Abstract
This paper presents a systematic review of the literature focused on the lung nodule detection in chest computed tomography (CT) images. Manual detection of lung nodules by the radiologist is a sequential and time-consuming process. The detection is subjective and depends on the radiologist’s experiences. Owing to the variation in shapes and appearances of a lung nodule, it is very difficult to identify the proper location of the nodule from a huge number of slices generated by the CT scanner. Small nodules (< 10 mm in diameter) may be missed by this manual detection process. Therefore, computer-aided diagnosis (CAD) system acts as a “second opinion” for the radiologists, by making final decision quickly with higher accuracy and greater confidence. The goal of this survey work is to present the current state of the artworks and their progress towards lung nodule detection to the researchers and readers in this domain. This review paper has covered the published works from 2009 to April 2018. Different nodule detection approaches are described elaborately in this work. Recently, it is observed that deep learning (DL)-based approaches are applied extensively for nodule detection and characterization. Therefore, emphasis has been given to convolutional neural network (CNN)-based DL approaches by describing different CNN-based networks.
Keywords: Lung nodule, Lung cancer, Early detection, Nodule detection, Feature engineering, Deep learning
Introduction
Cancer is a disease that has several manifestations and it is primarily associated with abnormal cell groups. These cancer cells continue to divide and grow to produce tumors. Among all types of cancer, lung cancer is the most life-threatening disease all over the world. According to the World Health Organization [1], lung cancer is the leading cause of death worldwide. In 2008, 1.37 million deaths caused by lung cancer occurred throughout the world [2]. Available data shows that the lung cancer is the largest among new cancer diagnoses worldwide (1,350,000 new cases and 12.4% of total new cancer cases) and it is the largest cause of death from cancer globally (1,180,000 deaths and 17.6% of total cancer deaths) [3].
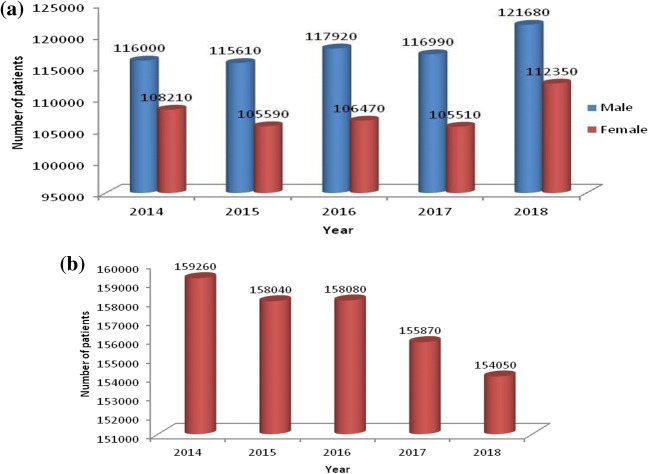
It is the most common cancer in men worldwide (1.1 million cases, 16.5% of the total). Among females, it was the fourth (516,000 cases, 8.5% of all cancers) most commonly diagnosed cancer and the second (427,000 deaths, 12.8% of the total) leading cause of cancer death [4]. Last 5 years (2014-2018) estimated new lung cancer cases and total estimated lung cancer deaths found in the USA [5] have been portrayed in Fig. 1. A report by National Cancer Centre Singapore [6] has shown that 1.2 million people were diagnosed with cancer and 7700 people died of cancer on average in the year 2015 and 158,080 cases were expected by the end of 2016. In Brazil, an average of 28,220 cases found in 2016 with 17,330 males and 10,890 females [7].
Fig. 1.
Last 5 years (2014–2018) lung cancer statistics in USA. a Nos. of estimated new cases. b Total nos. of estimated deaths. Source: American Cancer Society (ACS) [5]
Lung cancer can be divided into two broad categories, viz., small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). SCLC accounts for 15% of lung cancer cases and highly malignant. Non-small cell lung cancer (NSCLC) accounts for the remaining 85% of cases [3].
A pulmonary nodule refers to the lung tissue abnormality mostly found in lung cancer patients. In 1984, a glossary of terms published by Fleischner Society [8] for thoracic radiology. The society has defined lung nodule as “any pulmonary or pleural lesion represented in a radiograph by a sharply defined, discrete, nearly circular opacity 2-30 mm in diameter.” Twelve years later, the society defined lung nodule as “round opacity, at least moderately well marginated and no greater than 3 cm in maximum diameter.”
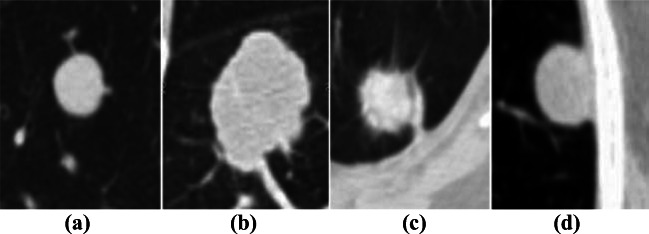
The solitary pulmonary nodule (SPN) is defined as a radiographic opacity with a diameter of up to 30 mm and at least two-thirds of its margins surrounded by lung parenchyma. SPNs are of the four types: (a) well-circumscribed: the nodule is located centrally in the lung without significant connections to vasculature, (b) vascularized: the nodule is located centrally in the lung, but has significant vascularization (connections to neighboring vessels), (c) juxtapleural: a significant proportion of the nodule periphery is connected to the pleural surface, and (d) pleural tail: The nodule is near the pleural surface, connected by a thin structure called “pleural tail” [9]. Figure 2 shows four types of SPNs. On the other hand, if the diameter is greater than 30 mm, it is called lung mass. Lung masses are generally cancerous.
Fig. 2.
Typical SPNs for different types. a Well-circumscribed nodule. b Juxtavascular nodule. c Nodule with a pleural tail. d Juxtapleural nodule (Dhara et al. [17])
It is observed from different research works that early detection of lung nodules can improve the 5-year survival rate. Among different types of cancer, the 5-year survival rate of lung cancer is the lowest with an estimated value of 18% of the diagnosed cases for years 2004-2010. The prognosis of lung cancer depends on the extent of disease at the time of diagnosis. Early-stage detection of lung cancer can improve the survival rate. The 5-year survival rate is 54% for a small and localized lesion [10]. According to Cancer Research UK [11], the survival rate is 87% for stage I, and 19% for stage IV lung cancer patients.
Many pitfalls of recent medical imaging techniques have been investigated by the work of Godoy et al. [12] for nodule detection and characterization. Computed tomography is the most recent dominated imaging tool used to capture the lung images. The National Lung Screening Trial (NLST) [13] reported that a reduction of 20% in mortality can be achieved with low-dose computed tomography (LDCT) image than chest X-ray. Therefore, computer-aided detection and computer-aided diagnosis has become the most prominent and valuable tool for lung nodule detection and characterization.
A complete guideline for small pulmonary nodule management including its growth information is provided by the work of MacMahon et al. [14]. Recent guidelines and management of pulmonary nodules are described in [15]. A deep and extensive study of patients with previous cancer history was presented by the work of Rena et al. [16] for SPN characterization.
The morphological appearances such as border, shape, and location characteristics of a lung nodule in CT image are described in [18-20]. The information about ground glass opacity (GGO) nodule is available in [21]. Different types of GGOs are shown in Fig. 3.
Fig. 3.
Examples of a sub-solid and b pure ground-glass nodules (Lederlin et al. [15])
Reviews of the pulmonary detection techniques are not new but as the computational techniques change from time to time, a thorough review is necessary to understand the technology shipment towards automated nodule detection systems. Different review works with objectives to find out the technological changes exist in the literature [17, 22-27].
However, owing to the developments of new techniques, it has been noticed that a recently deep learning-based approach has introduced for nodule detection. Therefore, a detailed survey is required to cope up with the technological changes and present consistent information to the thirsted readers, the aim of this survey work.
This survey work primarily conducted by focusing on the “nodule detection” process from CT images. Therefore, different works based on nodule detection have been searched and collected from IEEE, Elsevier, SPIE, and Springer. The following sentences were used as search terms for handcraft- (HC) or feature engineering-based works: (1) lung nodule detection, (2) nodule detection, (3) lung nodule detection using filters, (4) lung nodule segmentation, and (5) nodule candidate detection. On the other hand, sentences such as (1) lung nodule detection using deep learning, (2) lung nodule detection using deep features, (3) lung nodule detection using convolutional neural network, and (4) lung nodule detection using CNN have been used for deep learning-based works.
It is observed that, apart from nodule detection, CNN-based nodule characterization/classification tasks are also available in the literature. CNN acts as an automatic feature extractor, owing to this reason the nodule characterization becomes much easier. The following search terms have been used to collect different works based on the nodule characterization, viz., (1) lung nodule characterization, (2) lung nodule classification, (3) lung nodule characterization using CNN, and (4) lung nodule classification using CNN.
This review paper is organized as follows. The “Database Description” section presents a detail description of the available databases. The “Traditional/Handcraft/Feature Engineering-Based Approaches Towards Lung Nodule Detection” section describes different nodule detection frameworks based on handcraft/feature engineering approach. The “Deep Learning-Based Approach Towards Lung Nodule Detection” section is devoted to the deep learning-based approach towards nodule detection and characterization. Finally, the reported survey work has been concluded in the “Conclusion” section.
Database Description
In this step, CT images are collected for evaluation of the CAD systems. In most of the nodule detection works, the images used have dimension 512 × 512 pixels in the form of a 2-D slice. A number of slices are varying in different scans. All images use Digital Imaging and Communications in Medicine (DICOM) file format. The high-resolution CT (HRCT) images can be acquired from the few publicly available databases, e.g., LIDC, ANODE, and from different hospitals (private database). Below, a brief introduction has been given for the two publicly available databases.
(a) LIDC-IDRI
The Lung Image Database Consortium-Image Database Resource Initiative [28] is the world’s largest publicly available database that acts as a leading source of nodule reference database. This image repository was initiated and developed by five institutions in USA, viz., Weill Cornell Medical College, University of California (Los Angeles), University of Chicago, University of Iowa, and University of Michigan [29]. Readers are referred to the work of Armato III et al. [29] and Tan et al. [30] for the historical perspective of the LIDC-IDRI database. The database contains 1018 CT scans of 1010 patients. The thickness of each slice varies from 1.25 to 2.5 mm range with the pixel size ranges between 0.48 and 0.72 mm [31]. Most of the work used the nodule size between 3 to 30 mm (11-113 pixels). A detailed description of the LIDC-IDRI can be found in [29, 32]. The works of Gupta et al. [31], Hancock and Magnan [33], Han et al. [34], Hancock and Magnan [35], Messay et al. [36], and Paul et al. [37] have presented the marking techniques, evaluation methods, and guidelines to use of this database.
(b) ANODE09
The Automatic Nodule Detection 2009 (ANODE09) is another publicly available database consists of 55 CT scans, annotated by two radiologists. Out of 55 scans, 5 were provided as training cases, and the rest 50 cases were provided as the testing case. The details of this database can be found by the work of Ginneken et al. [38].
Apart from LIDC and ANODE09, three more databases, viz., TIME [39], ELCAP [40], and LISS [41], are available in the public domain for lung nodule research. These databases are small in size w.r.to LIDC.
Traditional/Handcraft/Feature Engineering-Based Approaches Towards Lung Nodule Detection
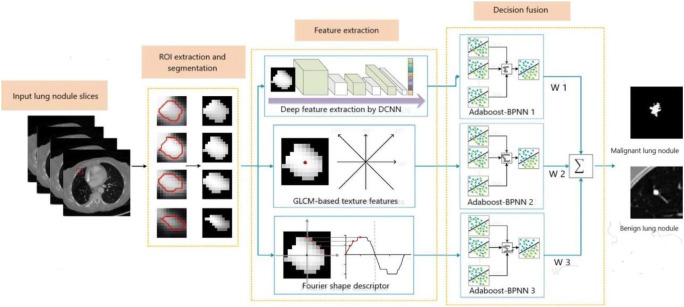
This section describes the workflow of a computer-aided detection (CADe) system by explaining different works based on “Feature Engineering” for nodule detection. This traditional framework consists of four main steps: (1) pre-processing, (2) lung segmentation, (3) nodule detection, and finally (4) classification (Fig. 4). Each step has been described precisely by considering the related works that have been found so far.
Fig. 4.
