pre-trained network (CaffeNet) and linear regression were employed as a feature extractor and classifier, respectively. The features were extracted from the last three layers only, and performance was evaluated for the possible combination of layers in conjunction with 1, 4, and 16 patches from the input image. In [34], the same trend was followed as in [31], but differences lie in the number of patches extracted from the images (12 non-overlapped patches), the dataset (BACH [35]), and employed CNN. They proposed their own CNN for feature extraction and used SVM as a classifier.
Fine-tuning of the pre-trained network is also one of the key aspects of transfer learning according to which the parameters of pre-trained networks (like learning rate, weights, and the layers of the network) are fine-tuned over the new task. In the study [36], the concept of fine-tuning was incorporated. To fine-tune the Inception and ResNet pre-trained networks, first, they considered only the last layer of the network and then considered the entire layers of the network. The obtained results from their study showed that the ResNet approach performed effectively in cancer-type classification but with some specific settings related to the data augmentation approach. In [37], transfer learning was implemented as a feature extractor for the magnification-independent binary classification using logistic regression as a final classifier. Through their experiments, the authors determined the ability of transfer learning in the classification of histopathological images. In some studies, new models were also proposed to conduct multi-classification of the breast cancer histopathological dataset, for example [38, 39]. In [38], the authors proposed a new deep network named as class structure–based deep convolutional neural network (CSDCNN) which is capable to learn discerning features in a hierarchy from the different level of representations. The authors compared the performance of the proposed model with the LeNet and AlexNet (pre-trained CNNs that were trained from scratch) and obtained the accuracy in the range of 92 to 95% for the different magnification factors in BreakHis dataset at both image and patient level.
In [39], a new classification framework was proposed which consists of three stages: patch-level classification, heat map–based post-processing and refinement model. In order to obtain a normalized dataset, the authors employed two different normalization techniques, (1) Macenko and (2) Vahadane normalization, and the patches were extracted from the normalized dataset. To obtain patch-level classification, Inception V3 was utilized and the obtained predictions were then fused to get image-level predictions by an ensemble framework composed of majority voting (MV), gradient boosting machine (GBM), and LR. Further, to improve the sensitivity of the system over two classes, normal and benign, dual path network (DPN) was employed as feature extractor and extracted features were ensemble using GBM, LR, and SVM. The experimental results of this study [39] showed an accuracy of 87.5%. A research group from Australia utilizes the combination of CNN with different techniques of local features extraction (LBP, contourlet transform (CT), histogram (H), discrete Fourier transform (DFT), and discrete cosine transform (DCT)) and demonstrated that the CNN with CT and H jointly that provides the best results over the BreakHis dataset with 200× magnification factor [40]. Despite the great clinical significance of multi-classification in providing a reliable diagnosis, most of the research works have been carried out only for binary classification [12, 17, 20, 31, 33, 40]. From the abundant studies on BC histopathological images classification, a very small portion is devoted to multi-classification [34, 36, 38, 39].
Contributions
In this paper, experimental work has been performed systematically and leads to the following contributions:
The handcrafted approach is applied to extract the features from the histopathological images and analyzed the classification performance for different conventional machine learning techniques to identify the best classifier in multi-classification of BC with the computed handcrafted features.
Transfer learning is employed for determining the possibility of knowledge transfer from natural images to histopathological images in multi-classification of BC. Transfer learning is applied in two different ways: as a feature extractor and as a baseline model. The utilization of a pre-trained model as feature extractor makes this study different from [36, 38]. In [36], single layer and the entire layers of a pre-trained network were subjected to fine-tuning for a magnification-independent multi-classification. However, in [38] only architecture of the pre-trained models was employed for magnification-dependent multi-classification and compared with the proposed technique.
The present study compares the performance of handcrafted approaches with the transfer learning approach. Additionally, we have demonstrated the influence of balanced and unbalanced training samples on the performance of the classification model, where the data is utilized in raw form without any augmentation.
Handcrafted Features
The properties which are derived by exploiting the information present in the image through a computational algorithm are termed as handcrafted features. The classification system based on handcrafted features is mainly composed of two stages: the three most important attributes color, shape, and texture are accounted to quantify the BC histopathological images for the feature extraction stage and the conventional classifiers such as RF, SVM, and LDA are considered for the classification stage.
Color (Colored Histogram)
In digital pathology, the staining process is performed before the digitization of tissue samples to provide a detailed view of the structures (nuclei, stroma, or cytoplasm) present in the tissue. Hematoxylin and eosin (H&E) is a standard staining protocol in which nuclei were dyed in blue or purple color with hematoxylin and the remaining structures dyed in pink color with eosin [41]. Therefore, color is a significant feature which needs to be considered in the classification of histopathological images. Color histogram is an approach which helps in determining the distribution of colors in an image [42]. Color histogram represents the number of pixels in each bin which have the same color for a fixed list of color ranges. The feature vector is obtained by adding the frequency of occurrence for each color. For instance: a colored image consists of three channels: red, green, and blue. If we considered a histogram of 8 bins for each channel, then the length of the obtained features vector would be 8 × 8 × 8 = 512. Instead of great importance of the color histogram in classification, this feature alone is not adequate to quantify BC histopathological images due to appearance variability. The factors responsible for appearance variability in histopathological images are differences in light sources or detectors employed in the scanner, variations in protocols used for fixation and staining process in different labs, utilization of different reagents, delays in fixation, and discrepancy in staining conditions [7]. Thus, it is necessary to consider other features besides color histogram to design a robust classification system.
Shape (Hu Moment)
In pathology, the shape of cells is another important parameter in determining the nature of cells whether cancerous or normal [43]. Prominent and darker nucleoli, scarcity in the cytoplasm, the chaotic arrangement of chromosomes, abnormal growth of cells, and non-uniform shape and size of the cell are some morphologic characteristics of cancer cells [10]. The Hu moment invariant is a widely used global shape feature descriptor in computer vision [44]. In the present work, Hu moment is employed to extract the features associated with the shape of cells. The speciality of these invariants lies in their ability to identify patterns or shapes independent of size, position, and orientation as well as their capability to learn new patterns [44]. In image processing, geometric moments can be defined as [45–47]:
| 1 |
where Mpq is a moment of a function f(x, y) of order (p + q) and (p, q = 0, 1, 2, …., ∞). xpyq is the basic function for which the moment is defined. Moment invariants comprise nonlinear combinations of rotational invariant central moments. Hu defines seven such moments, derived from the central moment of order three and collectively known as Hu moment [48]. A feature vector of size 7-d is obtained when Hu moment is applied to an image.
Texture (Haralick Textures)
The texture is the property of an image which provides information concerning the surface and appearance of the object present in the image. On the basis of the degree of randomness, the texture can be categorized into two categories regular and stochastic [49]. A regular texture consists of a periodic arrangement of the elementary parts of an object, whereas these elementary parts are organized in a random fashion for a stochastic texture [49].
The histopathological images always come up with a stochastic texture due to a random distribution of cells in the tissue which requires computation of statistical features in order to perform the texture analysis of histopathological images. GLCM is one of the methods to compute these statistical features by considering the spatial relationship of pixels [50]. In 1973, Haralick et al. suggested how to employ the GLCM in the quantification of texture [51]. The authors rendered 14 statistical matrices (angular second moment, contrast, correlation, variance, inverse difference moment, sum average, sum variance, sum entropy, entropy, difference variance, difference entropy, info. measure of correlation 1, info. measure of correlation 2, and maximum correlation coefficient) that were computed from GLCM. However, all features were taken into account except maximum correlation coefficient due to its computational instability [51]. Therefore, a feature vector of size 13-d is obtained when Haralick texture analysis is applied to an image.
Convolutional Neural Networks (CNNs) and Transfer Learning
CNNs are a subtype of the deep neural network, which consists of mainly two parts: convolutional base and classifier. The convolutional base is composed of a pile of convolutional, sub-sampling, and activation layers to extract the features from the data. The convolutional base is further followed by the classifier which is usually composed of dense layers. Finally, a softmax layer is added on the top of the classifier to provide the required results [23, 52]. The sub-sampling layers in CNN reduce the number of computations, learning parameters as well as the problem of overfitting by reducing the spatial size of the network. On the other hand, the activation layers make the network computationally efficient by activating few nodes per time [53]. In addition, the concept of parameter sharing and local connectivity makes CNNs computationally efficient. The stride value, presence or absence of zero-padding, filter size, and number of channels are some important parameters used to define a convolutional layer. Similar to the simple neural network, the gradient descent (GD) algorithm is used to train CNNs by minimizing the loss function. Stochastic gradient descent (SGD) is a variant of the GD approach wherein for every iteration; the cost gradient for one data sample is computed to minimize the loss by updating the weights. If a mini-batch of data samples is used instead of one; the approach is known as mini-batch GD which also helps in the enhancement of training speed. Extensively, cross-entropy loss (CEL) is used when softmax employed as the output layer in the network, given as
| 2 |
where N is the number of classes, yc and pc denote the correct classification and predicted probability for the particular observation, respectively. After the computation of loss, the weights are updated to further minimize the loss and make improvement in the classification.
CNNs exhibit significant performance for the visual data processing [24, 54, 55]. It typically requires high-performance graphical processing units for fast training, as the copious annotated dataset is needed by CNNs to acquire high classification accuracy. However, the requirement of high-performance systems has been solved to an extent with the following techniques like filter/channel pruning [56], kernel sparsity, tensor decomposition [57, 58], and by developing efficient architecture design [26, 59]. But in the medical domain, the collection of a large annotated dataset is very challenging due to an intricate and pricey procedure of data collection from the patient [60]. A lot of medical dataset consists of limited samples due to which the task to train CNNs from scratch become somewhat tedious. To address these challenges, the concept of transfer learning has been employed in which “off-the-shelf” features from a standard pre-existing network (like ResNet, Vgg-16, Alex-Net, LeNet) are used in solving the cross-domain but the related problem. In transfer learning, pre-existing networks can be utilized in three manners, namely, baseline model, feature extractor, and fine-tuning of the pre-trained network.