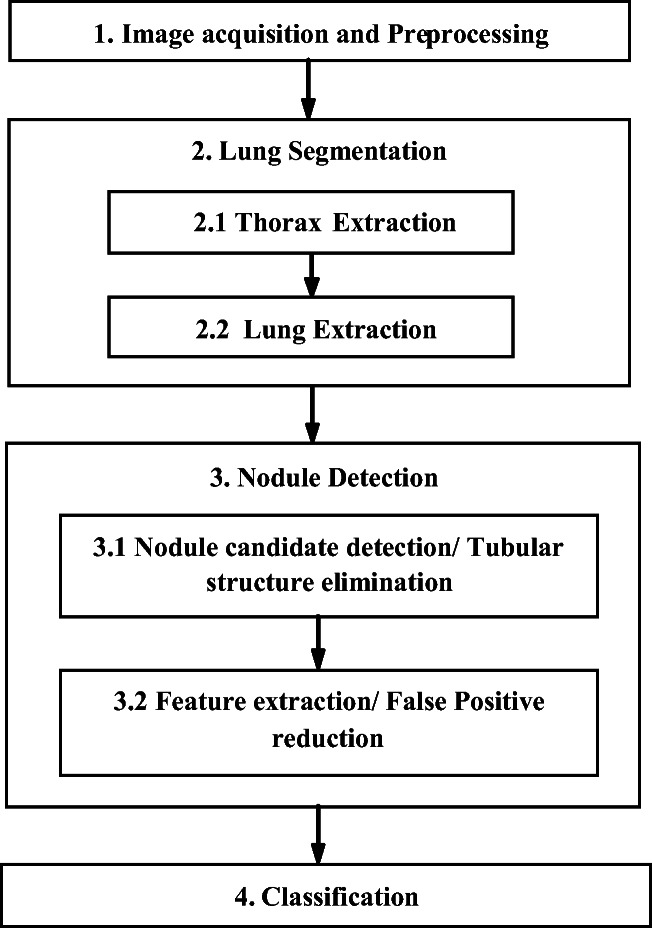
A simple CADe framework
Pre-processing
The aim of this step is to reduce noise and enhancement of nodule-like structures by applying different filters to the input lung CT image. Different filters can be found in literature, among them median filter, dot enhancement filter, log filter, and filters based on histogram equalization are widely used in this stage. Median filter has been used in [42-46]. The work of Sun et al. [47] and Choi and Choi [48] used dot enhancement filter. Log filter was used by the work of Nagata et al. [49], Jirapatnakul et al. [50], and Kubo et al. [51]. Wiener filter was applied in [52]. CLAHE and Wiener filter have been used in [53]. Multi-scale filter was examined by the work of Pei et al. [54] and Takemura et al. [55]. Selective enhancement filter has been used by Miyajima et al. [56] and Dhara and Mukhopadhyay [57]. Dhara et al. [58] applied a multi-scale selective enhancement filter. Gabor filter was used by the work of Hadavi et al. [59]. Filho et al. [45] and Namin et al. [60] have used Gaussian filter in the pre-processing stage. Apart from that, Iris filter [61], shell filter [62], sequential filter [63], fuzzy filter [64], and LBP filter [65] have been used as a pre-processing step.
Lung Segmentation
The second step of the nodule detection system is the lung segmentation. This stage aims to reduce the search space by extracting only the lung regions. This step mainly includes the following sub-steps thorax extraction and parenchyma structure extraction/lung extraction.
Thorax Extraction
This stage processes and removes all artifacts external to the patient’s body, e.g., bed sheets, the air that involves him, and the surface on which he lies [66].
Lung Extraction
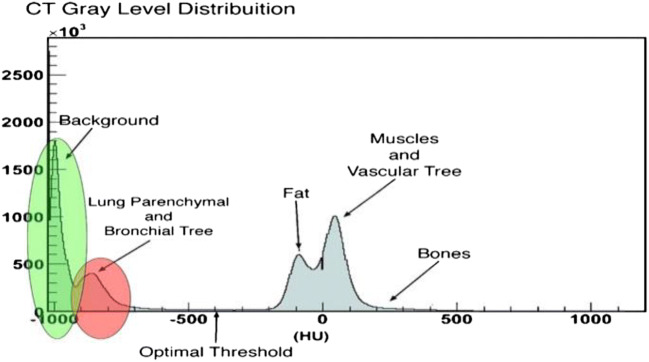
The overall aim of this module is the identification of the left and right lung from a CT slice. The histogram of the lung CT image with two sharp peaks from left to right has been shown in Figs. 5 and 6, respectively.
Fig. 5.
Gray-level distribution of lung CT image (Cascio et al. [67])
Fig. 6.
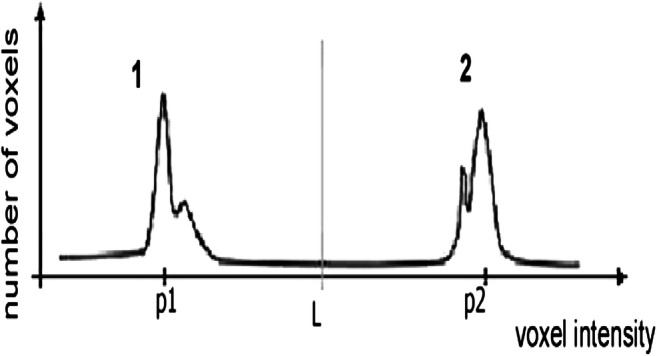
Threshold may be obtained by choosing the valley of the histogram (Sousa et al. [66])
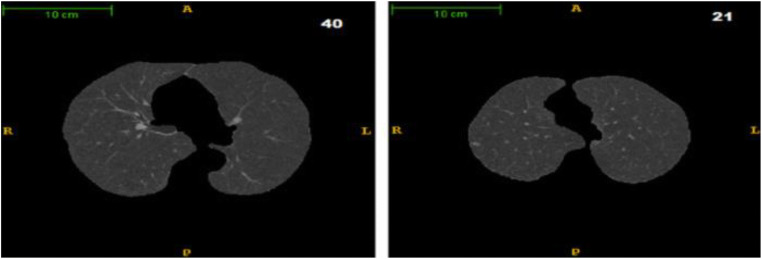
A typical thoracic CT slice contains voxel intensities in Hounsfield unit (HU) scale. It is observed from the above histograms that low-density lung tissues are visible in the intensity range of − 910 to − 500 HU, whereas high-density structures such as chest wall, blood, bone, fat, and muscles contain intensity value above − 500 HU. In most of the works, researchers have incorporated threshold-based segmentation, as there is a sharp valley in the histogram (Fig. 6). Experimentally, a value of − 500 HU may be chosen as the ideal threshold value. Otherwise, an iterative optimal threshold algorithm may be used to determine an optimal threshold. Therefore, by designing a proper mask, it is possible to extract the left and right lung region from CT image. A region-growing algorithm with a threshold as stopping criteria may be used for lung segmentation as mention in [66]. A 3-D region-growing algorithm has been used by several studies for lung segmentation. After segmentation, a 3-D labeling algorithm labels the image. Finally, two lung regions have been extracted as the biggest and next biggest connected regions from the labeled image and setting the rest of the segmented regions as background [68, 69]. Lung segmentation based on threshold can be found by the work of Leemput et al. [62], Cuifang et al. [64], Suárez-Cuenca et al. [61], Sousa et al. [66], Santos et al. [68], Zhang et al. [70], Fu et al. [71], Gopi and Selvakumar [72], Paing and Choomchuay [73, 74], Javaid et al. [75], Rendon-Gonzalez and Ponomaryov [76], Xu et al. [65], Saien et al. [77], Choi and Choi [78, 79], Pu et al. [80], and Messay et al. [81]. Segmentation based on iterative threshold [48, 82, 83], Otsu threshold [73, 84], adaptive threshold [61, 85], and 3-D adaptive fuzzy threshold [86] exist in the literature. Lung segmentation based on region and 3-D region growing can be found in [42, 67, 87-92]. Zhang et al. [70] have introduced a global active contour model for lung segmentation. Figure 7 shows an example output of the lung segmentation stage.
Fig. 7.
Output of “lung segmentation” stage. Segmented left and right lung appear in slices 40 and 21, respectively (Santos et al. 2014 [66])
Nodule Detection
This section describes the main module of a CADe-based system. The aim of this module is the detection of potential region of interests (ROIs), here lung nodule using image processing techniques. The term, “nodule detection” has been used in a broad sense and consists of the following two sub-stages, viz., (1) nodule candidate detection/tubular structure elimination and (2) feature extraction/false-positive reduction. The complete framework for the CADe system has been shown in Fig. 4. Lee et al. [25] have categorized the nodule detection scheme as (a) segmentation detection, (b) classification detection, (c) segmentation-template detection, and (d) segmentation-classification detection. It has been observed that in the traditional nodule detection approach, almost all the works are following the segmentation-classification-based approach. The aim of nodule detection module is to identify true nodule candidates from the false positives (FPs), such as blood vessels, shades, bronchioles, bifurcation points, and ribs by using different image processing and pattern recognition techniques. Next, use or invent different discriminating features so that nodules can be detected (Fig. 8) with higher classification accuracy. In this section, different journal works and few conference-based works have been described elaborately for nodule detection found so far from the time period 2009 to April 2018.
Fig. 8.
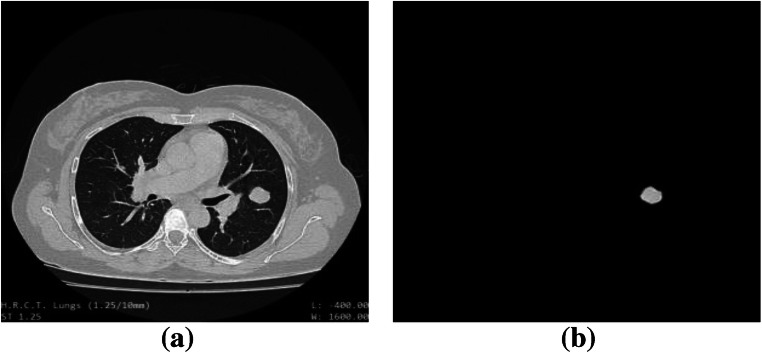
Output of “nodule detection” stage. a Original lung CT image, input to the CADe system. b Detected nodule
Nodule Candidate Detection/Tubular Structure Elimination
Image segmentation divides an image into different disjoint regions. This process changes the representation of an image into some meaningful image regions for better understanding and analysis in the subsequent stages. Therefore, this process reduces the search space within an image and at the same time finds the location of the desired objects for identification purpose. A good survey on the segmentation category and medical image segmentation can be found by the work of Pal and Pal [93], Haralick and Shapiro [94], Phamy et al. [95], and Sharma and Aggarwal [96]. The segmentation approaches can be divided into three generations. The first-generation approach includes different algorithms depending on the pixel intensity and different low-level image processing techniques. Segmentation involves edge and thresholding and region-based approaches are examples of this category. First-generation algorithms do not incorporate any prior information of the image. Second-generation algorithms introduce uncertainty models and different optimization techniques for image segmentation. Segmentation based on clustering, genetic algorithm (GA), neural network (NN), and partial differential equations are examples of second-generation segmentation. Third-generation algorithms incorporate high-level knowledge such as “a piece of prior” information; expert knowledge and knowledge about the object shape, orientation, continuity, elasticity, or smoothness are included in the segmentation process. Segmentation based on deformable model, active appearance model (AAM) and atlas-based approaches are examples of third-generation segmentation algorithm. Different segmentation algorithms have been introduced for accurate nodule segmentation or nodule candidate detection or both. Research works focused on nodule segmentation can be found in [36, 86, 97-101]. Our aim of this survey is to emphasize on the nodule detection process. In this subsection, different segmentation and detection frameworks have been discussed for nodule candidate detection.
The frameworks have been categorized based on different segmentation approaches. Segmentation based on thresholding [66, 78, 79, 81, 102-104], region-based [105], clustering-based [45, 75, 106-109], mixed or hybrid segmentation algorithm-based [110], partial differential equation-based [99, 111], model-based [67, 112] and atlas-based [113] methods were reported in the literature for nodule candidate detection. Apart from that, some other approaches, e.g., template matching based [88, 114], filtering based [48, 57, 62, 115-118], feature based [60, 119], concept based [42, 80, 120, 121], and works based on specialized algorithms [122-125] are available in the literature for complete nodule detection framework.
A. Thresholding-Based Methods
This approach is the simplest one among different image segmentation approaches. Image histogram is used to partition the image. Each peak of the histogram represents a particular region and the intensity value between two peaks called the “threshold” is selected to separate desired classes. Next, the segmentation process is done by grouping all pixel intensities w.r.to. threshold value. A good survey on thresholding-based segmentation can be found by the work of Sahoo et al. [126]. Sousa et al. [66] segmented internal structures of the lung by choosing appropriate threshold values. After that, tubular structures were eliminated by applying the 3-D skeletonization algorithm. In their work, researchers have observed that tubular structure, e.g., vessel shows a low variation of the average depth w.r.to medial axis. On the other hand, nodules show an abrupt increase of values from the nodule border. Therefore, by choosing appropriate threshold value with skeletonized segmentation algorithm nodules has been separated from vessels. Messay et al. [81] used intensity thresholding and morphological processing techniques for nodule candidate detection. Choi and Choi [78] have identified initial nodule candidates by using thresholding and shape information. Narayanan et al. [102] have segmented and detected candidate nodules simultaneously using intensity-based thresholding combined with morphological processing. Aresta et al. [103] developed a nodule detection framework using Otsu’s threshold method. Different solid, sub-solid, non-solid, and juxtapleural nodules have been detected by their proposed system. Solid nodules were searched slice-wise inside a fixed-width sliding window with stride 1. Next, the Otsu threshold method has been applied inside each window for candidate generation. Finally, all windows were combined using logical OR operation. The sub-solid and non-solid nodule candidates were detected using a Laplacian-of-Gaussian (LoG) filter combined with Otsu’s method. Mehre et al. [104] combined intensity threshold with structuring elements (SEs) for candidate nodule detection. Morphological opening operation was performed by using six different intensity values combined with radii of SEs, viz., (− 400, 2), (− 500, 2), (− 600, 2), (− 400, 3), (− 500, 3), (− 600, 3) for nodule candidate detection. Choi et al. [79] applied optimal multiple thresholding and rule-based pruning method for nodule candidate detection and segmentation.
It is observed that the detection sensitivity varies from 82.66% with an average 3 FPs/scan/case [81] to 94.1% at 5.54 FPs/scan [79] by using threshold-based approach. The juxtapleural nodule candidates were also detected with a sensitivity of 57.4% at 4 FPs/scan by using this approach [103].
The thresholding approach is entirely based on pixel intensity of an image. It highly depends on the histogram peaks of an image. Finding an appropriate threshold is a challenging task; as pixel intensity is more sensitive to noise and intensity inhomogeneities. Apart from these, spatial characteristics of an image are not taken into account by this approach and not good for images with wide plane valleys and unclear histogram peaks.
B. Region-Based Methods
This approach partitions an image into different regions by satisfying some homogeneity criteria. Gray-level pixel values are used as homogeneity criteria. Region-based segmentation includes different techniques, viz., region merging, region splitting, and split and merge. “Split and Merge” is a hybrid approach, taking the advantages of both methods. In this approach, pixels are grouped by satisfying some similarity criterion. The selection of the similarity criterion plays an important role in accurate segmentation. A survey on region-based segmentation is presented in Freixenet et al. [127]. Suárez-Cuenca et al. [105] developed a 3-D region-growing algorithm for accurate nodule segmentation. Initially, the nodule candidates were detected by a selective enhancement filter and thresholding approach. The researchers have detected 71.8% of nodules at 0.8 false positives per case, 75.5% at 1.6 FPs/case, and 80% at 3.4 FPs/case, respectively. The region-based algorithm gives poor results as compared with a threshold-based nodule detection approach.
The primary disadvantage of region-based approach is that it requires careful manual interaction for seed point initialization as well as proper seed planning for the regions that need further processing, although the split and merge technique does not require a seed point. Apart from that, different extracted regions may contain holes or even disconnected. These algorithms are also sensitive to noise.
C. Clustering-Based Methods
This method is based on the division of pixels into homogeneous clusters. This is an unsupervised approach to image segmentation method. It finds the natural grouping of pixels by executing certain criterion function. Different clustering algorithms, e.g., K-means or ISODATA algorithm, fuzzy c-means (FCM), and the expectation-maximization (EM) algorithm, have been applied extensively for medical image segmentation. Filho et al. [45] proposed a CADe system based on the quality threshold (QT) algorithm and the diversity index. The QT algorithm was used for nodule candidate detection. Javaid et al. [75] have used the K-means algorithm for nodule detection. Initially, the number of classes has been chosen as three. Next, the class with the highest mean intensity value was chosen as the nodule cluster. Juxtavascular nodules are attached to the blood vessels. Therefore, the separation of blood vessels from the Juxtavascular nodule is a crucial task. The researchers have categorized these attachments (connection) as 2-D and 3-D type. If vessels were connected to a nodule in a single slice, it was called “2-D connection.” On the other hand, a“3-D connection” was defined as the branches and nodules that appear to be separated objects on a single slice but were attached in any previous or some next slice. Researchers have used a shape-specific morphological opening operation to break the 2-D connection. A structuring element “line” with parameters length and angle has been used by their study. Removal of 3-D connected branches has been done by comparing the areas of 3-D connected regions on each slice. Finally, all nodule candidates were grouped into six classes by using the proposed equations, viz., “nodule inhomogeneities thickness” and “% of wall connectivity.” Santos et al. [68] developed a CADe system for automatic small nodule (2-10 mm) detection on CT images. After lung segmentation, the internal structures have been segmented using the Gaussian mixture model (GMM) and EM algorithm. Then a Hessian matrix was used for false-positive reduction. However, it has been observed that the presence of bronchi bifurcations could not be removed by using only a Hessian matrix. Nithila and Kumar [106] have presented a new technique that incorporates particle swarm optimization (PSO) algorithm with back-propagation neural network (BPNN). Nodules were initially detected by using FCM clustering. The data points have been clustered into three different intensity levels; low, medium, and high. Spherical objects have been identified using roundness rule, and tubular structures were removed using elongation rule. Farahani et al. [107] developed a type-II fuzzy algorithm for the quality improvement of raw CT images. Then, modified spatial kernelized fuzzy c-means (MSFCM) clustering algorithm has been used for internal structure segmentation. After that, morphological opening, closing, and filling operations have been used for nodule candidate detection. Netto et al. [108] developed a CADe system based on growing neural gas (GNG) clustering algorithm. Initially, candidate nodules were detected using GNG algorithm. Next, nodule candidates were further regrouped using 3-D region-growing algorithm for volume reduction. Hosseini et al. [109] proposed an automatic system that can learn and tune Gaussian interval type-2 membership functions (IT2MFs). Finally, the system has been applied for lung nodule detection.
It is observed that the detection sensitivity varies from 85.91% with 1.82FPs/exam [45] to 93.2% [107] by using the clustering-based method. An accuracy of 98% (for solid nodule), 99.5% (for part-solid nodule), and 97.2% (for non-solid nodule) respectively has been achieved by the work of Nithila and Kumar [106]. Farahani et al. [107] also have achieved an accuracy of 96.5% by using this approach. As compared with the previous threshold and region-based detection approaches, the detection accuracy of clustering approach is higher with lower false positives.
Most of the clustering algorithms require initial parameters. The EM algorithm has greater sensitivity to initialization than the K-means or FCM algorithm. This approach does not consider spatial modeling and can be sensitive to noise and intensity inhomogeneities. The lack of information and uncertainty in data can be dealt with the FCM algorithm by using a membership function. Fuzzy if-then rules could be utilized to do approximate inference. However, choosing or designing an appropriate membership function is not a trivial task and calculations involved in fuzzy approaches could be intensive.
D. Mixed or Hybrid Segmentation Algorithm-Based Methods
This approach consists of combining different segmentation approaches by using some criterion function. Antonelli et al. [110] designed a new CADe system that includes three sub-modules (1) image segmentation, whose aim is to extract pulmonary regions; (2) VOI (voxel of interest) detector, looking for VOIs inside the pulmonary regions; and (3) nodule detector, classify the VOIs into nodules and non-nodules. Three segmentation algorithms, viz., robust fuzzy c-means (RFCM), iterative threshold, and region growing, have been used in parallel to the initial segmented image for VOIs detection. A combination scheme (VOI detector combiner) has been used to merge the outputs of the algorithms to form the VOIs list. This VOI list acts as a nodule candidate locator in their proposed system. Finally, five classifiers, viz., linear classifier (ldc), quadratic classifier (qdc), logistic classifier (loglc), decision tree (treec), and a radial basis function neural network (rbnc), have been used for nodule detection. The proposed system has achieved a sensitivity of 92.5%. It is observed that the sensitivity value is high for mixed algorithm-based system but choosing the proper combination criteria is difficult as different segmentation algorithm belongs to a different generation and follows their philosophy.
E. Partial Differential Equation-Based Methods
This approach uses partial differential as its basic set of equations. Active contour model (ACM) and level-set method (LSM) belong to this category. Another name for ACM is snake. A snake is an energy minimizing, deformable spline. It is defined by two energy terms, viz., internal elastic energy and the external edge-based energy. Internal energy term controls the deformations applied to the snake, and the external energy term controls the fitting of the contour onto the image. The finally used energy function is the sum of the external energy and internal energy terms; it acts like a snake. Level-set methods are a conceptual framework and act as a tool for numerical analysis of surfaces and shapes. Keshani et al. [99] have presented a technique using ACM. In the first step, binary lung CT images were obtained by adaptive fuzzy thresholding. After that, false positives were reduced by applying two different sized windows, immediately followed by another two rotational windows. Next, two threshold values were used to improve the result and making an initial mask for the ACM model. A global region-based energy function that uses mean intensities has been used by their work for nodule candidate segmentation and detection. Gong et al. [111] have used a unique 3-D tensor-filtering algorithm combined with local features for nodule candidate detection. Next, a 3-D level-set method has been used to correct and refine the boundaries of nodule candidates. A sensitivity of 79.3% was achieved by their work. An overall detection rate of 89% with 7.3 FPs/scan has been achieved by the work of Keshani et al. [99]. The disadvantages of ACM include it is not parameter free, sensitive to local minima states, and accuracy highly depends on the convergence policy. On the other hand, LSM is a parameter-free approach. In LSM technique, constructing appropriate velocities for a level-set function is difficult.
F. Model-Based Methods
This approach assumed that the structure of the organs has a repetitive form of geometry and then it is modeled probabilistically for variation of shape and geometry. Active shape model (ASM), active appearance model (AAM), and deformable models are examples of model-based segmentation methods. A survey on deformable models can be found by the work of McInerney and Terzopoulos [128]. Cascio et al. [67] proposed a new algorithm based on the 3-D mass-spring model. The proposed system can detect small-sized (3 mm diameter) nodules. The stable 3-D mass-spring model (MSM) combined with spline curves has been used for nodule segmentation. Farag et al. [112] have used AAM with a template matching technique. Four templates were obtained using AAM for four categories of nodule detection. Cascio et al. [67] have achieved a detection rate of 97% with 6.1 FPs/CT with a sensitivity of 88% with 2.5 FPs/CT.
In model-based segmentation, manual interaction is required to place an initial model and choose appropriate parameters, the main disadvantage of this approach. Apart from that, this approach gives poor convergence to concave boundaries [96].
G. Atlas-Based Methods
This approach incorporates prior information of the object. Different expert knowledge or statistical information extracted from the available examples. The a prior knowledge is incorporated in both atlas construction and atlas-fitting procedure. In this approach, different features are computed using an atlas or lookup table. Alam et al. [113] have used an atlas-based segmentation method for lung nodule detection. In the first step atlas, pre-segmented image has been selected using nine feature descriptors and the K-nearest neighbors (K-NN) algorithm. Then in the second step, nodule candidates were selected using a patch-based image segmentation method. Finally, the Laplacian of Gaussian (LoG) filter has been applied for accurate nodule segmentation and detection. Researchers have reported a sensitivity of 100% by using the atlas-based approach.
The main advantage of this approach is that it can perform segmentation and classifications in one go [96]. The disadvantage of an atlas-based approach is the time necessary for atlas construction.
H. Template Matching-Based Methods
This is the “brute-force” algorithmic approach for object detection process. A template is a sub-image contains the region of interest (ROI); slides over the entire input image for the search of desired objects. Cross-correlation, normalized cross-correlation (NCC), sum of absolute difference (SAD), and sum of squared differences (SSD) are the metrics, commonly used for the matching process. Gonga et al. [88] developed a new algorithm based on dynamic self-adaptive template matching and Fisher linear discriminant analysis (FLDA) classifier. Initially, the lung volume was segmented by using Otsu and the 3-D region-growing algorithm. Next, a multi-scale Gaussian smoothing operation has been operated for noise reduction and gradient calculation. After that, the 3-D dot enhancement filter has been applied over the smooth image. Finally, a threshold was applied on the dot-enhanced image to get the nodule candidate regions. Then the spherical ROIs have been obtained by using a 3-D dot-filtering algorithm. Next, a 3-D dynamic self-adaptive template matching algorithm has been used with the middle slice of 3-D ROI as the gray distribution of the 3-D template. Researchers have used the maxima of NCC and SSD matching methods for nodule candidate detection. Then a 3-D region-growing algorithm has been applied for accurate nodule segmentation. Farag et al. [112] proposed a template matching technique for nodule detection. Four templates were designed by their work. Each template represents the mean shape and texture of one of the four types of nodules. These models have been generated using an AAM method that allowed for obtaining a more realistic texture and shape descriptor of the nodules. NCC has been used as the similarity measurement. Assefa et al. [114] have combined a template matching algorithm with a multi-resolution feature analysis technique for nodule detection and false-positive reduction.
Initially, nodule candidates were detected by using a template matching approach. Finally, authors have used NCC as a similarity measure for nodule candidate detection. A sensitivity of 90.24% with 4.54 and FPs/scan in LIDC and 84.1% with 5.59 FPs/scan in ANODE09 database has been achieved by the work of Gonga et al. [88]. Farag et al. [112] also have achieved a sensitivity of 85%. It is observed that sensitivity in the range of 84-91% has been achieved by using template matching approach.
The main demerit of template matching approach is the proper choice of metric for matching with the target image. The algorithms, e.g., NNC, SSD, SAD, are mostly time consuming, taking more time to compute the correlation. NCC involves the time-consuming division, square root operations for correlation computation and hence is more complicated as compared with both SSD and SAD algorithms.
I. Filtering-Based Methods
In this approach, different filters have been designed and processed for accurate nodule segmentation and candidate detection. Most of the filters have been designed based on the eigenvalues of the Hessian matrix. Another name of the Hessian matrix is second-moment matrix; derived from the gradient of a function. It is a square matrix and can be decomposed using eigenvalue decomposition; gives a linear combination of three eigenvalues with three eigenvectors. These three components provide explicit structural information about the surfaces, curvedness, and pointedness of an object that can be used for nodule candidate detection. Choi and Choi [48] detected nodule candidates by using a proposed multi-scale dot enhancement filter. Hessian matrix has been used to calculate the dot values of the filter. The proposed system has shown a maximum accuracy of 97.4% with a high sensitivity value of 97.5%. Jaffar et al. [115] also have used multi-scale filter based on the eigenvalues of Hessian matrices for nodule candidate detection. Retico et al. [118] applied a multi-scale dot enhancement filter combined with peak detector algorithm for internal nodule candidate detection. Yokota et al. [129] developed a system that can detect GGO nodules from lung CT images. A 3-D line filter using the Hessian matrix has been used for vessel removal. Next, GGO candidate nodules were detected using density and gradient information. Finally, GGO candidate nodules were segmented using a suitable threshold. Ye et al. [86] have used volumetric shape index map along with the Hessian matrix for nodule detection. The proposed system has achieved a sensitivity of 91.3% for solid nodule and 83.3% for GGO nodule with an average detection rate of 90.2% with 8.2 FPs/scan. Novo et al. [116] also have used Hessian matrix for nodule candidate detection. A new technique named as central adaptive medialness has been used along with the Hessian matrix by their work. This technique has been applied over the bright object regions where the sum of the eigenvalues was less than zero. The main advantage of this method is that it does not require any Gaussian smoothing operation. The method is more sensitive to small nodules. Chen et al. [117] proposed two new features for false-positive reduction. A blob-like structure enhancement (BSE) filter has been introduced for nodule detection. The “blobness” feature was computed by utilizing Hessian matrix and has applied across multi-scale fashion in the lung CT image. It is observed that Hessian-based BSE filter has low sensitivity in the boundary region; results in under-segmentation. Therefore, a segmentation technique named as “fine segmentation” has been introduced for the inclusion of the boundary voxels. A LoG filter has been used in a bounded region for candidate segmentation. Dhara and Mukhopadhyay [57] have investigated a hybrid pre-processing technique using geometry based diffusion (GBD) filter and selective enhancement filter for nodule detection. In the first step, GBD filter has been used for FP reduction. Next, a selective enhancement filter has been used for false-negative reduction. Leemput et al. [62] have proposed a new filter for nodule detection, named as shell filter. After lung segmentation, gray-scale dilation operation was performed with a spherical shell structuring element. Next, voxel wise minimum was computed between the filtered image and original lung segmented image. Finally, candidate regions were detected by taking the difference between the resulted image and the original image.