Baseline Model
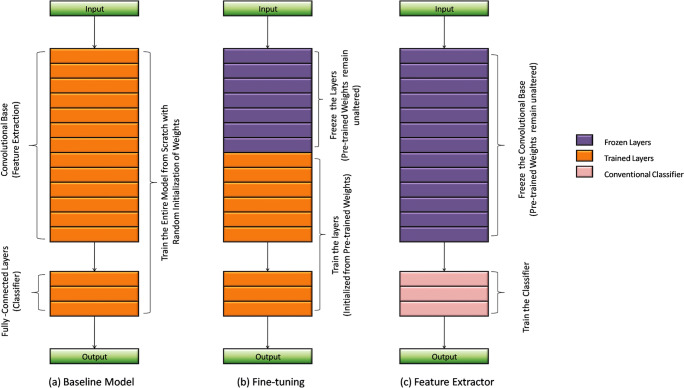
In this approach of transfer learning, only the architecture of the pre-existing model is utilized and the entire model is trained from scratch as per the new dataset by initializing the weights randomly as shown in Fig. 1a. However, the number of nodes in the last dense layer is made equivalent to the number of classes in the targeted application. Here, we have eight classes in the BreakHis dataset to classify, so the last dense layer of the network consists of eight nodes.
Fig. 1.
Strategies to implement transfer learning approach. a Baseline model. b Fine-tuning. c Feature extractor
Fine-Tuning of Pre-Trained Network
Weight initialization is a crucial step in the training of neural networks which determine the performance of the network. In fine-tuning, weights of a pre-trained network are transferred to the targeted network. The rationale behind the application of fine-tuning is the representation learning of CNNs as its early layers capture low-level features that are universal to most of the tasks related to computer vision and the high-level layers extract task-specific features from the samples. Thus, the fine-tuning of the few higher layers on the new application is sometimes adequate for good performance. There are two ways to fine-tune a pre-trained network: (1) layer-wise fine-tuning, and (2) partial training of the network. Layer-wise fine-tuning is an effective approach to test and determine what number of layers should be frozen and what number of layers should be trained? In the layer-wise approach, fine-tuning is initialized with the training of the last layer followed by other layers in a subsequent manner. In partial training of the network, weights of early layers are kept as it is, while the higher layers are undergone for training on the new dataset as illustrated in Fig. 1b.
Feature Extractor
In this form of transfer learning, we are going to use the convolutional base of the pre-trained network in its original form without altering their predefined weights as depicted in Fig. 1c. In contrast, the dense layers of the pre-trained network are replaced with a conventional classifier. The output of the convolutional base is fed directly to the classifiers. The conventional classifier is then trained on the extracted features to make a conclusive outcome. The pre-trained model is used as a fixed medium to extract the most significant features from the samples. The benefits to use the pre-trained network as a feature extractor lie in its ability to provide relevant combinations of features automatically. The feature extraction is a very time-consuming process in hand-designed representation as it demands domain expertise and firm decision on relevant feature selection. The consideration of pre-trained model as a feature generator is a good choice for conventional classifiers.
BreakHis Dataset
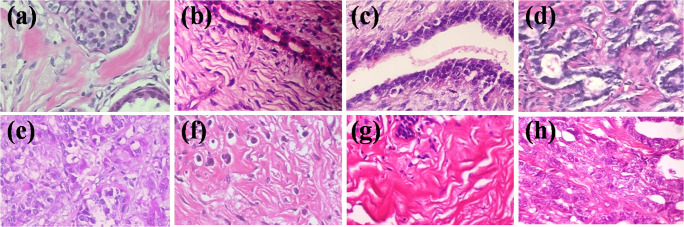
The BreakHis dataset is the largest dataset ever reported for histopathological images related to BC which is publicly available at https://web.inf.ufpr.br/vri/databases/breast-cancer/histopathological-database-breakhis/. The BreakHis dataset is a very challenging and unbalanced dataset that is composed of 7909 images containing two broad categories of BC: benign (B) and malignant (M) [20, 31]. These two categories are further divided into eight sub-categories of BC which includes four sub-classes of the benign class (i.e., adenosis (A), fibro-adenoma (FA), phyllods tumor (PT), and tubular adenoma (TA)) and four sub-classes of the malignant class (i.e., ductal carcinoma (DC), lobular carcinoma (LC), mucinous carcinoma (MC), and papillary carcinoma (PC)) as shown in Fig. 2. Each sub-class of BC consists of colored images of size 700 × 460 with four magnification levels: ×40, ×100, ×200, and ×400. The detailed distribution of the images for each sub-class of the benign and malignant class is summarized in Table 1.
Fig. 2.
Histopathological image samples from BreakHis dataset for eight categories of breast cancer for ×200 magnification factors a adenosis, b fibroadenoma, c phyllods tumor, d tubular adenoma, e ductal carcinoma, f lobular carcinoma, g mucinous carcinoma, and h papillary carcinoma
Table 1.
Distribution of images in BreakHis dataset for eight sub-classes of breast cancer with four magnification factors
| Category of BC/total images | Sub-category of BC | Magnification factor | |||
|---|---|---|---|---|---|
| ×40 | ×100 | ×200 | ×400 | ||
| B/2480 | A | 114 | 113 | 111 | 106 |
| F | 253 | 260 | 264 | 237 | |
| TA | 109 | 121 | 108 | 115 | |
| PT | 149 | 150 | 140 | 130 | |
| M/5429 | DC | 864 | 903 | 896 | 788 |
| LC | 156 | 170 | 163 | 137 | |
| MC | 205 | 222 | 196 | 169 | |
| PC | 145 | 142 | 135 | 138 | |
Methodology and Classification Approaches
The proposed methodology comprises three different approaches that are based on handcrafted feature and transfer learning technique. The performance of the network with the handcrafted feature extraction approach is evaluated by dint of box-plot analysis and plotted on the basis of accuracy acquired by a particular classifier. The performance of classifiers for transfer learning approaches is examined through accuracy, precision, recall, F1 score, receiver operating characteristic (ROC) curve, and the area under that curve (AUC). In each experiment, the labeling of the class is done in a manner that classes 0 to 7 have been assigned as A, DC, FA, LC, MC, PC, PT, and TA, respectively. For each set of experiments, the considered dataset is divided into 90% and 10% for training and testing respectively, and the training set is further divided into 80% for training purpose and 20% for validation purpose. The splitting of the dataset is carried out in a manner that the patients involved in the building of the training set are not included in the testing set. Table 1 shows the distribution of the instances in the sub-classes of BC which is uneven. In order to address this, an advanced version of the sampling technique termed as stratification has been applied on the training set. The stratification is a method of data sampling in which the available data is divided into non-overlapped units known as strata. The instances from each strata are selected in a way that each class is represented with the same frequency and has an equal probability for occurrence. The extra samples are randomly discarded from all the classes and made equal to the number of samples present in the class with least samples to analyze the influence of equal dispersion of instances on the performance of the network. In addition, a set of experiments with unequal distribution of instances (in every class) is also implemented to analyze the effect of the unbalanced dataset on the performance of the utilized networks. The same protocol is applied to all the magnification levels in the dataset.
In handcrafted feature-based approach, the labeled histological images are resized to the dimension of 224 × 224. This dimension is chosen for an unbiased comparison as all the other employed techniques considered the same dimension for an input image. Afterwards, the images are divided into training, validation and testing set as per the devised protocol. The Hu moment, Harallick texture, and colored histogram features were then extracted from the training set to produce semantic feature vectors and used to train some specific traditional classifiers to find out the best classifier. In most of the previous works [34, 61, 62], only SVM is considered to make a prediction on the data, but in the current investigation, RF, LR, LDA, kNN, CART, and NB classifiers are also included along with SVM to determine the performance in multi-classification of BC. In addition, some specific hyper-parameters related to SVM and RF have been considered and varied within a limit to control the frequency of the experiments. The number of trees (hyper-parameter) that were considered for the experiments with RF classifiers ranges from 50 to 4000. In the case of SVM, the results are obtained on three kernels: linear (L), radial basis function (RBF), and sigmoid (S). Besides that, the value of the penalty parameter (C) was also varied from 1 to 5. The remaining parameters for these classifiers were kept at their default settings. Throughout the paper, statistical comparison has been performed for all the classifiers but the results of classifiers that performed poorly have been summarized in the form of tables and figures and provided in the supplementary file.
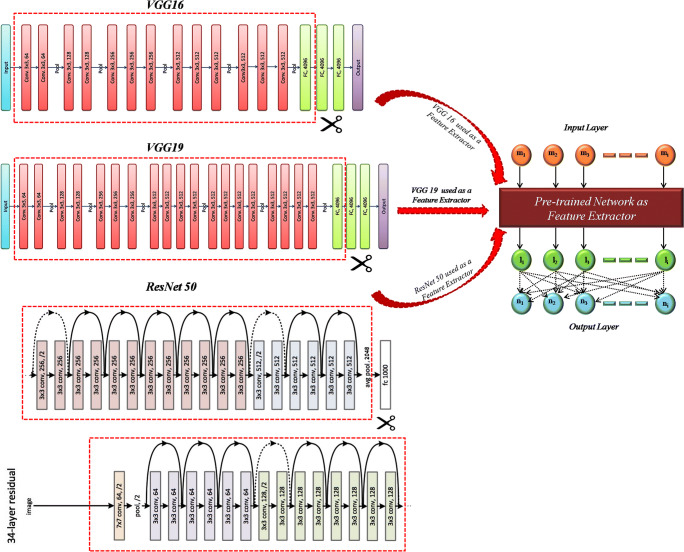
The experiments related to the transfer learning approach have been implemented using two widely used frameworks of deep learning, Keras and Tensorflow. Particularly, VGG16, VGG19, and Res-Net 50 are considered for this study along with the weights that are already trained on the ImageNet dataset. In the feature extractor approach of transfer learning, the last dense layer of the network is substituted by the same conventional classifiers that are used in the handcrafted approach for a coherent comparison. Since the features in case of VGG16 and VGG19 are extracted from the first dense layer of the network, the shape of the obtained feature vector is 1 × 7 × 7 × 512. Similarly, the shape of the feature vector for ResNet50 is 1 × 7 × 7 × 2048 due to extraction of features from the average pooling layer of the network (Fig. 3). Further, the extracted features are collapsed into a one-dimensional array as per the requirement of the conventional classifiers in the Sci-Kit Learn library by appending a flattened layer on the top of the layer from which the features are extracted. In the baseline scheme of transfer learning, the same pre-existing networks are considered without using pre-defined weights, while the last dense layer is replaced by a new dense layer of eight nodes. The weights are initialized randomly to train the network from scratch.