It is observed that sensitivity in the range of 88.65-97.5% has been achieved by filter-based approach. However, designing and choosing a good filter that can suppress vessel-like structure and enhance blob-like structure is a difficult task.
J. Feature-Based Methods
This approach is composed of computing different features for candidate nodule identification. Shape index (SI) and curvedness (CV) are the two most important features that have been used by feature-based methods. This strategy was originally proposed by Murphy et al. [119], where the researchers first down-sample the image and then apply a single Gaussian filter with σ = 1. Next, the SI and CV features were computed using the principal curvatures k1 and k2 for nodule candidate detection. The researchers have achieved a sensitivity of 80% with an average of 4.2 FPs/scan. Namin et al. [60] also applied shape index feature and achieved a sensitivity of 88% with 10.3FPs/subject. Ye et al. [86] used volumetric shape index feature for nodule candidate detection. Next, dot-like candidate regions were enhanced using a multi-scale dot enhancement filter. The proposed CAD system was able to detect both solid and GGO nodules from the CT image. The researchers have achieved a sensitivity of 91.3% for solid nodule and 83.3% for GGO nodule.
It has been observed that maximum sensitivity was achieved by the work of Ye et al. [86]. Therefore, these features can be combined with some other features, e.g., Hessian matrix based and texture based, for accurate nodule detection.
K. Concept-Based Methods
In this subsection, different concept-based approaches have been discussed for nodule candidate detection. Froz et al. [120] have used a concept based on artificial Life. A hierarchical vector quantization (VQ) scheme was adopted by the work of Han et al. [121]. Concepts based on the “Gestalt Psychology” principle and “Break-and-Repair” can be found in [42, 80]. Froz et al. [120] presented a new algorithm based on the concept of artificial crawlers (AC) and rose diagram (RD) for nodule detection. AC was defined as a model that uses two-dimensional images and lives as an individual in the pixels of gray-level images. The researchers have used candidate curve and template curve by employing the AC algorithm. In their work, nodule and non-nodules were used to generate the candidate curve, and only true nodules were used to generate the template curve. Next, a feature vector was formed by taking the differences between the template and candidate curve and employing area under the curve. Five distance functions, viz., Euclidean, simple matching, Jaccard, Chebyshev, and Manhattan have been used for each template. Next, different features, e.g., mean direction, circular variance, circular standard variation, mean resulting length, kurtosis, and skewness have been computed by employing the RD. Finally, nodules were detected by combining different features computed from the AC model and RD model. Han et al. [121] have introduced a new algorithm for the CADe system based on a hierarchical VQ scheme. Vector quantization was originally used in the field of data compression and signal processing and then become popular in the fields of speech recognition, face detection, image compression, classification, and segmentation, as mentioned by the researchers. In the reported work, the lung fields were segmented using a “high level” VQ methodology. The final lung mask was obtained using connected component analysis, followed by a flood-fill operation for filling up the holes and other disconnected regions. In the second step, a “low level” VQ algorithm has been used for nodule candidate detection and segmentation. Qiu et al. [42] proposed a CADe system based on the Gestalt psychology principle; states that people first focus on things as a whole and then understand the details view of things with the help of global understanding. Researchers have used comprehensive images, obtained by a linear combination of maximum intensity projection (MIP) image and the original lung CT image. Next, the axial, coronal, and sagittal comprehensive CT images have been combined for candidate detection. Pu et al. [80] have introduced a new methodology named as “Break-and-Repair.” The strategy consists of three steps: (1) “modeling,” (2) “break,” and (3) “repair.” In the first stage, the gross ROIs have been extracted and represented as a surface model. After that, non-anatomical structure regions were identified and removed in the second stage. Next, in the third stage, an implicit function has been used for “repairing” (estimating) the removed suspicious regions. In the “break” stage, the principal curvatures and the principal directions were calculated from the triangle mesh of the cube. Candidate nodules have been identified using Koenderink’s shape classification based on a “visibility” testing procedure that followed by curvature analysis. Finally, seed points satisfying the “visible” condition and have classified as convex-cylinder types by the curvature analysis were removed as false positives. Qiu et al. [42] have achieved an accuracy of 91.29%. An overall sensitivity of 82.7% with 4 FPs/scan and 89.2% at 4.14 FPs/scan has been reported in [121] for juxtapleural nodule detection. Pu et al. [80] achieved an overlapping ratio of 69.91 ± 9.43% by using the “Break-and-Repair” strategy.
L. Special Algorithm-Based Methods
This approach is based on designing some specific algorithms for nodule candidate detection. Taşci et al. [123] presented a new CADe system using the α-hull method for juxtapleural nodule detection. The candidate regions are defined as the region between lines obtained by applying α-hull and boundaries obtained by the lung segmentation. The accuracy of the proposed system was 95.88%. Retico et al. [125] reported a CADe system using 3-D directional-gradient concentration (DGC) algorithm followed by a morphological opening operation for nodule detection. Finally, a candidate list was prepared by using a peak detector algorithm. The FP rejection ability of the system was up to 99% as reported by the researchers. Yuan et al. [122] proposed a semi-supervised learning algorithm for automatic detection of GGO nodules from different CT slices. Radiologist only provides a mark in one slice, and the location of the GGO nodule in different adjacent slices will be detected automatically; reduce the time of GGO candidate nodule selection time significantly. Recognition rate of the proposed system was in the range from 91 to 100% as reported by the researchers. Wang et al. [124] reported support vector machine (SVM) classifier based on the 3-D matrix pattern technique that can prevent the loss of structural and local information. The researchers have achieved an overall sensitivity of 98.2% with 9.1 FPs per section. It has been observed that the average sensitivity of the systems using these specific algorithms is quite high and produces good results with higher detection accuracy. Table 1 summarizes different nodule detection systems using feature engineering with reported best performance in reverse chronological order.
Table 1.
Different nodule detection system using feature engineering approach in lung CT images
| Sl. No. | First author with publication year | Nodule detection process | Performance |
|---|---|---|---|
| 1. | Gong et al. (2018) [111] | 3-D tensor-filtering algorithm with local feature analysis followed by 3-D level-set segmentation | Sensitivity = 79.3% at an average of 4FPs/scan in LUNA16 database and sensitivity = 84.62% at an average of 2.8 FPs/scan in ANODE09 database |
| 2. | Farahani et al. (2018) [107] | Kernelized fuzzy c-means (MSFCM) clustering algorithm followed by morphological operation | Accuracy = 96.5%, sensitivity = 93.2%, specificity = 98.1% |
| 3. | Zhang et al. (2017) [70] | 3-D skeletonization based feature, voxel removal rate (VRR) | Avg. accuracy = 93.6%, sensitivity = 89.3% with 2.1 false-positive rate (FPR) per subject |
| 4. | Alam et al. (2017) [113] | Atlas-based segmentation method | Sensitivity = 100% |
| 5. | Narayanan et al. (2017) [102] | Intensity-based thresholding with morphological processing | Specificity of 3 false positives per case/patient on average and sensitivity in CT image is 87.86% |
| 6. | Jaffar et al. (2017) [115] | Multi-scale filter based on the eigenvalues of Hessian matrices | Accuracy = 98.7% and sensitivity = 97.5% |
| 7. | Aresta et al. (2017) [103] |
(1) Solid nodules are detected slice-wise inside a fixed-width sliding window with stride 1 followed by the Otsu threshold and logical OR operation. (2) Non-solid nodules are detected using LoG filter |
Sensitivity is 57.4% with 4 FPs/scan |
| 8. | Yuan et al. (2017) [122] | Semi-supervised based learning algorithm for automatic detection of GGO nodules | Recognition rate is in the range from 91 to 100% |
| 9. | Qiu et al. (2016) [42] | Gestalt psychology principle | Accuracy = 91.29% |
| 10. | Froz et al. (2016) [120] | Artificial crawlers and rose diagram–based method | Mean accuracy (mACC) = 94.30%, mean sensitivity (mSEN) = 91.86%, and mean specificity (mSPC) = 94.78% and mean area under the receiver operating characteristic (mROC) = 0.922 |
| 11. | Mehre et al. (2016) [104] | Combinations of thresholds and structuring elements | Sensitivity = 92.91% with 3 FP/scan |
| 12. | Nithila and Kumar (2016) [106] | Fuzzy c-means clustering with three clusters, viz., low, medium, and high | Accuracy is 98% for solid nodules, 99.5% for part-solid nodule, and 97.2% for non-solid nodules |
| 13. | Javaid et al. (2016) [75] | K-means algorithm | Accuracy = 96.22%, sensitivity = 91.65% with 3.19 FPs per case, and sensitivity = 83.33% for small size nodule |
| 14. | Gonga et al. (2016) [88] | 3-D dot filtering combined with dynamic self-adaptive template matching algorithm | Sensitivity = 90.24% with 4.54 FPs/scan in LIDC dataset, sensitivity = 84.1% with 5.59FPs/scan in ANODE09 dataset |
| 15. | Taşci and Uğur (2015) [123] | α-hull method | Accuracy = 95.88% and area under curve (AUC) = 0.9679 |
| 16. | Leemput et al. (2015) [62] | Shell filter | Overall system score was 0.336 |
| 17. | Novo et al. (2015) [116] | Central adaptive medialness technique with Hessian matrix | Sensitivity = 88.65%, |
| 18. | Yokota et al. (2015) [129] | Density and gradient information | Accuracy = 93.0% |
| 19. | Han et al. (2014) [121] | “Low level” vector quantization (VQ) algorithm | Overall sensitivity = 82.7%,specificity = 4 FPs/scan, and sensitivity = 89.2% at 4.14 FPs/scan for juxtapleural nodules |
| 20. | Ciompi et al. (2014) [133] | Bag-of-Frequencies (BoF) feature descriptor | Area under the ROC curve for spiculation (Az) = 0.907 (for axial),0.903 (for sagittal), and 0.911(for coronal) images |
| 21. | Santos et al. (2014) [68] | Hessian matrix with Tsallis’s and Shannon’s Q entropy features | Accuracy = 88.4%, sensitivity = 90.6%, specificity = 85% and false positives per exam is 1.17 |
| 22. | Filho et al. (2013) [45] | Quality threshold (QT) algorithm | Accuracy = 97.55%, sensitivity = 85.91%, specificity = 97.70%, with a false positive rate of 1.82 per exam and 0.008 per slice and area under the free-response operating characteristic is of 0.8062 |
| 23. | Choi and Choi (2013) [48] | Multi-scale dot enhancement filter with angular histogram of surface normals (AHSN) feature | Accuracy = 97.4%, overall sensitivity = 97.5% with 6.76 FPs/scan, and specificity = 97.7% |
| 24. | Keshani et al. (2013) [99] | ACM segmentation followed by 3-D averaging feature | Overall detection rate is 89% with 7.3 false positives per scan |
| 25. | Wang et al. (2013) [124] | 3-D matrix pattern technique | Overall sensitivity = 98.2% with 9.1 FPs per section |
| 26. | Assefa et al. (2013) [114] | Template matching algorithm with multi-resolution feature analysis technique | The detection rate is 81.212% |
| 27. | Cascio et al. (2012) [67] | Stable 3-D mass-spring model (MSM) is combined with spline curves | The detection rate of the system is 97% with 6.1 FPs/CT. A reduction to 2.5 FPs/CT is achieved at 88% sensitivity |
| 28. | Netto et al. (2012) [108] | Growing neural gas (GNG) clustering algorithm | The methodology ensures that nodules of reasonable size be found with sensitivity = 86%, specificity = 91%, and a mean accuracy of 91% |
| 29. | Choi et al. (2012) [79] | Optimal multiple thresholding and rule-based pruning | 94.1% sensitivity at 5.45 false positives per scan |
| 30. | Soltaninejad et al. (2012) [46] | 2-D stochastic features and 3-D anatomical features | 90% detection rate with 5.63FPs/scan |
| 31. | Dhara and Mukhopadhyay (2012) [57] | Geometry based diffusion (GBD) and selective enhancement filter | Volumetric overlap (VO) mean = 0.98 (solid nodule) and 0.93 for GGO nodule. Hausdorff distance (HD) mean = 9.4 (solid nodule) and 7.8 for GGO nodule |
| 32. | Chen et al. (2012) [117] | Blob-like structure enhancement (BSE) filter followed by an LoG filter | Average TPR = 93.6% with 12.3FPs/case |
| 33. | Farag et al. (2011) [112] | Template matching technique with AAM method | Sensitivity = 85% and specificity = 99% using SIFT feature descriptor |
| 34. | Hosseini et al. (2011) [109] | Gaussian interval type-2 membership functions (IT2MFs) | Average ROC accuracy is 95% with a 99% confidence interval (CI) of [92–99]% |
| 35. | Suárez-Cuenca et al. (2011) [105] | 3-D region-growing algorithm | 71.8% of nodules detected at 0.8 false positives per case, 75.5% at 1.6 FPs/case, and 80% at 3.4 FPs/case, respectively |
| 36. | Antonelli et al. (2011) [110] | Combined the output of three segmentation algorithms (1) robust fuzzy c-means (RFCM), (2) iterative threshold, and (3) region growing | Sensitivity = 92.5% and specificity = 83.5% |
| 37. | Pu et al. (2010) [80] | “Break-and-Repair” technique | RMS error (mm) 1.08 ± 0.45 and overlapping (%) 69.91 ± 9.43 |
| 38. | Messay et al. (2010) [81] | Combined intensity thresholding and morphological processing | Sensitivity = 82.66%, with an average of 3 FPs per CT scan/case |
| 39. | Namin et al. (2010) [60] | Shape index (SI) feature | Sensitivity = 88%, with 10.3 FPs per subject |
| 40. | Sousa et al. (2009) [66] | 3-D skeletonization algorithm | Sensitivity = 84.84%, specificity = 96.15%, and accuracy = 95.21% |
| 41. | Retico et al. (2009) [125] | 3-D directional-gradient concentration (DGC) algorithm, followed by a morphological opening operation | Sensitivity = 72% and the FP rejection ability of the system is up to 99% |
| 42. | Murphy et al. (2009) [119] | Shape index and curvedness features | Sensitivity = 80%, with an average of 4.2 FPs per scan |
| 43. | Ye et al. (2009) [86] | Shape index map and dot map | Average detection rate of 90.2%, with approximately 8.2 FP/scan |
| 44. | Retico et al. (2009) [118] | Dot enhancement filter and the “normals” to the pleura surface technique | Sensitivity in the 80–85% range is achieved with an average number of 6–9 FP per scan in their dataset and 67% sensitivity is achieved at 8 FP per scan in ANODE09 database |
| 45. | Choi and Choi (2009) [78] | Thresholding and shape information | 85.11% detection rate with 1.13 FPs per scan |
Feature Extraction and False-Positive Reduction
In this stage, different intensity-based, morphological /shape-based, and texture-based features are extracted as feature vector from the detected nodule candidate regions and fed to a classifier for true nodule detection. Generally, nodules have a circular shape in 2-D CT images and spherical shapes in 3-D CT image, i.e., nodules have a compact structure. It follows Gaussian distribution and the region is more or less convex. Therefore, different geometrical and intensity-based features have been extracted for true nodule detection. A list of features is available in [75, 81, 121, 130-132] used for nodule detection. Sousa et al. [66] applied different morphological, textural, and intensity-based features for nodule detection. Messay et al. [81] identified nodules by extracting 245 features from candidate nodules for false-positive reduction. After feature extraction, features were selected using sequential forward selection (SFS) technique. Finally, Fisher linear discriminant (FLD) classifier and a quadratic classifier have been used for nodule detection. Choi and Choi [78] have used principal component analysis (PCA) for false-positive reduction. Narayanan et al. [102] computed a list of 503 features for nodule detection. Mehre et al. [104] have reduced false positives by using different structural and intensity-based statistical features. Different structural features, e.g., XY-width, Z-extent, elongation, cube compactness, pondered radial distance, and Boyce-Clark radial shape index have been used by their work. Choi and Choi [79] have extracted a set of 2-D and 3-D features for nodule detection purposes. Filho et al. [45] have used two diversity indexes, viz., Simpson’s index and Shannon’s index as features for nodule detection. Javaid et al. [75] grouped nodules into six classes by using their proposed equations, viz., “nodule thickness” and “% of wall connectivity.” Next, different features were extracted specific to tiny, small, and big group nodules. Tiny and small group nodules were classified using if-then rules. An SVM with radial basis function (RBF) kernel was used for big nodules classification. Nithila and Kumar [106] computed different textural features from the candidate nodules. First-order statistics (FOS), second-order statistics (SOS), and gray-level co-occurrence matrix (GLCM) have been used as textural features for nodule detection. Chen et al. [117] proposed two new features, viz., “Neighbourhood Feature 1” and “Neighbourhood Feature 2” for false-positive reduction. Feature 1 was computed by taking the ratio between nodule candidate region and the surrounding region. If the value was less than or equal to one, then the candidate nodule will be treated as FP. Retico et al. [118] computed Gradient and Hessian matrix-based features for nodule detection. Choi and Choi [48] proposed a new feature, angular histogram of surface normals (AHSN), for nodule detection. In the lung images, a “wall” is a pleuron or part of another big object near the target. Researchers have proposed a wall detection and elimination algorithm with the help of AHSN feature. Authors have reported in their work that an improvement in accuracy has been achieved by applying the “wall” detection algorithm. Zhang et al. [70] have introduced a new feature named as voxel remove rate (VRR) for nodule detection. The 3-D VRR feature was computed along with four gray features, viz., mean value, standard deviation, skewness, and kurtosis, and five shape-based features, viz., effective radius, elongation, compactness, 1st normalized contour sequence moment, and 3rd normalized contour sequence central moments for nodule identification. Jaffar et al. [115] used multi-coordinate histogram of oriented gradients (MCHOG) feature descriptor and Intensity-based statistical features (IBSF) to discriminate nodules from non-nodules. Froz et al. [120] computed mean direction, circular variance, circular standard variation, mean resulting length, kurtosis, and skewness features for nodule detection. Gonga et al. [88] have used 11 sets of features with Fisher’s linear discriminant analysis (FLDA) classifier for false-positive reduction. Taşci and Uğur [123] have calculated 40 features for false-positive reduction. A feature selection method based on the ranking of features has been carried out after feature extraction. Ten classifiers were evaluated by using these features. Finally, 22 features were selected from 40 features by using a generalized linear model regression (GLMR) classifier. Han et al. [121] have reduced false positives by using rule-based filtering operation. Ciompi et al. [133] investigated a new feature, named as Bag-of-Frequencies (BoF). This feature was computed by analyzing the image intensity in a 3-D spherical neighborhood of a voxel of interest. BoF feature has been applied in two aspects: (1) the discrimination of ROI (nodule) with false positives, e.g., vessels and bifurcation points and (2) the characterization of the spiculated nodule. Intensity profiles were sampled in a multi-scale fashion on spherical surfaces from the center of the nodule. After that, a Fourier transform was used on each intensity profile to obtain a spectrum. Finally, “Bag-of Frequencies” (descriptor) have been created from the spectra. Santos et al. [68] used two texture descriptors, viz., Tsallis’s and Shannon’s Q entropy from the initial nodule candidate set for false-positive reduction. Assefa et al. [114] applied a multi-resolution technique for false-positive reduction. The researchers have used one 2-D separable scaling function along with three separable 2-D wavelets, viz., horizontal, vertical, and diagonal for nodule detection. Cascio et al. [67] computed different 2-D and 3-D features for false-positive reduction. Netto et al. [108] used a 3-D distance transformation for false-positive reduction. Finally, different shape- and intensity-based features have been extracted for nodule detection. Soltaninejad et al. [46] have used 2-D stochastic features and 3-D anatomical features for nodule detection and classification. 3-D averaging was used to differentiate between nodule and vessel. Active contour model has been used for nodule contour extraction that also acts as an FP reducer. Finally, detected small nodules were visualized using the surface rendering (SR) technique. Farag et al. [112] applied two geometric feature descriptors, viz., scale-invariant feature transform (SIFT) and local binary patterns (LBP) for false-positive reduction. Suárez-Cuenca et al. [105] have calculated 18 features for nodule classification. Finally, six classifiers, viz., LDA, QDA, ANN, SVM-dot, SVM-poly, and SVM-ANOVA, were combined using five combination methods. It is reported in their work that the majority-vote rule gives the highest performance among other combination methods. Namin et al. [60] used different intensity features combined with morphological features, e.g., volume size, sphericity, elongation, border variation, and effective radius for false-positive reduction. Murphy et al. [119] applied two successive K-nearest neighbor (KNN) classifiers for false-positive reduction.
Classification
This is the final stage for any CADe system. The output of this stage is yes/no decision. In this stage, recognition of lung nodule has been done with the help of different classifiers. Sensitivity, specificity, and accuracy are the three main parameters to measures the performance of the CADe-based system. In Table 1, the performance of different proposed CADe systems has been given. The choice of a good classifier is an important aspect for any CADe system and varies with the design approach of the system. Most of the works have used SVM as a classifier for nodule detection. Vapnik [134] introduced the original SVM algorithm for classification problems. An SVM finds the maximum-margin hyper-plane that divides the data points into different distinct regions. SVM was used by the works of Choi and Choi [48], Sousa et al. [66], Zhang et al. [70], Han et al. [121], Keshani et al. [99], and Wang et al. [124]. SVM with radial basis function (RBF) kernel has been used in Javaid et al. [75], Froz et al. [120], Aresta et al. [103], Mehre et al. [104], Han et al. [121], and Netto et al. [108]. Qiu et al. [42] used SVM with radial basis Gaussian kernel function. Wang et al. [124] used SVM as a 3-D matrix pattern. Ye et al. [86] have used a weighted SVM. Apart from SVM, Nithila and Kumar [106] have used a neural network (NN). Retico et al. [118] applied NN with 12 input, 14 hidden layers, and 1 output unit, trained with the back-propagation learning algorithm for nodule detection. The works of Farahani et al. [107], Taşci and Uğur [123], Suárez-Cuenca et al. [105], and Antonelli et al. [110] can find ensemble of classifiers for nodule detection. Choi et al. [79] presented a genetic program (GP) technique to optimize the classifier parameters. Gong et al. [111] used a random forest-based classifier. Leemput et al. [62] also have used a random forest classifier with 100 decision trees. Messay et al. [81] have used FLD classifier and quadratic classifier. The authors reported a performance comparison study between the two classifiers and finally, FLD classifier was selected for nodule detection. Murphy et al. [119] used a KNN classifier. Retico et al. [125] have used a voxel-based neural approach (VBNA) based classifier for nodule detection.
Deep Learning-Based Approach Towards Lung Nodule Detection
Deep learning algorithms have become a valuable tool in the field of medical imaging, used for lesion detection, characterization, and analysis. This methodology consists of designing layer wise network architecture by keeping in mind the goal of higher classification accuracy. A survey on deep learning on different network architecture with applications in medical imaging can be found in [135]. Wong et al. [136] and Bruijne [137] presented recent developments and challenges of machine learning in medical imaging. Due to large variations and complexity in biomedical images, it is very difficult to recognize medical objects such as lesions and anatomies from the input image. Therefore, instead of searching for good handcraft-based features, emphasis has been given to the automated feature learning approach called “learning from examples.” In this technique, computers will learn the features automatically from the examples given as input. The learning methods have classified as supervised and unsupervised, in the view of machine learning paradigm. Auto-encoders (AEs) and stacked auto-encoders (SAEs) are examples of unsupervised network models because the labeled training data is absent and only the input image has given to the network. On the other hand, CNNs are the example of supervised network model [135]. Labeled training data is mandatory for CNN architecture. CNN and massive training artificial neural network (MTANN) are the two most recent approaches used in deep learning. Both models use pixel values in images directly as input information, instead of features calculated (handcrafted) from segmented ROIs [138]. A good comparison study of these two end-to-end computing techniques, viz., MTNN and CNN, can be found by the work of Tajbakhsh and Suzuki [139] for nodule detection. However, CNN is the most popular reference architecture used in medical image analysis. From Table 2, it is observed that almost all works have used CNN as the reference architecture for lung nodule detection and characterization purpose. Therefore, a precise and brief introduction to CNN in the context of deep learning has been presented in the “CNN as a Deep Learning Tool” section.
Table 2.
Different nodule detection and characterization system using CNN-based deep learning approach in lung CT images
| Sl. No. | First author with publication year | Method | Purpose | Performance |
|---|---|---|---|---|
| 1. | Paul et al. (2018) [37] | Pre-trained vgg-s network with merge CNN | Nodule characterization | Accuracy = 76.79%, ROC = 0.87 |
| 2. | Liu et al. (2018) [157] | Multi-view multi-scale CNN | Nodule categorization | Overall accuracy = 92.3% (LIDC dataset) and 90.3% (ELCAP dataset) |
| 3. | Yuan et al. (2017) [155] | Pre-trained vgg network followed by CNN-based features fused with handcraft-based features (multi-view multi-scale CNN) | Nodule categorization | Overall accuracy = 93.1% (LIDC dataset) and 93.9% (ELCAP dataset) |
| 4. | Xie et al. (2017) [156] | Applied CNN at decision level | Nodule characterization | AUC of 96.65%, 94.45%, and 81.24% |
| 5. | Silva et al. (2017) [154] | Genetic algorithm-based CNN | Nodule characterization | Sensitivity = 94.66%, specificity = 95.14%, accuracy of 94.78%, and area under the ROC curve of 0.949. |
| 6. | Wang et al. (2017) [153] | Data-driven-based machine learning model using central focused convolutional neural network (CF-CNN) | Nodule segmentation | Dice score = 82.15% (LIDC dataset) and 80.02% (GDGH dataset) |
| 7. | Dou et al. (2016) [152] | Three 3-D CNN applied with fusion technique | Nodule detection | Sensitivity = 94.4% and sensitivity = 92.2% at 8FPs/scan |
| 8. | Shen et al. (2016) [149] | Multi-crop convolutional neural network (MC-CNN) | Nodule characterization | Accuracy = 87.14%, AUC = 0.93, sensitivity = 77%, specificity = 93% |
| 9. | Setio et al. (2016) [150] | Multiple streams of 2-D ConvNets (multi-view architecture) | Nodule detection | Sensitivity = 85.4% with 1 FP/scan and 90.1% with 4 FPs/scan |
| 10. | Ciompi et al. (2015) [151] | 2-D convolution-based architecture named as “OverFeat” | Peri-fissural nodules (PFNs) detection | Area under curve (AUC) = 0.868 |
CNN as a Deep Learning Tool
This type of architecture initially appeared in the works of Hasegawa et al. [140], Lin et al. [141], and Lo et al. [142]. Lin et al. [141] have applied CNN for FP reduction and achieved a cited accuracy. However, owing to limited computational resources, CNN did not receive popularity at that time. In recent years, with the advent of powerful graphics processing unit (GPUs) such as TitanX (8 GB, 12 GB), CNNs have gained popularity, especially in the field of biomedical image processing. CNN is the most dominated architecture in deep learning and acts as a powerful feature extractor and classifier. The convolution operation is the heart of the CNN architecture. The layers adapting the “convolution operation” are called convolutional layer and responsible for detecting local features in all locations of the input image. CNNs are like multi-layer perceptrons (MLPs) but there are two key differences. First, in CNNs, weights in the network are shared and second, a typical CNN may contain several pooling layers, which is absent in MLPs. Therefore, in CNN, fewer weight parameters require as compared with MPL. The detail of convolution operations and working principle of CNN architecture is available in Dumoulin and Visin [143]; Lecture Series and tutorials from Stanford University [144, 145]. Several CNN-based architecture exists in literature, e.g., the LeNet (1998), AlexNet (2012), VGG16/19 (2014), GoogLeNet/Inception, and ResNet. LeNet and AlexNet are examples of shallow network as it consists of two and five layers, respectively. On the other hand, GoogLeNet and ResNet architecture are examples of deep networks. The CNN architecture used for nodule detection and characterization is somehow deep. Tajbakhsh and Suzuki [139] have mentioned that increasing the depth of the architecture with large training dataset gives better performance. Authors have applied five CNNs separately, viz., a shallow CNN (sh-CNN), the LeNet architecture, a relatively deep (rd-CNN), AlexNet, and FT AlexNet over lung dataset for nodule detection and characterization. FT Alexnet with a large training dataset has given the best performance from the rest of the CNNs for lung nodule detection. Apart from detection, ROIs can be segmented using CNN. Jiang et al. [146] address the medical image semantic segmentation problem by applying the modern CNN-based model. Yan et al. [147] performed a comparative study of slice-level 2-D CNN, nodule-level 2-D CNN, and nodule-level 3-D CNN for nodule detection. Tan et al. [148] have addressed the recent progress and challenges in CNN.