Fig. 3.
The pre-trained networks: VGG16, VGG19 and ResNet50 as feature extractor with conventional classifiers
Moreover, the data augmentation techniques are also utilized to determine the impact of more training data samples on the classification performance of the model. In this context, flipping, translation, scaling, and rotation technique are utilized to expand the training set as the histopathological images are invariant to rotation [63]. Each image in the training set is rotated with 90°, 180°, and 270° to avoid any background noise in the image. Left-right, up-down, and transpose flipping besides translation and central scaling of 90%, 80%, and 70% of the original image is applied to remove the biasness regarding the presence of certain lesions at specific locations in the image. The entire experimental work has been performed on the Intel system with Core i7-7500U CPU @ 2.90 GHz using a 64-bit Windows 10 operating system (OS) and 8 GB onboard memory.
Results
In this section, the results of employed classification approaches are elaborated to provide a systematic and consistent explanation.
Handcrafted Feature-Based Classification
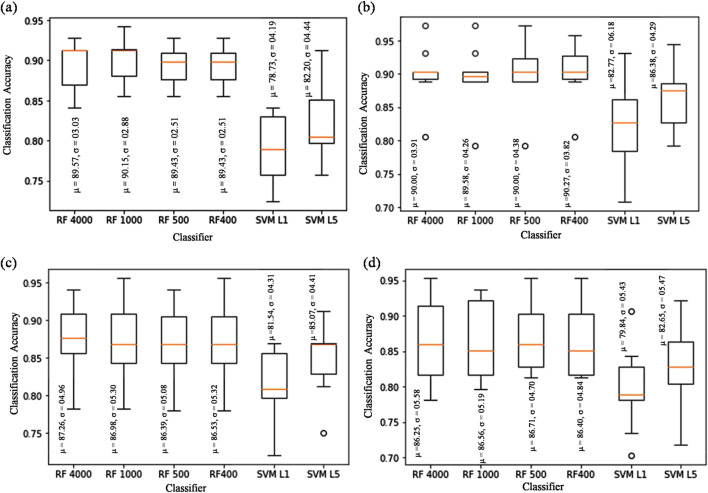
The performance of six best combinations of conventional classifiers with the extracted handcrafted features has been demonstrated. Table 2 shows the classification performance in term of accuracies obtained by RF and SVM under different settings of hyper-parameters (hyper-parameters that varied during the execution of experiments are embraced within parenthesis). Furthermore, a box-plot analysis has been performed to evaluate the performance of the selected combinations of the classifiers at a particular magnification level. Figure 4 shows the box-plots for handcrafted approaches, in which classification accuracy is obtained from the 10-fold cross-validation on the test set for ×40, ×100, ×200, and ×400 magnification levels. In each box-plot, the mean (μ) and standard deviation (σ) are computed for an easy understanding and the plotted whiskers are based on the Tukey method. It has been observed that the RF with 1000 number of trees provided the highest classification accuracy for the ×40 magnification level (median = 90.33%, μ = 90.15, and σ = 02.88) which is comparable to the accuracy obtained with the 50, 200, 400, 500, and 4000 numbers of trees as well as with the SVM for linear kernel and the penalty parameter (C = 5) (Fig. 4a). However, alteration in the value of C from 5 to 1 in SVM with a linear kernel showed a significant fall in overall accuracy which is comparable with that obtained by employing kNN, LR, LDA, LA, and CART (ranges from 71.20 to 79.60%), tabulated in Table S1 (Supporting Information). The unsatisfactory results are obtained from NB and SVM (with RBF and S kernel) classifier, where NB provides an accuracy of 51.80%, while the use of SVM with RBF and S kernel resulted in the lowest accuracy (ranges from 11.28 to 38.50%). From the performance of SVM, it can be anticipated that the features obtained from this handcrafted approach are linearly separable and the increment in C helped in avoiding misclassification of samples. In a similar manner, the analysis of classifiers performance has also been carried out for the other magnification levels. A similar trend has been observed for the classifiers at ×100, ×200, and ×400 magnification factor in which RF showed the best performance. The only difference lies in the number of trees involved in the RF classifier which further depends on the requirement of tuning for hyper-parameters.
Table 2.
Classification accuracy from handcrafted features (HF) and conventional classifiers for a balanced dataset
| Classifiers | Model | Magnification level (accuracy) | |||
|---|---|---|---|---|---|
| ×40 | ×100 | ×200 | ×400 | ||
| RF | HF + RF (400) | 89.50 | 91.35 | 86.78 | 85.15 |
| HF + RF (500) | 89.50 | 91.15 | 86.78 | 86.78 | |
| HF + RF (1000) | 90.33 | 89.88 | 86.78 | 85.15 | |
| HF + RF (4000) | 90.28 | 90.10 | 87.43 | 86.55 | |
| SVM | HF + SVM(L, 1) | 79.15 | 82.77 | 81.88 | 78.90 |
| HF + SVM (L, 5) | 82.20 | 87.58 | 86.48 | 82.95 | |
Italicized number in the table represents the best classifier with the highest accuracy
Fig. 4.
Box-plots of classification accuracy at a ×40, b ×100, c ×200, and d ×400 magnification factor. Outliers are represented by circles
Another important observation noticed from Table S1 (Suppoting Information) is the effect of an unbalanced dataset on the performance of the considered classifiers for the same settings. The unbalanced dataset has a negative impact on the performance of the classifiers which diminishes the performance of classifiers by a substantial amount specifically for ×100, ×200, and ×400. A similar trend was identified for the entire magnification levels in case of unbalanced data. Thus, for the remaining experiments, only balanced data is considered.
Pre-Existing Network as Baseline Model
In order to test the performance of the pre-existing network as a baseline model, the weights of the network are initialized randomly and the network is trained from the scratch. The considered models took around 2 h in full training which further rely on the model capacity. All the results are represented in Tables S2, S3, S4, and S5 (Supporting Information) for the ×40, ×100, ×200, and ×400 magnification factors, respectively. The values for evaluation metrics (i.e., precision, recall and F1 score) are the weighted average of these metrics (computed for each class separately). The accuracies for all the magnification factors are summarized in Table 3 which shows the highest performance of the ResNet50 network for full training. However, the accuracies acquired by the networks for full training are less than 50% which is inapplicable. The main reason behind low performance of theses network is the small size of dataset in respect of the number of classes to classify.
Table 3.
Classification accuracy of pre-existing network as baseline (BL) model at the entire magnification factors (balanced data) & (augmented)
| Classifiers | Model | Magnification factor (accuracy in %) | |||
|---|---|---|---|---|---|
| ×40 | ×100 | ×200 | ×400 | ||
| VGG16 | VGG16 as BL | 15.58 | 11.25 | 9.21 | 8.33 |
| VGG19 | VGG19 as BL | 15.58 | 7.50 | 9.21 | 8.33 |
| ResNet50 | ResNet50 as BL | 38.96 | 33.75 | 39.47 | 33.33 |
| VGG16 | VGG16 as BL | 25.95 | 21.63 | 22.81 | 22.43 |
| VGG19 | VGG19 as BL | 25.95 | 17.46 | 22.81 | 22.43 |
| ResNet50 | ResNet50 as BL | 51.21 | 49.78 | 54.67 | 49.22 |
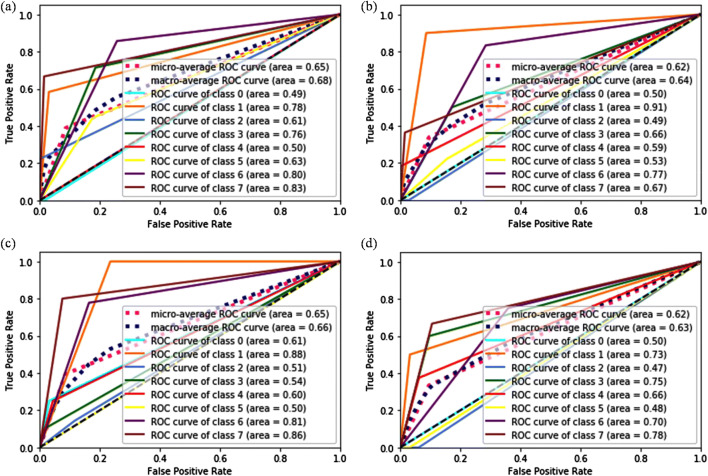
The ROC curve analysis has been executed to further evaluate the classifier’s performance, and the AUC has been computed to ensure the convergence of the network for all classes, illustrated in Figs. S1 and S2 (Supporting Information) and Fig. 5. In case of ResNet50 network, each class is converging with large AUC, whereas the AUC obtained by the VGG16 and VGG19 network lies near 0.50 for each class which confirmed their poor performance. This shows their inability to learn discerning representations from the images when undergone for full training. Despite the highest performance of Resnet50 for full training, the results obtained from this network are also inapplicable for the multi-classification application of BC images. Overfitting of the network is the major reason behind their unsatisfactory performances that arises due to the large capacity of these networks with respect to the size of the dataset. Therefore, the training of a network with deeper architecture from scratch is not a wise option for a dataset of limited size. The performance of the networks can be enhanced by augmenting the data which would help in generating more data sample. Although, the selection of the augmentation method should be in a way that it does not alter the inherent properties of the images. In this context, we have been applied rotation, flipping, scaling and translation technique to enlarge the dataset and performed the experiments under the same settings that have been utilized for the balanced data without augmentation. It has been observed from the results that even after enlarging the dataset; the models are incapable to learn the discerning features from the data and achieved insignificant performance. It implies that the samples in training set are still not sufficient to tune a large numbers of model parameters. Consequently, the model is trying to over fit on the test data.
Fig. 5.