Works Based on CNN Architecture
Recently, deep learning approaches are applying widely owing to its higher detection and classification accuracy. In this subsection, we describe different CNN-based works found so far in the period 2009-April 2018 for nodule detection, categorization, and characterization. The works have been categorized based on (1) type of basic convolution operations used to implement network such as 2-D [149-151] or 3-D [152] or both [153], (2) evolutionary technique applied for CNN parameter optimization [154], (3) combining/fusing feature engineering features with automatic extracted CNN-based features [155, 156], and (4) use of pre-trained network combined with CNN [37]. Table 2 shows the performance of different proposed systems.
2-D Convolution-Based CNN
In this approach, a basic 2-D convolution operation has been used to find local features from the entire image. This is mostly used CNN architecture for its simplicity and computational efficiency. Most of the images are two-dimensional and contain intensity values in the form of a matrix. Several 2-D filters/features are automatically learned from the training dataset by this network. Setio et al. [150] used multiple streams of 2-D ConvNets. Initially, three candidate detector algorithms specially designed for solid, sub-solid, and large nodules have combined for nodule candidate detection. Then for each candidate, a set of 2-D patches of size 64 × 64 pixels have been extracted from differently oriented planes. The researchers have used nine views for this purpose. Then multiple 2-D ConvNets were used for feature extraction. Finally, the outputs of different networks have been merged by applying three fusion techniques, viz., committee fusion, late fusion, and mixed fusion. The authors have reported that late fusion technique gives better detection performance in comparison with committee fusion and mixed fusion techniques. Ciompi et al. [151] proposed a 2-D CNN architecture named as “OverFeat” for peri-fissural nodule detection. The architecture used six convolutional layers, with filter size ranges from 7 × 7 to 3 × 3. Next, three view images (axial, coronal, and sagittal) were fed to the “OverFeat” architecture for nodule feature extraction. Finally, researchers have used a two-stage classifier for peri-fissural nodule detection. Shen et al. [149] have designed a multi-crop convolutional neural network (MC-CNN) by introducing a “multi-crop” pooling strategy for automatic lung nodule characterization. Researchers have used a single network that was capable of producing multi-scale features, instead of using multiple CNN. They proposed a multi-crop-based pooling operation strategy. The multi-crop pooling strategy consists of three pooling operations. Input to the first pooling operation was convolutional features R0, obtained either from the original input image or from the pooled features. R1 was the center region cropped from R0, and R2 was the center region cropped from R1. After that, R0 has max pooled twice and R1 has max pooled one time to generate the pooled feature maps f0 and f1, respectively. R2 serves as f2 without a pooling operation. The final multi-crop features were computed by concatenating f0, f1, and f2. The work has used randomized leaky rectified linear units (RReLU) as an activation function for SGD learning algorithm that minimize the cross-entropy loss. The proposed architecture was also able to perform semantic label prediction by computing two attributes, viz., nodule subtlety and margin, and can estimate nodule diameter with promising results.
3-D Convolution-Based CNN
In this network, the basic convolution operation has been applied in three directions (x, y, z) at the same time. The convolutional filters used are 3-D, and have applied over the input 3-D data for automatic feature extraction. These networks are more expensive in terms of computation efficiency than 2-D-based network. Convolution operations based on 3-D need more calculations and more memory because 3-D matrix is required to store the extracted features in computer memory. The main advantage of 3-D CNN is that it produces multi-view features with the help of 3-D filters. Dou et al. [152] proposed a 3-D CNN-based architecture for lung nodule detection. Researchers have used three 3-D CNNs to encode spatial information and representative features using hierarchical architecture. For the first architecture, a receptive field of size 20 × 20 × 6 has been used, for second architecture, the size was 30 × 30 × 10, and for third architecture, it was 40 × 40 × 26. Finally, the three CNNs have been merged and act as a feature extractor for nodule detection. The proposed architecture was validated by participating in LUng Nodule Analysis 2016 (LUNA16) challenge and achieved a sensitivity of 94.4%, for nodule detection.
2-D-3-D-Based CNN
The 2-D and 3-D convolution operations can be combined and used to design a hybrid network; we named it as 2-D-3-D CNN. The network is capable of capturing both 2-D and 3-D features from the lung nodule image. In general, the 3-D patch branch is capable of learning multi-view features from multiple CT slices whereas the 2-D patch branch learns multi-scale features. Wang et al. [153] developed a central focused convolutional neural network (CF-CNN) learning model by emphasizing nodule segmentation. The architecture consists of two paths, viz., 3-D CNN-based path and a 2-D CNN-based path. This architecture is a good example of patch-based nodule segmentation approach. Variations in the lesion have been captured using both 2-D and 3-D features from the dataset. The voxels close to center of the image patch were more relevant to the target voxel than the voxels close to edges. Owing to this reason, the researchers have proposed a centrally focused pooling operation. This type of pooling operation reserves location information and many features around the patch center that were not captured in conventional max pooling operation. In central pooling operation, the pooling kernel size varies according to the pooling position and non-uniformly distributed on an input image. On the other hand, in traditional max pooling operation, the pooling kernels are of the same size and uniformly distributed. A parametric rectified linear unit (PReLU) has been used as activation function for more effective and promising results than the conventional ReLU. Researchers have achieved a dice score of 82.15% in LIDC dataset.
Evolutionary Algorithm-Based CNN
This type of network uses evolutionary-based algorithms to optimize different network parameters. Silva et al. [154] designed an evolutionary-based CNN for lung nodule characterization. A good pre-processing step was carried out before applying the classifier. Initially, the Otsu algorithm based on PSO has been applied over the dataset. The goal of this step is to divide each nodule into two sub-regions with maximum inter-class variance. Next, the two-dimensional CT slices were resized and used as an individual sample and three databases each having 21,631 slices have been prepared. Finally, three CNNs were used for nodule characterization. Genetic algorithm was used to optimize the CNN hyperparameters in the convolutional layer. Researchers also have chosen an optimum number of neurons in the fully connected layers with the help of GA. The fitness function has been defined in terms of sensitivity, specificity, and accuracy. A higher weight value was assigned to the sensitivity metric so that the network can classify malignant nodules effectively. The researchers have reported a sensitivity of 94.66% for nodule characterization.
Fusion-Based CNN
In this network, handcraft-based features can be combined with the CNN-based extracted features to form the final feature vector. We have named the network, using this approach, as fused-based CNN. Yuan et al. [155] have categorized five types of lung nodule, viz., well-circumscribed, juxtapleural, juxtavascular, pleural tail, and GGO using CNN. Researchers have used a multi-view multi-scale CNN pre-trained with Visual Geometry Group (VGG) network. Different statistical features were extracted from the nodule image. Next, SIFT feature descriptor was computed and encoded in the form of Fisher vector (FV). The Gaussian mixture model has been used to compute the Fisher geometrical features. Finally, all the handcrafted features were fused with CNN extracted features by using multiple kernel learning (MKL). Yuan et al. [155] achieved an overall accuracy of 93.1%. Liu et al. [157] applied almost the same approach for nodule categorization. Xie et al. [156] have fused handcrafted textural and shape-based features with automated deep features (Fuse-TSD) by using deep convolution neural network for nodule characterization (Fig. 9). The fused features were GLCM texture (T), Fourier shape descriptor (S), and deep learning-based features (D). The authors fused the extracted features in the decision level (decision fusion) which was the output of a fully connected layer. The same features were also fused on the feature extractor level (feature fusion), i.e., before feeding to the classifier. Authors have reported that fusion at decision level was given better performance than at the feature level. Therefore, Fuse-TSD can be used as a higher level decision fusion-based classifier over both manual and automated segmentation process.
Fig. 9.
A CNN-based nodule characterization framework that fuses handcraft features with CNN-based features (Xie et al. [156])
Pre-trained CNN
“Transfer Learning” is one most important concepts in deep learning. In this scenario, the CNN is trained on a large and different dataset. The final weights of the CNN were fined tuned and then used for object detection or classification. This is very helpful as the availability of the medical imaging data is limited, so a network trained with different datasets and modality can be used for the task of deep feature extraction. Paul et al. [37] proposed a new CNN architecture (Fig. 10) for lung nodule characterization. Researchers have used a pre-trained VGG network trained with the Imagenet dataset. Three networks, viz., vgg-f, vgg-m, and vgg-s, have been used for this purpose. Here, the f, m, and s stand for fast, medium, and slow and refer to training time. Vgg-s has been chosen as final pre-trained architecture, as it gives higher classification accuracy. Next, 4096 deep features have been extracted by using vgg-s. Next, three CNN architectures have been trained independently by using the cancerous dataset from NLST. Next, “Architecture-3,” a cascaded CNN, has been chosen for its higher classification accuracy and 1024 deep features were extracted. Finally, nodules were characterized by using all the deep features extracted by using “Architecture-3,” fused with 219 radiomics (handcrafted/feature engineering) features for higher classification accuracy. Researchers have reported an accuracy of 76.79% using pre-trained CNN.
Fig. 10.
A CNN-based nodule characterization framework that pre-trained with VGG network (Paul et al. [37])
It has been observed that different researchers have emphasized different aspects, e.g., some of them on nodule detection, some on nodule characterization, and others have used for FP reduction, by using CNN as basic network architecture. Table 2 gives different CNN-based works in reverse chronological order.
Conclusion
Although there are many studies with the same purpose as described in this paper, it may conclude that a definitive methodology does not exist currently, that could be considered efficient in every aspect of lung nodule detection. Failure of different methodologies include in the parenchyma extraction stage, accurate nodule segmentation stage, e.g., segmentation of juxtavascular nodule, and mostly in the false-positive reduction stage. Apart from that, nodule characterization is very important for cancer studies. Different algorithms may be proposed for nodule characterization based on growth rate, which will ultimately help early detection of cancer and its diagnosis. Automatic analysis of lung cancer staging is another vital finding for a patient. Unfortunately, very few works with significant success rates reported in this direction [22]. In summary, other alternatives must be investigated and at the same time, previous works should be revisited so that an increase in 5-year survival rate, (which is at present only 10-15%) may be achieved.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. [Online] http://www.who.int/en/. Accessed 05 Sept 2017
- 2.Global Cancer Observatory. [Online] http://gco.iarc.fr/. Accessed 12 Oct 2017
- 3.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clinics in Chest Medicine. 2011;32(4):605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ABID Larbi: Lung cancer around the world and Arab countries. AMAAC Workshop, Algiers2011
- 5.American Cancer Society (ACS). [Online] https://www.cancer.org/. Accessed 20 Dec 2018
- 6.National Cancer Centre Singapore. [Online] https://www.nccs.com.sg/Pages/Home.aspx/. Accessed 12 Oct 2017
- 7.National Cancer Institute (INCA). [Online] https://www.inca.gov.br/. Accessed 16 Oct 2017.