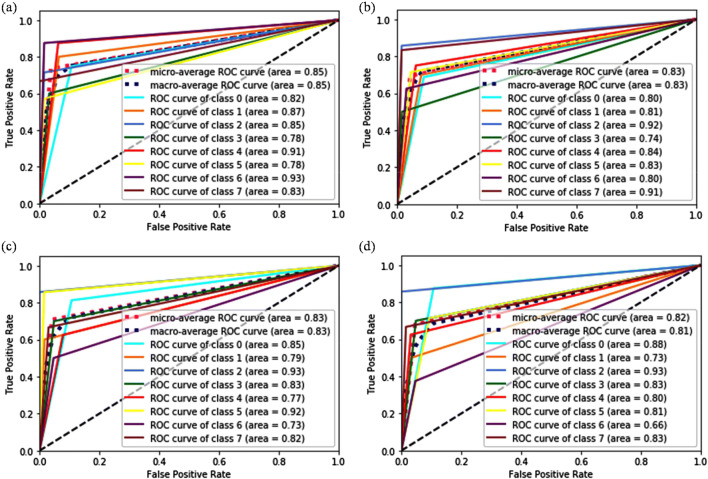
ROC curve analysis obtained for ResNet50 network when used as baseline model at a ×40, b ×100, c ×200, and d ×400 magnification factor
One more observation noticed from Fig. 5 is that instead of utilizing the same network for all the different magnification factors, the distribution of AUC is different. The AUC is minimal for class 0 and class 4 at ×40 resolution. At ×100 resolution, the computed AUC is very less for class 0 and class 2. The scenario is slightly different for ×200 and ×400 resolutions in which AUC for class 2 and class 5 is the least one. It shows that the extraction of useful representation from the images of class 0 and class 2 are very tedious and require special care to improve the overall accuracy of the classifiers.
Pre-trained Network as Feature Extractor
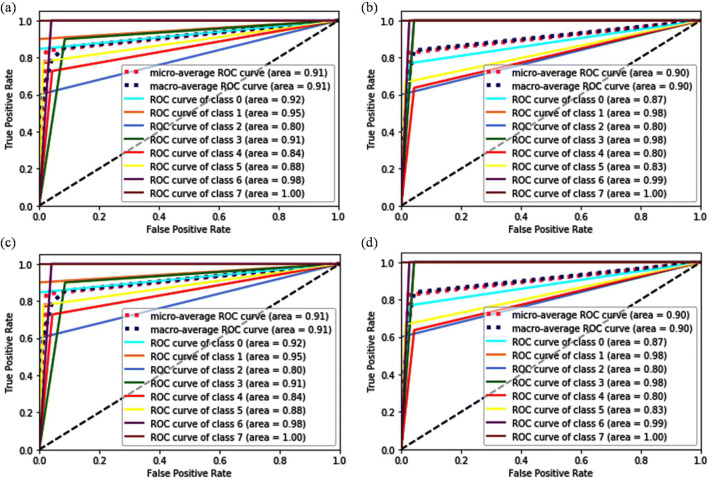
The evaluation of results for this section utilizes the same metrics that considered for the baseline approach. The results of all the classifiers for the ×40 magnification level are given in Table S6 (Supporting Information). By comparing the performances of classifiers, it has been found that the pre-trained network VGG19 provided the maximum accuracy (91.21%) with SVM (L, 1) and SVM (L, 5), followed by VGG16 + LR and VGG16 + SVM (L, 1) classifier (89.61%) (Table 4). Since the value of precision, recall, and F1 score for these three classifiers is almost similar, still the acquired accuracies are different. Figure 6 a–d show the ROC curve analysis for the VGG16 + LR, VGG 16 + SVM(L, 1), VGG19 + SVM(L,1), and VGG19 + SVM(L, 5), respectively. These curves validated that the VGG16 + LR and VGG16 + SVM (L, 1) classifier are less sensitive to all the classes whereas VGG19 + SVM (L, 1) and VGG19 + SVM (L, 5) classifiers are highly sensitive to all the classes and the acquired AUC for the micro and macro-average curve is 0.95. The VGG16 + LR classifier obtained the minimum AUC for class 0 (0.87) and class 2 (0.89), while the VGG16 + SVM (L, 1) classifier acquired very less AUC for class 3 (0.79) which confirm their low sensitivity towards these classes. Hence the poor sensitivity in the case of VGG16 + LR and VGG16 + SVM (L, 1) classifiers becomes the prime reason behind their degraded performance as compared to VGG19 + SVM classifier.
Table 4.
Performance metrics of pre-trained network as feature extractor with conventional classifiers for the ×40 magnification level (balanced dataset)
| Pre-trained network | Model | Performance metrics | |||
|---|---|---|---|---|---|
| Accuracy (%) | Precision | Recall | F1-score | ||
| VGG16 | VGG16 + RF(200) | 81.82 | 0. 85 | 0.82 | 0.81 |
| VGG16 + SVM(L, 1) | 89.61 | 0.90 | 0.88 | 0.88 | |
| VGG16 + SVM(L, 5) | 88.31 | 0.90 | 0.88 | 0.88 | |
| VGG16 + LR(L2) | 89.61 | 0.91 | 0.90 | 0.90 | |
| VGG16 + kNN | 81.82 | 0.84 | 0.82 | 0.82 | |
| VGG16 + LDA | 81.82 | 0.83 | 0.82 | 0.82 | |
| VGG19 | VGG19 + RF(200) | 79.22 | 0.78 | 0.78 | 0.77 |
| VGG19 + SVM(L, 1) | 91.21 | 0.91 | 0.90 | 0.90 | |
| VGG19 + SVM (L, 5) | 91.21 | 0.91 | 0.90 | 0.90 | |
| VGG19 + LR(L2) | 88.31 | 0.89 | 0.88 | 0.88 | |
| VGG19 + kNN | 76.62 | 0.81 | 0.81 | 0.80 | |
| VGG19 + LDA | 77.92 | 0.79 | 0.78 | 0.78 | |
Italicized number in the table represents the best classifier with the highest accuracy
Fig. 6.
ROC curve analysis at 40X magnification factor for a VGG16 + LR(L2), b VGG 16 + SVM(L, 1), c VGG19 + SVM(L, 1), d VGG19 + SVM(L, 5)
On the other hand, the VGG19 + LR classifier has shown a comparable performance to the VGG16 + LR classifier by obtaining an accuracy of 88.31%. The performance dropped by a considerable amount (from ~ 6 to ~18%) when the pre- trained model (VGG16 and VGG19) are used in combination with SVM (RBF), RF, kNN, and LDA. Similarly, NB and CART classifiers have shown an inadequate performance with the marginal accuracies ranging from 51.95 to 63.64%. However, the lowest performance is given by SVM with S kernel. S kernel makes SVM a non-linear classifier, which requires further tuning of kernels’ hyper-parameters for its better working. We have also conducted experiments with the ResNet50 pre-trained network, but observed no significant increment in the classification accuracy due to the least sensitivity of this network towards all the classes, given in Table S6 (Supporting Information).
At ×100 magnification factor, SVM (L, 1) and RF (4000) classifier surpassed all the classifiers by obtaining an accuracy of 89.75% with VGG16 and VGG19, given in Table S7 (Supporting Information). The performance of the linear SVM gets deteriorated by 2.25% when the penalty parameter C changed from 1 to 5 (Table 5). This happened due to the alteration in the decision boundary which divides the training data. On the other hand, the use of SVM with RBF kernel offered an accuracy of ~ 78% which is much less than that obtained from SVM (L, 1), whereas SVM with sigmoid kernel showed the lowest performance at this magnification which describes its incapability in the classification of feature vectors obtained from the pre-trained networks. This happens because of the non-linear behavior of the sigmoid function. In the case of the RF classifier, a direct relationship between the accuracy and the number of trees has been observed. The performance of the RF classifier increases with the increase in the number of trees due to a proportional increment in the number of tunable hyper-parameters. The accuracy of the RF classifier with 50, 200, 400, and 500 numbers of trees lies in the range of 75 to 81.25% which is comparable with the accuracies obtained in case of the LR, LDA, and KNN classifiers, while the CART and NB classifiers gained marginal accuracy. It is noteworthy that RF, SVM, KNN, and LDA classifiers provide the same accuracy when combined with VGG16 and VGG19. Additionally, we have not found any difference in the value of precision, recall, and F1 score (Table S7).
Table 5.
Performance metrics of the re-trained network as feature extractor with conventional classifiers for the ×100 magnification factor (balanced dataset)
| Pre-trained network | Model | Performance metrics | |||
|---|---|---|---|---|---|
| Accuracy (%) | Precision | Recall | F1-score | ||
| VGG16 | VGG16 + RF(500) | 81.25 | 0.86 | 0.81 | 0.82 |
| VGG16 + RF(1000) | 83.75 | 0.87 | 0.84 | 0.84 | |
| VGG16 + RF(4000) | 89.75 | 0.89 | 0.87 | 0.87 | |
| VGG16 + SVM(L, 1) | 89.75 | 0.87 | 0.85 | 0.85 | |
| VGG16 + SVM(L, 5) | 87.50 | 0.87 | 0.85 | 0.85 | |
| VGG16 + LR(L2) | 81.25 | 0.83 | 0.81 | 0.81 | |
| VGG19 | VGG19 + RF(500) | 81.25 | 0.86 | 0.81 | 0.82 |
| VGG19 + RF(1000) | 83.75 | 0.87 | 0.84 | 0.84 | |
| VGG19 + RF(4000) | 89.75 | 0.89 | 0.87 | 0.87 | |
| VGG19 + SVM(L, 1) | 89.75 | 0.87 | 0.85 | 0.85 | |
| VGG19 + SVM(L, 5) | 87.50 | 0.87 | 0.85 | 0.85 | |
| VGG19+ LR(L2) | 81.25 | 0.83 | 0.81 | 0.81 | |
Italicized number in the table represents the best classifier with the highest accuracy.
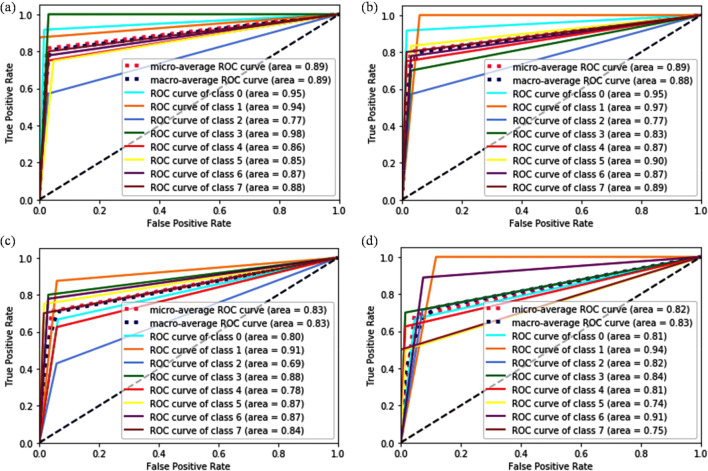
Further, the performance and the sensitivity of classifier towards a particular class has been analyzed through the ROC curve for ×100 magnification factor and shown in Fig. 7. It is observed in Fig. 7 a and c that the RF (4000) classifier with VGG16 and VGG19 shows the same sensitivity. Similarly, the SVM (L, 1) classifier also shows the same sensitivity graph with VGG16 and VGG19, illustrated in Fig. 7 b and d. The pre-trained models with the SVM (L, 1) classifier shows lower sensitivity towards class 0, class 4 and class 5 in comparison with the RF (4000) classifier, while both the classifiers are least sensitive to class 2. The overall performance of RF (4000) classifier is better as compared with SVM (L, 1) for the ×100 magnification factor.
Fig. 7.
ROC curve analysis at ×100 magnification factor for a VGG16 + RF (4000), b VGG16 + SVM (L, 1), c VGG19 + RF (4000), and d VGG19 + SVM (L, 1)
In case of the ×200 magnification level, the VGG16 + LR classifier provided the highest accuracy, followed by VGG16 + LDA classifier, shown in Table 6. The accuracy falls substantially by 9.63% when linear SVM is used with the VGG16 model. Therefore, we changed the kernel from linear to radial but obtained further decrement in the accuracy by 7.9% which determined that the feature vectors obtained from the VGG16 model for the images with ×200 magnification are linearly separable. From Table S8 (Supporting Information), it has been analyzed that the VGG16 + RF classifier rendered insignificant results (ranges from 59.21 to 69.74%) even when the number of trees is increased. However, the RF classifier provides accuracy in the range of 67.11 to 71.05% when VGG19 is used as feature extractor. The VGG19 + KNN classifier provides a maximum accuracy of 72.37% which is also inapplicable. These results confirm the inability of the VGG19 pre-trained model in extracting more discerning representations from the images with ×200 resolution.
Table 6.
Performance metrics of the pre-trained network as a feature extractor with conventional classifiers for the ×200 magnification level (balanced dataset)
| Pre-trained network | Model | Performance metrics | |||
|---|---|---|---|---|---|
| Accuracy (%) | Precision | Recall | F1 score | ||
| VGG16 | VGG16 + SVM(L, 1) | 78.95 | 0.81 | 0.82 | 0.81 |
| VGG16 + SVM(L, 5) | 78.95 | 0.81 | 0.82 | 0.81 | |
| VGG16 + SVM(R, 5) | 71.05 | 0.69 | 0.68 | 0.67 | |
| VGG16 + LR(L2) | 88.58 | 0.89 | 0.89 | 0.89 | |
| VGG16 + kNN | 72.37 | 0.78 | 0.74 | 0.74 | |
| VGG16 + LDA | 87.26 | 0.88 | 0.86 | 0.86 | |
| VGG19 | VGG19 + RF(200) | 71.05 | 0.73 | 0.71 | 0.71 |
| VGG19 + RF(400) | 71.05 | 0.72 | 0.71 | 0.71 | |
| VGG19 + RF(500) | 69.74 | 0.74 | 0.72 | 0.73 | |
| VGG19 + RF(1000) | 69.74 | 0.71 | 0.70 | 0.70 | |
| VGG19 + RF(4000) | 69.74 | 0.69 | 0.68 | 0.68 | |
| VGG19 + KNN | 72.37 | 0.78 | 0.74 | 0.74 | |
Italicized number in the table represents the best classifier with the highest accuracy
Figure 8 shows the ROC curve for analyzing the sensitivity of the best classifiers for the eight classes of BC in case of ×200 resolution. The maximum AUC for the macro-average ROC curve is covered by the VGG16 + LR classifier (0.89) which is the major reason behind the best performance of this classifier. The VGG19 + RF classifier provides a very low AUC for class 2 (0.69) and class 4 (0.78), while VGG19 + KNN classifier provides the lowest AUC for class 5 (0.74) and class 7 (0.75) in comparison with the other three classifiers which degrades their performance by a significant amount. The images belonging to class 2 are more complex at ×200 magnification level as the AUC obtained for class 2 is very low (Fig. 8). It is hard to extract the important features from the images of class 2 (F). The ResNet 50 network also fails to learn discriminating features from the images, and the worst performance has been noticed in the classification of BC histopathology images. The results are tabulated in Table S8 (Supporting information).
Fig. 8.
ROC curve analysis at ×200 magnification factor for a VGG16 + LR(L2), b VGG16 + LDA, c VGG19 + RF(400), and d VGG19 + KNN
The comparison of classifiers’ performances for multi-classification of images with ×400 magnification level is presented in Tables 7 and S9 (Supporting Information). It has been found that the VGG16 + SVM with linear kernel provide the highest accuracy of 80.00%. The accuracies obtained in the case of KNN, LDA, RF, radial SVM and LR classifiers lie in the range of 62.50 to 69.44% (Table S9), which are not much considerable for the multi-classification application. On the other hand, the accuracies obtained by all the classifiers with the ResNet50 network are less than 50%. The large capacity of the ResNet50 network is the rationale behind its worst performance due to which degrees of freedom involved in the parameters increased, consequently leading to over fitting.
Table 7.
Performance metrics of pre-trained network as feature extractor with conventional classifiers for the ×400 magnification level (balanced dataset)
| Pre-trained network | Model | Performance metrics | |||
|---|---|---|---|---|---|
| Accuracy (%) | Precision | Recall | F1-score | ||
| VGG16 | VGG16 + RF (400) | 69.44 | 0.70 | 0.69 | 0.69 |
| VGG16 + RF (500) | 69.44 | 0.70 | 0.69 | 0.69 | |
| VGG16 + SVM(L, 1) | 80.00 | 0.80 | 0.79 | 0.79 | |
| VGG16 + SVM (L, 5) | 80.00 | 0.80 | 0.79 | 0.79 | |
| VGG16 + LR(L2) | 69.44 | 0.71 | 0.69 | 0.69 | |
| VGG16 + kNN | 68.06 | 0.75 | 0.72 | 0.72 | |
| VGG19 | VGG19 + RF (1000) | 63.89 | 0.65 | 0.64 | 0.64 |
| VGG19 + RF (4000) | 63.89 | 0.65 | 0.64 | 0.64 | |
| VGG19 + SVM(L, 1) | 69.44 | 0.72 | 0.71 | 0.71 | |
| VGG19 + SVM (L, 5) | 66.67 | 0.72 | 0.71 | 0.71 | |
| VGG19 + LR(L2) | 68.06 | 0.69 | 0.68 | 0.68 | |
| VGG19 + kNN | 63.89 | 0.66 | 0.62 | 0.63 | |
Italicized number in the table represents the best classifier with the highest accuracy
The ROC curve is shown in Fig. 9 for the best classifiers in case of ×400 magnification in which the maximum AUC is covered by the VGG16 + SVM with a linear kernel. It is further noticed that the AUC achieved by VGG16 + SVM for class 3 (0.78) and class 5 (0.78) is the lowest one as compared with other classes. The VGG16 + LR classifier provided the least AUC for class 3 (0.74) only. However, VGG19 + SVM and VGG19 + LR classifier has acquired the minimum AUC for class 6 (0.73 and 0.66, respectively). Therefore, the classification of BC histopathological images for class 3, class 5, and class 6 becomes more complicated at ×400 magnification level due to major involvement of small-grained appearances in the images at higher magnification.
Fig. 9.
ROC curve analysis at ×400 magnification factor for a VGG16 + SVM (L, 1 and 5), b VGG16 + LR(L2), c VGG19 + SVM(L, 1), and d VGG19 + LR(L2)
Pre-Trained Network as Feature Extractor with Augmented Dataset
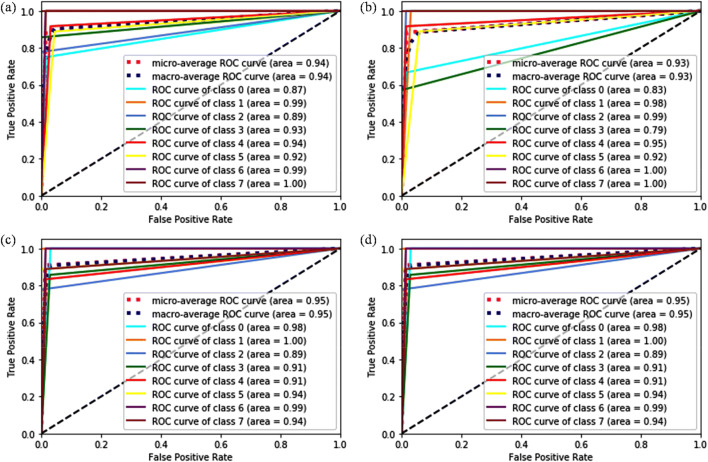
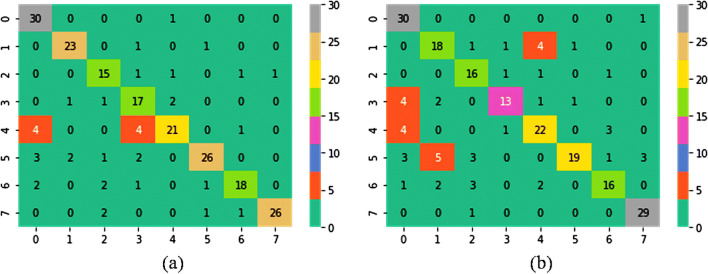
Following the devised topology for the use of pre-trained networks as feature extractor, the highest obtained accuracies are 91.21%, 89.75%, 88.58%, and 80.00% for images with magnification factors ×40, ×100, ×200, and ×400, respectively. As a matter of fact, the classification of histopathological images is a very complex problem and requires proper tuning of parameters to map an image correctly into a label. Therefore, an adequate amount of image samples is required to tune the parameters appropriately in order to get good performance through the classification model. By applying rotation, translation, scaling, and flipping technique, a tremendous increment in the classification accuracy has been observed for all magnification factors. The performance metrics for the best possible combinations at each magnification factor is presented in Table 8. The VGG16 pre-trained model with linear SVM surpassed all the classifiers and helped in achieving the highest accuracies at all magnification levels. This is due to sufficient tuning of model parameters after employing the data augmentation techniques. To further demonstrate the performance of outstanding CNN as feature extractor (VGG16 + SVM (L, 1)) in multi-classification of BreakHis dataset for BC detection, the patient-based accuracy [31] is also evaluated, i.e., 93.25%, 91.87%, 91.5%, and 92.31% for ×40, ×100, ×200, and ×400, respectively. Figures 10 and 11 demonstrate the ROC curves and confusion matrix to validate the performance of the best classification model. On the other hand, VGG19 with linear SVM provides a comparable performance to the best classifier (VGG16 with linear SVM) for all the magnification levels except ×400. The major rationale behind the low performance of VGG19 + SVM (L, 1) lies in their inability to represent intended output for the given input. The extracted features from augmented dataset create confusion for the classification model due to their similar clinical expressions. In particular, Fig. 12 a and b shows the confusion matrix of the VGG19 + SVM (L, 1) classifier for the balanced data at ×400 magnification without considering augmentation and with augmentation, respectively. The numbers of samples which have been classified correctly in balanced dataset are 23, 17, 26, and 18 for ductal carcinoma, lobular carcinoma, papillary carcinoma, and phyllods tumor, respectively. However, after enlarging the dataset by employing augmentation technique, the number of correctly classified samples dropped to 18, 13, 19, and 16. Four samples of ductal carcinoma are misclassified as mucinous carcinoma and one sample as fibro-adenoma. Similarly, four samples of lobular carcinoma are misclassified as adenoma after data augmentation and lower the performance of VGG19 + SVM (L, 1) classifier.