- 8.Fleischner Society. [Online] https://fleischner.memberclicks.net/. Accessed 16 Oct 2017
- 9.Kostis WJ, Reeves AP, Yankelevitz DF, Henschke CI. Three-dimensional segmentation and growth-rate estimation of small pulmonary nodules in helical CT images. IEEE Transaction on Medical Imaging. 2003;22(10):1259–1274. doi: 10.1109/TMI.2003.817785. [DOI] [PubMed] [Google Scholar]
- 10.Golan R, Jacob C, Denzinger J: Lung nodule detection in CT images using deep convolutional neural networks. In: IEEE International Joint Conference on Neural Networks (IJCNN), pp. 243–250, 2016. 10.1109/IJCNN.2016.7727205
- 11.Cancer Research UK. [Online] https://www.cancerresearchuk.org/. Accessed 12 Oct 2017
- 12.Godoy MC, Truong MT, Carter BW, Viswanathan C, De Groot P, Ko JP. Pitfalls in pulmonary nodule characterization. Seminars in Roentgenology. 2015;50(3):164–174. doi: 10.1053/j.ro.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 13.National Lung Screening Trial (NLST).[Online]https://www.cancer.gov/types/lung/research/nlst/. Accessed 25 Oct 2017
- 14.MacMahon H, Austin JH, Gamsu G, Herold CJ, Jett JR, Naidich DP, Patz EF, Jr, Swensen SJ. Guidelines for management of small pulmonary nodules detected on CT scans: a statement from the Fleischner Society. Radiology. 2005;237(2):395–400. doi: 10.1148/radiol.2372041887. [DOI] [PubMed] [Google Scholar]
- 15.Lederlin M, Revel MP, Khalil A, Ferretti G, Milleron B, Laurenta F. Management strategy of pulmonary nodule in 2013. Diagnostic and Interventional Imaging. 2013;94(11):1081–1094. doi: 10.1016/j.diii.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Rena O, Davoli F, Boldorini R, Roncon A, Baietto G, Papalia E, Turello D, Massera F, Casadio C. The solitary pulmonary nodule in patients with previous cancer history: results of surgical treatment. European Journal of Surgical Oncology. 2013;39(11):1248–1253. doi: 10.1016/j.ejso.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Dhara AK, Mukhopadhyay S, Khandelwal N. Computer-aided detection and analysis of pulmonary nodule from CT images: a survey. Technical Review IETE. 2012;29(4):265–275. [Google Scholar]
- 18.Attili AK, Kazerooni EA: Imaging of the solitary pulmonary nodule. In: Evidence-Based Imaging. New York: Springer, 2006. 10.1007/0-387-31216-1_23
- 19.Sharma SK, Mohan A: Solitary Pulmonary Nodule: How and How Much to Investigate? The Association of Physicians of India, Ch. 109:824–832, medicine update, 2008
- 20.Brandman S, Ko JP. Pulmonary nodule detection, characterization, and management with multi detector computed tomography. Journal of Thoracic Imaging. 2011;26(2):90–105. doi: 10.1097/RTI.0b013e31821639a9. [DOI] [PubMed] [Google Scholar]
- 21.De Hoop B, Gietema H, Van de Vorst S, Murphy K, Van Klaveren RJ, Prokop M. Pulmonary ground-glass nodules: increase in mass as an early indicator of growth. Radiology. 2010;255(1):199–206. doi: 10.1148/radiol.09090571. [DOI] [PubMed] [Google Scholar]
- 22.Sluimer I, Schilham A, Prokop M, Van Ginneken B. Computer analysis of computer tomography scan of the lung: a survey. IEEE Transactions on Medical Imaging. 2006;25(4):385–405. doi: 10.1109/TMI.2005.862753. [DOI] [PubMed] [Google Scholar]
- 23.Doi K: Computer-aided diagnosis in radiological imaging: current status and future challenges. In: SPIE Proceedings of Medical Imaging .7497, pp 1A-1–1A-8, 2009. 10.1117/12.851178
- 24.Devaki K, MuraliBhaskaran V: Study of computed tomography images of the lungs: a survey. In: IEEE International Conference on Recent Trends in Information Technology (ICRTIT), pp 837–842, 2011. 10.1109/ICRTIT.2011.5972308
- 25.Lee SLA, Kouzani AZ, Hu EJ. Automated detection of lung nodules in computed tomography images: a review. Machine Vision and Applications. 2010;23(1):151–163. [Google Scholar]
- 26.Valente IR, Cortez PC, Neto EC, Soares JM, De Albuquerque VH, Tavares JM. Automatic 3D pulmonary nodule detection in CT images: a survey. Computer Methods and Programs in Biomedicine. 2015;124:91–107. doi: 10.1016/j.cmpb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Lee N, Laine AF, Mrquez G, Levsky JM, Gohagan JK. Potential of computer-aided diagnosis to improve CT lung cancer screening. IEEE Reviews in Biomedical Engineering. 2009;2:136–146. doi: 10.1109/RBME.2009.2034022. [DOI] [PubMed] [Google Scholar]
- 28.LIDC-IDRI. [Online] https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI. Accessed 16 Oct 2017
- 29.Armato III Samuel G., McLennan Geoffrey, McNitt-Gray Michael F., Meyer Charles R., Yankelevitz David, Aberle Denise R., Henschke Claudia I., Hoffman Eric A., Kazerooni Ella A., MacMahon Heber, Reeves Anthony P., Croft Barbara Y., Clarke Laurence P. Lung Image Database Consortium: Developing a Resource for the Medical Imaging Research Community. Radiology. 2004;232(3):739–748. doi: 10.1148/radiol.2323032035. [DOI] [PubMed] [Google Scholar]
- 30.Tan M, Deklerck R, Cornelis J, Jansen B. Phased searching with NEAT in a time-scaled framework: experiments on a computer-aided detection system for lung nodules. Artificial Intelligence in Medicine. 2013;59(3):157–167. doi: 10.1016/j.artmed.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Gupta A, Märtens O, Moullec YL, Saar T: A tool for lung nodules analysis based on and morphological pperation. In: IEEE International Symposium on Intelligent Signal Processing Intelligent Signal Processing (WISP) Proceedings, pp 1–5, 2015. 10.1109/WISP.2015.7139186
- 32.Armato SG, III, et al. The lung image database consortium (LIDC) and image database resources initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical Physics. 2011;38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock MC, Magnan JF: Predictive capabilities of statistical learning methods for lung nodule malignancy classification using diagnostic image features: an investigation using the Lung Image Database Consortium dataset. In: SPIE Proceedings of Medical Imaging, 10134, 25-1–25-12, 2017. 10.1117/12.2254446
- 34.Han F, Song B, Ma H, Qian W, Liang Z: Risk prediction of small pulmonary nodules based on novel CT image texture markers. In: SPIE Proceedings of Medical Imaging, 10134, 3Q-1–3Q-6, 2017. 10.1117/12.2253486
- 35.Hancock MC, Magnan JF. Lung nodule malignancy classification using only radiologist- quantified image features as inputs to statistical learning algorithms: probing the Lung Image Database Consortium dataset with two statistical learning methods. SPIE Journal of Medical Imaging. 2016;3(4):044504-1-044504-1. doi: 10.1117/1.JMI.3.4.044504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messay T, Hardie RC, Tuinstra TR. Segmentation of pulmonary nodules in computed tomography using a regression neural network approach and its application to the Lung Image Database Consortium and Image Database Resource Initiative dataset. Medical Image Analysis. 2015;22(1):48–62. doi: 10.1016/j.media.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Paul R, Hawkins SH, Schabath MB, Gillies RJ, Hall LO, Goldgof DB. Predicting malignant nodules by fusing deep features with classical radiomics features. SPIE Journal of Medical Imaging. 2018;5(1):011021-1–011021-11. doi: 10.1117/1.JMI.5.1.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Ginneken B, et al. Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: the ANODE09 study. Medical Image Analysis. 2010;14(6):707–722. doi: 10.1016/j.media.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 39.TIME. [Online] http://cmp.felk.cvut.cz/projects/LungTIME/, 2009. Accessed 14 Nov 2017
- 40.ELCAP. [Online] http://www.via.cornell.edu/databases/lungdb.html/, 2013. Accessed 25 Nov 2017
- 41.LISS. [Online] http://isc.cs.bit.edu.cn/MLMR/LISS.html/, 2015. Accessed 26 Nov 2017
- 42.Qiu S, Wen D, Cui Y, Feng J. Lung Nodules Detection in CT Images Using Gestalt-Based Algorithm. Chinese Journal of Electronics IET. 2016;25(4):711–718. [Google Scholar]
- 43.Sangamithraa PB, Govindaraju S: Lung tumour detection and classification using EK- mean clustering. In: IEEE International Conference on Wireless Communications,Signal Processing and Networking (WiSPNET), pp 2201–2206, 2016. 10.1109/WiSPNET.2016.7566533
- 44.Aggarwal T, Furqan A, Kalra K: Feature extraction and LDA based classification of lung nodules in chest CT scan images. In: IEEE International Conference on Advances in Computing, Communications and Informatics (ICACCI), pp 2201–2206, 2015. 10.1109/ICACCI.2015.7275773
- 45.De Carvalho Filho AO, De Sampaio WB, Silva AC, De Paiva AC, Nunes RA, Gattass M. Automatic detection of Solitary lung nodules using quality threshold clustering, genetic algorithm and diversity index. Artificial Intelligence in Medicine. 2013;60(3):165–177. doi: 10.1016/j.artmed.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Soltaninejad S, Keshani M, Tajeripour F: Lung nodule detection by KNN classifier and active contour modelling and 3D visualization. In: IEEE International Symposium on Artificial Intelligence and Signal Processing (AISP), pp 440–445, 2012. 10.1109/AISP.2012.6313788
- 47.Sun S, Gu W, Fan L, Ren H: Detect GGO nodule based on dot filter. In: IEEE International Conference on Biomedical Engineering and Biotechnology (iCBEB), pp 765–767, 2012. 10.1109/iCBEB.2012.129
- 48.Choi WJ, Choi TS. Automated pulmonary nodule detection based on three-dimensional shape- based feature descriptor. Computer Methods and Programs in Biomedicine. 2013;113(1):37–54. doi: 10.1016/j.cmpb.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Nagata R, Kawaguchi T, Miyake H: Automated detection of lung nodules in chest radiographs using a false-positive reduction scheme based on template matching. In: IEEE International Conference on BioMedical Engineering and Informatics (BMEI), pp 216–223, 2013. 10.1109/BMEI.2012.6512916
- 50.Jirapatnakul AC, Fotin SV, Reeves AP, Biancardi AM, Yankelevitz DF, Henschke CI: Automated nodule location and size estimation using a multi- scale Laplacian of Gaussian filtering approach. In: IEEE Conference on Engineering in Medicine and Biology Society, pp 1028–1031, 2009. 10.1109/IEMBS.2009.5334683 [DOI] [PubMed]
- 51.Kubo M et al.: A cad system for lung cancer based on low dose single-slice CT image. In: SPIE Proceedings of Medical Imaging 4684:1262–1269, 2002. 10.1117/12.467086
- 52.Shao H, Cao L, Liu Y: A detection approach for solitary pulmonary nodules based on CT images In: IEEE Proceedings of 2012 2nd International Conference on Computer Science and Network Technology, pp 1253 1257, 2013. 10.1109/ICCSNT.2012.6526151
- 53.Punithavathy K, Ramya MM, Poobal S: Analysis of statistical texture features for automatic lung cancer detection in PET/CT images. In: IEEE International Conference on Robotics, Automation, Control and Embedded Systems (RACE), pp 1–5, 2015. 10.1109/RACE.2015.7097244
- 54.Pei X, Guo H, Dai J: Computerized detection of lung nodules in CT images by use of multiscale filters and geometrical constraint region growing. In: IEEE International Conference on Conference on Bioinformatics and Biomedical Engineering, pp 1–4, 2010. 10.1109/ICBBE.2010.5517771
- 55.Takemura S, Han X, Chen YW, Ito K, Nishikwa I, Ito M: Enhancement and detection of lung nodules with multiscale filters in CT images. In: IEEE International Conference on Intelligent Information Hiding and Multimedia Signal Processing, pp 717 720, 2008. 10.1109/IIH-MSP.2008.303
- 56.Miyajima T, Tokisa T, Maeda S, Kim H, Tan JK, Ishikawa S, Murakami S, Aoki T: Classification of lung nodules on temporal subtraction image based on statistical features and improvement of segmentation accuracy. In: IEEE International Conference on Control, Automation and Systems, pp 1814–1817, 2012
- 57.Dhara AK, Mukhopadhyay S: A hybrid preprocessing method using geometry based diffusion and selective enhancement filtering for pulmonary nodule detection. In: SPIE Proceedings of Medical Imaging, 8315, pp 2Y-1–2Y-6, 2012. 10.1117/12.911644
- 58.Dhara AK, Mukhopadhyay S, Alam N, Khandelwal N: Measurement of spiculation index in 3D for solitary pulmonary nodules in volumetric lung CT images. In: SPIE Proceedings of Medical Imaging, 8670, pp 0K-1–0K-6, 2013. 10.1117/12.2006970
- 59.Hadavi N, Nordin MJ, Shojaeipour A: Lung cancer diagnosis using CT-scan images based on cellular learning automata. In: IEEE International Conference on Computer and Information Sciences (ICCOINS), pp 1–5, 2014. 10.1109/ICCOINS.2014.6868370
- 60.Namin ST, Moghaddam HA, Jafari R, Esmaeil-Zadeh M, Gity M: Automated detection and classification of and classification of pulmonary nodules in 3D thoracic CT images. In: IEEE International Conference on Systems, Man and Cybernetics, pp 3774–3779, 2010. 10.1109/ICSMC.2010.5641820
- 61.Suárez-Cuenca JJ, Tahoces PG, Souto M, Lado MJ, Remy-Jardin M, Remy J, Vidal JJ. Application of the iris filter for automatic detection of pulmonary nodules on computed tomography images. Computers in Biology and Medicine. 2009;39(10):921–933. doi: 10.1016/j.compbiomed.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Leemput SVD, Dorssers F, Bejnordi BE: A novel spherical shell filter for reducing false positives in automatic detection of pulmonary nodules in thoracic CT scans. In: SPIE Proceedings of Medical Imaging, 9414, pp 2P-1–2P-6, 2015. 10.1117/12.2082298
- 63.Wang D, Wang J, Du Y, Tang P. Adaptive solitary pulmonary nodule segmentation for digital radiography images based on random walks and sequential filter. IEEE Access. 2017;5:1460–1468. [Google Scholar]
- 64.Cuifang Li, Shengdong N, Yuanjun W, Xiwen S: Experimental investigation of fuzzy enhancement for nonsolid pulmonary nodules. In: IEEE Symposium on Robotics and Applications (ISRA), pp 756–759, 2012. 10.1109/ISRA.2012.6219301