Table 8.
Performance metrics of pre-trained network as feature extractor with conventional classifiers for augmented dataset
| Model | ×40 | ×100 | ×200 | ×400 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc (%) | Pre | Rec | F1 | Acc (%) | Pre | Rec | F1 | Acc (%) | Pre | Rec | F1 | Acc (%) | Pre | Rec | F1 | |
| VGG16 + SVM(L,1) | 93.97 | 0.94 | 0.93 | 0.94 | 92.92 | 0.92 | 0.91 | 0.91 | 91.23 | 0.92 | 0.92 | 0.92 | 91.79 | 0.92 | 0.91 | 0.91 |
| VGG16 + SVM (L,5) | 93.07 | 0.93 | 0.93 | 0.93 | 92.92 | 0.93 | 0.91 | 0.91 | 91.23 | 0.92 | 0.92 | 0.92 | 91.79 | 0.92 | 0.91 | 0.91 |
| VGG16 + LR(L2) | 91.34 | 0.92 | 0.91 | 0.91 | 91.25 | 0.92 | 0.91 | 0.91 | 86.84 | 0.84 | 0.82 | 0.82 | 85.51 | 0.86 | 0.86 | 0.86 |
| VGG19 + SVM(L,1) | 92.64 | 0.92 | 0.92 | 0.92 | 91.25 | 0.91 | 0.91 | 0.91 | 89.42 | 0.90 | 0.89 | 0.89 | 84.11 | 0.83 | 0.82 | 0.82 |
| VGG19 + SVM (L,5) | 92.21 | 0.92 | 0.92 | 0.92 | 91.67 | 0.91 | 0.91 | 0.91 | 89.42 | 0.90 | 0.89 | 0.89 | 83.64 | 0.83 | 0.82 | 0.82 |
| VGG19 + LR(L2) | 88.74 | 0.89 | 0.89 | 0.89 | 90.00 | 0.90 | 0.90 | 0.90 | 83.87 | 0.84 | 0.83 | 0.83 | 80.84 | 0.82 | 0.81 | 0.81 |
Italicized number in the table represents the best classifier with the highest accuracy
Fig. 10.
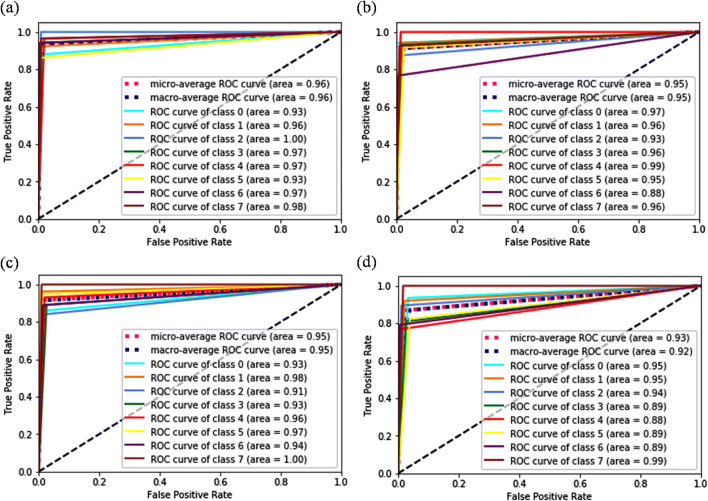
ROC curve analysis of VGG16 + SVM (L, 1) classifier applied to augmented data at a ×40, b ×100, c ×200, and d ×400
Fig. 11.
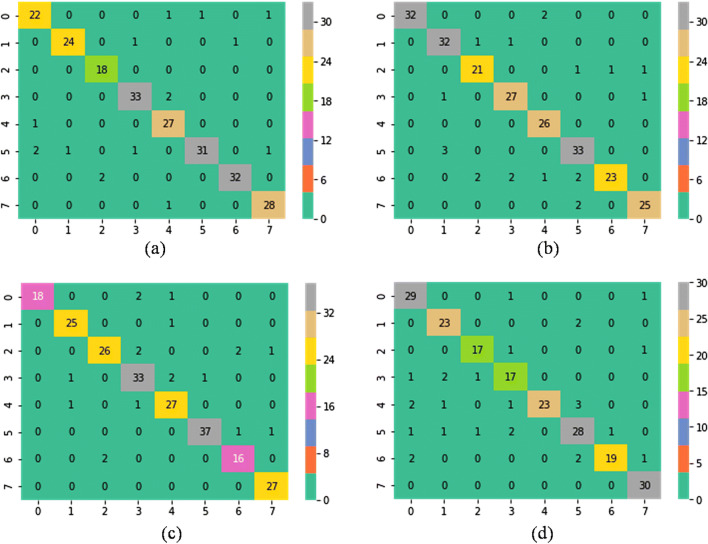
Confusion matrixes of VGG16 + SVM (L, 1) classifier for augmented data at a ×40, b ×100, c ×200 and d ×400
Fig. 12.
Confusion matrixes for VGG19 + SVM (L, 1) classifier at ×400 magnification: a balanced data without augmentation, b augmented data
Discussion
This study emphasizes the conventional machine learning and transfer learning approach for multi-classification of BC histopathological images. In “Results”, the performances of proposed approaches are evaluated on the BreakHis dataset and a noticeable improvement in accuracy figure has been observed. In the case of the handcrafted approach, the RF classifier has shown remarkable performance for the entire magnification scales (accuracy of 90.28% for ×40, 90.10% for ×100, 87.43% for ×200, and ×86.55% for ×400). It is well known that the RF classifier is inherently suited for multi-classification problem and require very less tuning of hyper-parameters to provide robust performance. It is reported for the first time that a handcrafted feature–based approach achieved accurate and reliable performance for such a challenging dataset of histopathological images. Even for a higher magnification, the discriminating ability of our handcrafted approach is better than other existing conventional models, tabularized in Table 9.
Table 9.
Comparison of existing handcrafted approaches and the present handcrafted approach for the entire magnification level
| Model | Features | Magnification Factor | Accuracy (%) | Ref. |
|---|---|---|---|---|
| BOW + SVM | DSIFT |
×40 ×100 ×200 ×400 |
18.77 17.28 20.16 17.49 |
[64] |
| SURF |
49.65 47.00 38.84 29.50 |
|||
| LLC + SVM with SPL(0) | DSIFT |
×40 ×100 ×200 ×400 |
48.46 49.44 43.97 32.60 |
[64] |
| SURF |
55.80 54.24 40.83 37.20 |
|||
| LLC + SVM with SPL(1) | DSIFT |
×40 ×100 ×200 ×400 |
47.44 44.32 44.46 32.86 |
[64] |
| SURF |
54.61 53.92 48.10 38.12 |
|||
| LLC + SVM with SPL(2) | DSIFT |
×40 ×100 ×200 ×400 |
44.54 51.68 44.30 35.54 |
[64] |
| SURF |
53.75 44.30 45.30 40.88 |
|||
| BOW + CNN | DSIFT |
×40 ×100 ×200 ×400 |
41.80 38.56 49.75 38.67 |
[64] |
| SURF |
53.07 60.80 70.00 51.01 |
|||
| LLC + CNN | DSIFT |
×40 ×100 ×200 ×400 |
60.58 57.44 70.00 46.96 |
[64] |
| SURF |
80.37 63.84 74.54 54.70 |
|||
| Fractal dimension + SVM | Fractal dimension | ×40 | 55.60 | [61] |
| (Hu moment + colored histogram + Haralick texture) + RF (4000) | Hu moment, Colored histogram, and Haralick texture |
×40 ×100 ×200 ×400 |
90.28 90.10 87.43 86.55 |
Present Work |
| (Hu moment + colored histogram + Haralick texture) + SVM (linear kernel and C = 5) | Hu moment, colored histogram, and Haralick texture |
×40 ×100 ×200 ×400 |
82.20 87.58 86.48 82.95 |
Bardou et al. performed multi-classification using DSIFT and SURF features separately [64]. The features were encoded with a coding model named as bag of words (BOW) and classified using SVM. The reported accuracy with the BOW+SVM model for DSIFT features at ×40, ×100, ×200, and ×400 is 18.77%, 17.28%, 20.16%, and 17.49%, respectively. Concurrently, the accuracies obtained for SURF features are 49.65%, 47.00%, 38.84%, and 29.50% for the same order of magnification factors. Further, locality constrained linear coding (LLC) was used as a coding model besides spatial pyramid matching with 3 different levels to enhance the performance [64]. The accuracies acquired by LLC + SVM model for DSIFT features are lying in the range of 32.60 to 49.44% at level 0, 32.86 to 47.44% at level 1, and 35.54 to 51.68% at level 2, for the different magnification factors. However, the range of accuracy obtained for SURF features using the LLC + SVM model at levels 0, 1, and 2 for the different magnification factors are 37.20 to 55.80%, 38.12 to 54.61%, and 40.88 to 53.75%, respectively. The authors succeeded in increasing the accuracy at all the magnification levels using the LLC + SVM model, but still the obtained accuracies are insufficient. The marginal performance of these models confirmed their inadequate performance for the multi-classification application.
Further, the authors introduced an alternative approach in which a set of handcrafted features is classified using CNN instead of a conventional classifier. The topology used by the authors for this work consisted of three dense layers in which the first two were followed by the ReLU activation layer with a 50% dropout. To train the network, the weights were initialized by Gaussian distribution and the hyper-parameters like weight decay, learning rate, batch size and iterations were set to 0.1, 0.001, 32 and 20,000, respectively. Although the proposed approach (BOW + CNN and LLC + CNN) produced better results, the performance is still less than our handcrafted approach employed for multi-classification. On the other hand, Chan et al. extracted a set of features using the fractal dimension technique and fed as input to the SVM classifier [61]. They reported an accuracy of 55.6% for ×40 magnification only. Our handcrafted approach outperforms all the handcrafted approaches proposed in [61, 64]. Although the performance of the handcrafted approach is outstanding, it requires deep knowledge about the morphology of cancerous cells and staining protocol used in histopathology procedures in advance. Therefore, deep learning techniques are a better alternative to these requirements as the deep learning models are capable to extract the useful representations directly and automatically from the data. The deep learning models require copious of data for efficient learning and consume a lot of time in the training process.
In order to overcome the problem of less data availability and large training time, we have examined the ability of “transfer learning” technique on histopathological images of BC for multi-classification using three pre-existing models, namely, VGG16, VGG19, and ResNet50. The utilization of pre-existing networks as baseline model showed very poor performance and provides accuracy in the range of 7.50 to 39.47% for the different magnifications. There are several reasons behind the worst performance of baseline models: (a) overfitting and (b) difficulty in extracting the distinctive features from the images belonging to different classes due to similar clinical expressions in class A, F, and LC at ×40, class F, MC, and PC at ×100 and ×200, and class DC, MC, and PT at ×400, (c) limited instances per classes, and (d) a large number of classes to classify. The pre-trained network as feature generator showed an accuracy of 91.21% for ×40, 89.75% for ×100, 88.58% for ×200, and 80.00% for ×400 using VGG19 + SVM (L, 1), VGG16 + SVM (L, 1), and VGG16 + LR (L2) classifiers. These three classifiers performed in a different manner for all four magnification levels according to which VGG19 + SVM (L, 1) gives the best performance for ×40 and ×100, VGG16 + SVM (L, 1) for ×100 and ×400, while VGG16 + LR (L2) for ×200 when applied to the balanced data without augmentation. The classifiers are not equally sensitive to all the classes for the entire magnification levels which can be easily validated by computing the AUC for each classifier. This happens because the trainable parameters of the classifiers are not tuned properly in the lack of sufficient data samples. However, after augmenting the data, a considerable improvement in the classification accuracy of VGG16 + SVM (L, 1) has been observed for all the magnification levels, i.e., 93.97% for ×40, 92.92% for ×100, 91.23% for ×200, and 91.79% for ×400 and attained the best performance.
Bardou et al. designed their own CNN which composed of five convolutional layers and two dense layers [64]. They have reported an accuracy ranging from 79.74 to 86.34% for different magnifications with the original dataset. To further enhance the performance, they employed data augmentation technique (rotation and horizontal flip) and obtained accuracy within the range of 80.83 to 84.48% for different resolutions, but an enhanced performance was observed for all the resolutions except ×40. A hybrid approach “CNN + SVM” was also considered by the authors to improve the classification performance. However, the classification performance deteriorated for this configuration. Further, an ensemble model was applied to the augmented data for an improvement in the classification performance. They utilized 10 predictive models with the highest accuracies. This ensemble model showed the best result with this CNN topology (ranging from 83.31 to 88.23% for different resolutions). However, the same CNN showed an inadequate performance when employed as feature extractor in conjunction with KNN, RBF SVM, linear SVM, and RF classifier (ranging from 65.12 to 75.43%).
Han et al. proposed the CSDCNN model for achieving a remarkable performance in multi-classification of BC histopathological images. They have reported the accuracies in the range of 87.6 to 90.8% for the raw data and 92.8 to 93.9% for the augmented data at different magnifications [38]. However, the transfer learning approach as feature generator with augmented data outperforms all the CNN-based approaches used in [64] as well as to the CSDCNN approach (when applied to the original data) [38], while the results of [38] with the augmented dataset are comparable (Table 10). During comparison, it is found that the proposed transfer learning approach is good enough to learn distinguishing features from the complex data of histopathological images at all the magnification levels.
Table 10.
Comparison of existing approaches and the present work based on deep learning at image level for the entire magnification levels
| Technique | Model | Magnification factor | Accuracy (%) | Precision | Recall | F1 score | Ref. |
|---|---|---|---|---|---|---|---|
| Deep learning | CNN + original data |
×40 ×100 ×200 ×400 |
86.34 84.00 79.83 79.74 |
– | – | – | [64] |
| Deep learning | CNN + augmented data |
×40 ×100 ×200 ×400 |
83.79 84.48 80.83 81.03 |
84.27 84.29 81.85 80.84 |
83.79 84.48 80.83 81.03 |
83.74 84.31 80.48 80.63 |
[64] |
| Deep learning | CNN + SVM |
×40 ×100 ×200 ×400 |
82.89 80.94 79.44 77.94 |
– | – | – | [64] |
| Deep learning | CNN + ensemble model |
×40 ×100 ×200 ×400 |
88.23 84.64 83.31 83.98 |
– | – | – | [64] |
| Deep learning | CNN features + KNN |
×40 ×100 ×200 ×400 |
70.48 68.00 70.08 66.38 |
– | – | – | [64] |
| Deep learning | CNN features + RBF SVM |
×40 ×100 ×200 ×400 |
75.43 71.20 67.27 65.12 |
– | – | – | [64] |
| Deep learning | CNN features + linear SVM |
×40 ×100 ×200 ×400 |
72.35 67.68 66.45 64.95 |
– | – | – | [64] |
| Deep learning | CNN features + RF |
×40 ×100 ×200 ×400 |
66.38 65.12 69.80 67.96 |
– | – | – | [64] |
| Deep learning | CSDCNN + original data |
×40 ×100 ×200 ×400 |
89.4 ± 5.4 90.8 ± 2.5 88.6 ± 4.7 87.6 ± 4.1 |
– | – | – | [38] |
| Deep learning | CSDCNN + augmented data |
×40 ×100 ×200 ×400 |
92.8 ± 2.1 93.9 ± 1.9 93.7 ± 2.2 92.9 ± 1.8 |
– | – | – | [38] |
| Deep learning |
VGG19 + SVM (L, 1) (balanced data) |
×40 ×100 ×200 ×400 |
91.21 89.75 68.42 69.44 |
91.00 89.00 69.00 72.00 |
90.00 87.00 68.00 71.00 |
90.00 87.00 68.00 71.00 |
Present Work |
| Deep learning |
VGG19 + SVM (L, 1) (balanced + augmented data) |
×40 ×100 ×200 ×400 |
92.64 91.25 81.42 80.84 |
92.00 91.00 82.00 82.00 |
92.00 91.00 82.00 81.00 |
92.00 91.00 82.00 81.00 |
|
| Deep learning |
VGG16 + SVM (L, 1) (balanced data) |
×40 ×100 ×200 ×400 |
89.61 89.75 78.95 80.00 |
90.00 89.00 81.00 80.00 |
88.00 87.00 82.00 79.00 |
88.00 87.00 81.00 79.00 |
|
| Deep learning |
VGG16 + SVM (L, 1) (balanced + augmented data) |
×40 ×100 ×200 ×400 |
93.97 92.92 91.23 91.79 |
94.00 92.00 92.00 92.00 |
93.00 91.00 92.00 91.00 |
94.00 91.00 92.00 91.00 |
Conclusions
In this paper, handcrafted feature–based approach and transfer learning approach with different configurations (as a feature extractor and as a baseline model) for the multi-classification of breast cancer histopathological images have been compared. It has been observed throughout our study that the transfer learning approach as feature extractor provides a remarkable performance in contrast to other employed approaches. Among different combinations of classifiers, VGG16 + SVM (L, 1) provides the best result for all magnification factors (×40, ×100, ×200, and ×400). For this classifier, the process of feature extraction is very efficient as it utilizes pre-trained weights that are obtained through the training of VGG16 network on a very large ImageNet dataset. Due to this fact, VGG16 + SVM (L, 1) has considered to be more robust and strong classifier for the present configuration. Additionally, the data augmentation techniques are also employed which help in further improving the classification accuracy by tuning the parameters in an appropriate manner. The impact of magnification level on the classification accuracy relies on the complexity level of histopathological images which goes hand to hand with the rise in the level of magnification.
In the near future, the layer-wise fine-tuning approach and ensemble modeling with pre-trained networks can be investigated to determine the overall impact on the accuracy of the models for the multi-classification of histopathological images. Moreover, the embedding of pre-trained CNN’s as a classifier in the handcrafted feature–based approach instead of conventional classifier could be additional strand to solve the problem of breast cancer multi-classification.
Electronic Supplementary Material
(DOCX 1.72 MB)
(PDF 620 kb)
Acknowledgements
The authors are immensely thankful to Dr. S. S. Patnaik, director NITTTR, Chandigarh, India, for providing all the necessary facilities and support during the execution of the work. Also, the author Shallu Sharma would like to thank Dr. Sumit Kumar, Associate Professor, Lovely Professional University, Jalandhar, Punjab, India, for every scientific discussion and constant support.
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Breast Cancer. Available at http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/.
- 2.Breast Cancer Facts & Figures 2017-2018. Available at https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2017-2018.pdf.
- 3.Aubreville M, et al. Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Scientific reports. 2017;7:11979. doi: 10.1038/s41598-017-12320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson ML, Fleming KA, Kuti MA, Looi LM, Lago N, Ru K: Access to pathology and laboratory medicine services: A crucial gap. The Lancet, 2018 [DOI] [PubMed]
- 5.Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, Crawford JM, Clark JR, Cantor-Weinberg J, Joshi MG, Cohen MB, Prystowsky MB, Bean SM, Gupta S, Powell SZ, Speights VO Jr, Gross DJ, Black-Schaffer WS. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Archives of Pathology and Laboratory Medicine. 2013;137:1723–1732. doi: 10.5858/arpa.2013-0200-OA. [DOI] [PubMed] [Google Scholar]
- 6.Pöllänen I, Braithwaite B, Haataja K, Ikonen T, Toivanen P: Current analysis approaches and performance needs for whole slide image processing in breast cancer diagnostics. Proc. Embedded Computer Systems: Architectures, Modeling, and Simulation (SAMOS), 2015 International Conference on: City
- 7.Veta M, Pluim JP, Van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: A review. IEEE Transactions on Biomedical Engineering. 2014;61:1400–1411. doi: 10.1109/TBME.2014.2303852. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS, Varmus H. A new initiative on precision medicine. New England Journal of Medicine. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reardon S. Precision-medicine plan raises hopes: US initiative highlights growing focus on targeted therapies. Nature. 2015;517:540–541. doi: 10.1038/nature.2015.16774. [DOI] [PubMed] [Google Scholar]
- 10.Baba AI, Câtoi C. Comparative oncology: Publishing House of the Romanian Academy Bucharest. 2007. [PubMed] [Google Scholar]
- 11.Yn S, Wang Y, Sc C, Wu L, Tsai S. Color-based tumor tissue segmentation for the automated estimation of oral cancer parameters. Microscopy Research and Technique. 2010;73:5–13. doi: 10.1002/jemt.20746. [DOI] [PubMed] [Google Scholar]
- 12.Alhindi TJ, Kalra S, Ng KH, Afrin A, Tizhoosh HR: Comparing LBP, HOG and Deep Features for Classification of Histopathology Images. arXiv preprint arXiv:180505837, 2018
- 13.Belsare A, Mushrif M, Pangarkar M, Meshram N: Classification of breast cancer histopathology images using texture feature analysis. Proc. TENCON 2015–2015 IEEE Region 10 Conference: City
- 14.Rublee E, Rabaud V, Konolige K, Bradski G: ORB: An efficient alternative to SIFT or SURF. Proc. Computer Vision (ICCV), 2011 IEEE international conference on: City
- 15.Keskin F, Suhre A, Kose K, Ersahin T, Cetin AE, Cetin-Atalay R. Image classification of human carcinoma cells using complex wavelet-based covariance descriptors. PloS one. 2013;8:e52807. doi: 10.1371/journal.pone.0052807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dheeba J, Singh NA, Selvi ST. Computer-aided detection of breast cancer on mammograms: A swarm intelligence optimized wavelet neural network approach. Journal of biomedical informatics. 2014;49:45–52. doi: 10.1016/j.jbi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Wan S, Huang X, Lee H-C, Fujimoto JG, Zhou C: Spoke-LBP and ring-LBP: New texture features for tissue classification. Proc. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on: City
- 18.Zhang Y, Zhang B, Coenen F, Xiao J, Lu W. One-class kernel subspace ensemble for medical image classification. EURASIP Journal on Advances in Signal Processing. 2014;2014:17. doi: 10.1186/1687-6180-2014-17. [DOI] [Google Scholar]
- 19.Boyd S, El Ghaoui L, Feron E, Balakrishnan V. Linear matrix inequalities in system and control theory: Siam. 1994. [Google Scholar]
- 20.Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Transactions on Biomedical Engineering. 2016;63:1455–1462. doi: 10.1109/TBME.2015.2496264. [DOI] [PubMed] [Google Scholar]
- 21.Suykens JA, Vandewalle J. Least squares support vector machine classifiers. Neural processing letters. 1999;9:293–300. doi: 10.1023/A:1018628609742. [DOI] [Google Scholar]
- 22.Breiman L. Random forests. Machine learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 23.Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. Proc. Advances in neural information processing systems: City
- 24.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556, 2014
- 25.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
- 26.Szegedy C, et al.: Going deeper with convolutions. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
- 27.Zeiler MD, Fergus R: Visualizing and understanding convolutional networks. Proc. European conference on computer vision: City
- 28.Lin M, Chen Q, Yan S: Network in network. arXiv preprint arXiv:13124400, 2013
- 29.Lakhani P, Gray DL, Pett CR, Nagy P, Shih G. Hello world deep learning in medical imaging. Journal of digital imaging. 2018;31:283–289. doi: 10.1007/s10278-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 31.Spanhol FA, Oliveira LS, Petitjean C, Heutte L: Breast cancer histopathological image classification using convolutional neural networks. Proc. Neural Networks (IJCNN), 2016 International Joint Conference on: City
- 32.BreakHis Dataset. Available at https://web.inf.ufpr.br/vri/databases/breast-cancer/histopathological-database-breakhis/).
- 33.Spanhol FA, Oliveira LS, Cavalin PR, Petitjean C, Heutte L: Deep features for breast cancer histopathological image classification. Proc. Systems, Man, and Cybernetics (SMC), 2017 IEEE International Conference on: City
- 34.Araújo T, et al. Classification of breast cancer histology images using convolutional neural networks. PloS one. 2017;12:e0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.BACH Dataset. Available at https://iciar2018-challenge.grand-challenge.org/Dataset/.
- 36.Motlagh NH, et al.: Breast Cancer Histopathological Image Classification: A Deep Learning Approach. bioRxiv:242818, 2018
- 37.Sharma S, Mehra R. Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express. 2018;4:247–254. doi: 10.1016/j.icte.2018.10.007. [DOI] [Google Scholar]
- 38.Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S. Breast cancer multi-classification from histopathological images with structured deep learning model. Scientific reports. 2017;7:4172. doi: 10.1038/s41598-017-04075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vang YS, Chen Z, Xie X: Deep Learning Framework for Multi-class Breast Cancer Histology Image Classification. Proc. International Conference Image Analysis and Recognition: City
- 40.Nahid A-A, Kong Y. Histopathological breast-image classification using local and frequency domains by convolutional neural network. Information. 2018;9:19. doi: 10.3390/info9010019. [DOI] [Google Scholar]
- 41.Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B. Histopathological image analysis: A review. IEEE reviews in biomedical engineering. 2009;2:147. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeong S: Histogram-based color image retrieval. Psych221/EE362 project report, 2001
- 43.Shukla K, Tiwari A, Sharma S. Classification of Histopathological images of Breast Cancerous and Non Cancerous Cells Based on Morphological features. Biomedical and Pharmacology Journal. 2017;10:353–366. doi: 10.13005/bpj/1116. [DOI] [Google Scholar]
- 44.Hu M-K. Visual pattern recognition by moment invariants. IRE transactions on information theory. 1962;8:179–187. [Google Scholar]
- 45.Lin H, Si J, Abousleman GP. Orthogonal rotation-invariant moments for digital image processing. IEEE Trans Image Processing. 2008;17:272–282. doi: 10.1109/TIP.2007.916157. [DOI] [PubMed] [Google Scholar]
- 46.Sonka M, Hlavac V, Boyle R: Image processing, analysis and machine vision London. England: Chapman & Hall Computing:423–431, 1993
- 47.Tsai W-H, Chou S-L. Detection of generalized principal axes in rotationally symmetric shapes. Pattern Recognition. 1991;24:95–104. doi: 10.1016/0031-3203(91)90080-O. [DOI] [Google Scholar]
- 48.Huang Z, Leng J: Analysis of Hu's moment invariants on image scaling and rotation. Proc. Computer Engineering and Technology (ICCET), 2010 2nd International Conference on: City
- 49.Lin W-C, Hays J, Wu C, Kwatra V, Liu Y: A comparison study of four texture synthesis algorithms on regular and near-regular textures. Tech Rep, 2004
- 50.Hua B, Fu-Long M, Li-Cheng J. Research on computation of GLCM of image texture [J] Acta Electronica Sinica. 2006;1:155–158. [Google Scholar]
- 51.Haralick Robert M., Shanmugam K., Dinstein Its'Hak. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973;SMC-3(6):610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 52.LeCun Y: LeNet-5, convolutional neural networks. URL: http://yannlecuncom/exdb/lenet:20, 2015
- 53.Zhang W: Shift-invariant pattern recognition neural network and its optical architecture. Proc. Proceedings of annual conference of the Japan Society of Applied Physics: City
- 54.Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City [DOI] [PubMed]
- 55.Girshick R, Donahue J, Darrell T, Malik J: Rich feature hierarchies for accurate object detection and semantic segmentation. Proc. Proceedings of the IEEE conference on computer vision and pattern recognition: City
- 56.He Y, Zhang X, Sun J: Channel pruning for accelerating very deep neural networks. Proc. International Conference on Computer Vision (ICCV): City
- 57.Rabanser S, Shchur O, Günnemann S: Introduction to Tensor Decompositions and their Applications in Machine Learning. arXiv preprint arXiv:171110781, 2017
- 58.Lebedev V, Ganin Y, Rakhuba M, Oseledets I, Lempitsky V: Speeding-up convolutional neural networks using fine-tuned cp-decomposition. arXiv preprint arXiv:14126553, 2014
- 59.Howard AG, et al.: Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:170404861, 2017
- 60.Yamashita R, Nishio M, Do RKG, Togashi K: Convolutional neural networks: an overview and application in radiology. Insights into imaging:1–19, 2018 [DOI] [PMC free article] [PubMed]
- 61.Chan A, Tuszynski JA. Automatic prediction of tumour malignancy in breast cancer with fractal dimension. Royal Society open science. 2016;3:160558. doi: 10.1098/rsos.160558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nahid A-A, Mehrabi MA: Kong Y: Histopathological Breast Cancer Image Classification by Deep Neural Network Techniques Guided by Local Clustering. BioMed research international 2018, 2018 [DOI] [PMC free article] [PubMed]
- 63.Veeling BS, Linmans J, Winkens J, Cohen T, Welling M: Rotation equivariant cnns for digital pathology. Proc. International Conference on Medical image computing and computer-assisted intervention: City
- 64.Bardou D, Zhang K, Ahmad SM. Classification of Breast Cancer Based on Histology Images Using Convolutional Neural Networks. IEEE Access. 2018;6:24680–24693. doi: 10.1109/ACCESS.2018.2831280. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1.72 MB)
(PDF 620 kb)