- 65.Xu F, Zhang WJ, Li XY, Xiao H, Peng SH, Nam HD, Zhang M-F: A 3D multi-scale block LBP filter for lung nodule enhancement based on the CT images. In: IEEE International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), pp 1376–1380, 2017. 10.1109/CISP-BMEI.2016.7852931
- 66.De Silve Sousa JRF, Silva AC, De Paiva AC, Nunes RA. Methodology for automatic detection of lung nodules in computerized tomography images. Computer Methods and Programs Programs in Biomedicine. 2009;98(1):1–14. doi: 10.1016/j.cmpb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 67.Cascio D, Magro R, Fauci F, Iacomi M, Raso G. Automatic detection of lung nodules in CT datasets based on stable 3D mass-spring models. Computers in Biology and Medicine. 2012;42(11):1098–1109. doi: 10.1016/j.compbiomed.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Santos AM, De C, Filho AO, Silva AC, De Paiva AC, Nunes RA, Gattass M. Automati detection of small lung nodules in 3D CT data using Gaussian mixture models. Tsallis entropy and SVM. Engineering Applications of Artificial Intelligence. 2014;36:27–39. [Google Scholar]
- 69.Nunzio GD, Tommasi E, Agrusti A, Cataldo R, Mitri ID, Favetta M, Maglio S, Massafra A, Quarta M, Torsello M, Zecca I, Bellotti R, Tangaro S, Calvini P, Camarlinghi N, Falaschi F, Cerello P, Oliva P. Automatic lung segmentation in CT images with accurate handling of the Hilar region. Journal of Digital Imaging. 2011;24(1):11–27. doi: 10.1007/s10278-009-9229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W, Wang X, Li X, Chen J. 3D skeletonization feature based computer-aided detection system for pulmonary nodules in CT datasets. Computers in Biology and Medicine. 2017;92:64–72. doi: 10.1016/j.compbiomed.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 71.Fu L, Ma J, Ren Y, Han Y S, Zhao J: Automatic detection of lung nodules: false positive reduction using convolution neural networks and handcrafted features. In: SPIE Proceedings of Proceedings of Medical Imaging, 10134, pp 0A-1–0A-8, 2017. 10.1117/12.2253995
- 72.Gopi K, Selvakumar J: Lung tumor area recognition and classification using EK- mean clustering and SVM. In: IEEE International Conference on Nextgen Electronic Technologies: Silicon to Software (ICNETS2), pp 97–100, 2017. 10.1109/ICNETS2.2017.8067906
- 73.Paing MP, Choomchuay S: Classification of margin characteristics from 3D pulmonary nodules. In: IEEE International Conference on Biomedical Engineering International (BMEiCON), pp 1–5, 2017. 10.1109/BMEiCON.2017.8229104
- 74.Paing MP,Choomchuay S: Ground glass opacity (GGO) nodules detection from lung CT scans. In: IEEE International Symposium on Electronics and Smart Devices (ISESD), pp 230–235, 2018. 10.1109/ISESD.2017.8253338
- 75.Javaid M, Javid M, Rehman MZ, Shah SI. A novel approach to CAD system for the detection of lung nodules in CT images. Computer Methods and Programs in Biomedicine. 2016;135:125–139. doi: 10.1016/j.cmpb.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 76.Rendon-Gonzalez E, Ponomaryov V: Automatic lung nodule segmentation and classification in CT images based on SVM. In: IEEE International Kharkiv Symposium on Physics and Engineering of Microwaves, Millimeter and Submillimeter Waves (MSMW), pp 1–4, 2016. 10.1109/MSMW.2016.7537995
- 77.Saien S, Pilevar AH, Moghaddam HA. Refinement of lung nodule candidates based on local geometric shape analysis and Laplacian of Gaussian kernels. Computers in Biology and Medicine. 2014;54:188–198. doi: 10.1016/j.compbiomed.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 78.Choi WJ, Choi TS: False positive reduction for pulmonary nodule detection using two- dimensional principal component analysis. In: SPIE Proceedings of Applications of Digital Image Processing, 7443, pp 22-1–22-8, 2009. 10.1117/12.827252
- 79.Choi WJ, Choi TS. Genetic programming-based feature transform and classification for the automatic detection of pulmonary nodules on computed tomography images. Information Sciences. 2012;212:57–78. [Google Scholar]
- 80.Jianto P, Paik DS, Meng X, Roos JE, Rubin GD. Shape “break-and-repair” strategy and its application to automated medical image segmentation. IEEE Transactions on Visualization and Computer Graphics. 2010;17(1):115–124. doi: 10.1109/TVCG.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Messay T, Hardie RC, Rogers SK. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Medical Image Analysis. 2010;14(3):390–406. doi: 10.1016/j.media.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 82.Kamra P,Vishraj R, Kanica, Gupta S: Performance comparison of image segmentation techniques for lung nodule detection in CT images. In: IEEE International Conference on Signal Processing, Computing and Control (ISPCC), pp 302–306, 2016. 10.1109/ISPCC.2015.7375045
- 83.Wei Y, Shen G, Li JJ. A fully automatic method for lung parenchyma segmentation and repairing. Journal of Digital Imaging. 2012;26(3):483–495. doi: 10.1007/s10278-012-9528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nurfauzi R, Nugroho HA, Ardiyanto I: Lung detection using adaptive border correction. In: IEEE International Conference on Science and Technology-Computer (ICST), pp 57–60, 2017. 10.1109/ICSTC.2017.8011852
- 85.Li XY, Xu F, Hu X, Peng SH, Nam HD, Zhao J-M: Self-adapting threshold of pulmonary parenchyma. In: IEEE International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), pp 1429–1434, 2017. 10.1109/CISP-BMEI.2016.7852941 [DOI]
- 86.Ye X, Lin X, Dehmeshki J, Slabaugh G, Beddoe G. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Transactions on Biomedical Engineering. 2009;56(7):1810–1820. doi: 10.1109/TBME.2009.2017027. [DOI] [PubMed] [Google Scholar]
- 87.El-Regaily SA, Salem MAM, Aziz MHA, Roushdy MI: Lung nodule segmentation and detection in computed tomography. In: IEEE International Conference on Intelligent Computing and Information Systems (ICICIS), pp 72–78, 2018. 10.1109/INTELCIS.2017.8260029
- 88.Gonga J, Liu JY, Wang LJ, Zheng B, Niea SD. Computer-aided detection of pulmonary nodules using dynamic self-adaptive template matching and a FLDA classifier. Physica Medica. 2016;32(12):1502–1509. doi: 10.1016/j.ejmp.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 89.Agarwal R, Shankhadhar A, Sagar RK: Detection of lung cancer using content based medical image retrieval. In: IEEE International Conference on Advanced Computing & Communication Technologies, pp 48–52, 2015. 10.1109/ACCT.2015.33
- 90.Farahani FV, Ahmadi A, Zarandi MHF: Lung nodule diagnosis from CT images based on ensemble learning. In: IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), pp 1–7, 2015. 10.1109/CIBCB.2015.7300281
- 91.Sun S, Li W, Kang Y: Lung nodule detection based on GA and SVM. In: IEEE International Conference on Biomedical Engineering and Informatics (BMEI), pp 96–100, 2016. 10.1109/BMEI.2015.7401480
- 92.Cerello P. The MAGIC-5 CAD for nodule detection in low dose and thin slice lung CTs. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Spectrometers, Detectors and Associated Equipment. 2010;623(2):832–835. [Google Scholar]
- 93.Pal NR, Pal SK. A review on image segmentation techniques. Pattern Recognition. 2003;26(9):1277–1294. [Google Scholar]
- 94.Haralick RM, Shapiro LG. Image Segmentation Techniques. Proceedings of Applications of Artificial Intelligence II. 1985;0548:39-1–39-6. [Google Scholar]
- 95.Phamy DL, Xu C, Prince JL. A survey of current methods in medical image segmentation. Annual Review of Biomedical Engineering. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 96.Sharma N, Aggarwal LM. Automated medical image segmentation techniques. Journal of Medical physics/Association of Medical Physicists of India. 2010;35(1):3–14. doi: 10.4103/0971-6203.58777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jung J, Hong H, Goo JM. Ground-glass nodule segmentation in chest CT images using asymmetric multi-phase deformable model and pulmonary vessel removal. Computers in Biology and Medicine. 2017;92:128–138. doi: 10.1016/j.compbiomed.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 98.Badura P, Pietka E. Soft computing approach to 3D lung nodule segmentation in CT. Computers in Biology and Medicine. 2014;53:230–243. doi: 10.1016/j.compbiomed.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 99.Keshani M, Azimifar Z, Tajeripour F, Boostani R. Lung nodule segmentation and recognition using SVM classifier and active contour modeling: a complete intelligent system. Computers in Biology and Medicine. 2013;43(4):287–300. doi: 10.1016/j.compbiomed.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 100.Sun S, Guo Y, Guan Y, Ren H, Fan L, Kang Y. Juxta-vascular nodule segmentation based on flow entropy and geodesic distance. IEEE Journal of Biomedical and Health Informatics. 2014;18(4):1355–1362. doi: 10.1109/JBHI.2014.2303511. [DOI] [PubMed] [Google Scholar]
- 101.Mukhopadhyay S. A segmentation framework of pulmonary nodules in lung CT images. Journal of Digital Imaging. 2015;29:86–103. doi: 10.1007/s10278-015-9801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Narayanan BN, Hardie RC, Kebede TM, Sprague MJ. Optimized feature selection-based clustering approach for computer-aided detection of lung nodules in different modalities. Pattern Analysis and Applications. 2017;22(2):559–571. [Google Scholar]
- 103.Aresta G, Cunha A, Campilho A: Detection of juxta-pleural lung nodules in computed tomography images. In: SPIE Proceedings of Medical Imaging, 10134, pp 3N-1–3N-7, 2017. 10.1117/12.2252022
- 104.Mehre SA, Mukhopadhyay S, Dutta A, Harsha NC, Dhara AK, Khandelwal N: An automated lung nodule detection system for CT images using synthetic minority oversampling. In: SPIE Proceedings of Medical Imaging, 9785, pp 0H-1–0H-8, 2016. 10.1117/12.2216357
- 105.Suárez-Cuenca JJ, Guo W, Li Q: Automated detection of pulmonary nodules In CT: false positive reduction by combining multiple classifiers. In: SPIE Proceedings of Medical Imaging, 7963, pp 38-1–38-6, 2011. 10.1117/12.878793 [DOI]
- 106.Nithila EE, Kumar SS. Automatic detection of solitary pulmonary nodules using swarm intelligence optimized neural networks on CT images. Engineering Science and Technology, an an International Journal. 2016;20(3):1192–1202. [Google Scholar]
- 107.Farahani FV, Ahmadi A, Zarandi MHF. Hybrid intelligent approach for diagnosis of the lung nodule from CT images using spatial kernelized fuzzy c-means and ensemble learning. Mathematics and Computers in Simulation. 2018;149:48–68. [Google Scholar]
- 108.Netto SMB, Silva AFC, Nunes RA, Gattass M. Automatic segmentation of lung nodules with growing neural gas and support vector machine. Computers in Biology and Medicine. 2012;42(11):1110–1121. doi: 10.1016/j.compbiomed.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 109.Hosseini R, Qanadli SD, Barman S, Mazinani M, Ellis T, Dehmeshki J. An automatic approach for learning and tuning Gaussian interval type-2 fuzzy membership functions applied to lung CAD classification system. IEEE Transactions on Fuzzy Systems. 2011;20(2):224–234. [Google Scholar]
- 110.Antonelli M, Cococcioni M, Lazzerini B, Marcelloni F. Computer-aided detection of lung nodules based on decision fusion techniques. Journal of Pattern Analysis and Applications. 2011;14(3):295–310. [Google Scholar]
- 111.Gong J, Liu JY, Wang LJ, Sun XW, Zheng B, Nie SD. Automatic detection of pulmonary nodules in CT images by incorporating 3D tensor filtering with local image feature analysis. Physica Medica. 2018;46:124–133. doi: 10.1016/j.ejmp.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 112.Farag A, Ali A, Graham J, Farag A, Elshazly S, Falk R: Evaluation of geometric feature descriptors for detection and classification of lung nodules in low dose CT scans of the chest. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp 169–172, 2011. 10.1109/ISBI.2011.5872380
- 113.Alam M, Sankaranarayanan G, Devarajan V: Lung nodule detection and segmentation using a patch-based multi-atlas method. In: IEEE International Conference on Computational Science and Computational Intelligence (CSCI), pp 23–28, 2017. 10.1109/CSCI.2016.0012
- 114.Assefa M, Faye I, Malik AS, Shoaib M: Lung nodule detection using multi- resolution analysis. In: IEEE International Conference on Complex Medical Engineering (CME), pp 457–461, 2013. 10.1109/ICCME.2013.6548290
- 115.Jaffar MA, Siddiqui AB, Mushtaq M. Ensemble classification of pulmonary nodules using gradient intensity feature descriptor and differential evolution. Cluster Computing. 2017;21(1):393–407. [Google Scholar]
- 116.Novo J, Gonçalves L, Mendonça AM, Campilho A: 3D lung nodule candidate detection in multiple scales. In: IEEE International Conference on Machine Vision Applications (MVA), pp. 61–64, 2015 doi: 10.1109/MVA.2015.7153133
- 117.Chen B, Kitasaka T, Honma H, Takabatake H, Mori M, Natori H, Mori K: Automatic segmentation of solitary pulmonary nodules based on Local intensity structure analysis and 3D neighborhood features in 3D chest CT Images. In: SPIE Proceedings of Medical Imaging, 8315, pp 38-1–38-8, 2012. 10.1117/12.911782
- 118.Retico A, Bagagli F, Camarlinghi N, Carpentieri C, Fantacci ME, Gori I: A voxel-based neural approach (VBNA) to identify lung Nodules in the ANODE09 study. In: SPIE Proceedings of Medical Imaging, 7260, pp 1S-1–1S-8, 2009. 10.1117/12.811721
- 119.Murphy K, Van Ginneken B, Schilham AM, De Hoop BJ, Gietema HA, Prokop M. A large-scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classification. Medical Image Analysis. 2009;13(5):757–770. doi: 10.1016/j.media.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 120.Froz BR, De Carvalho Filho AO, Silva AC, De Paiva AC, Nunes RA, Gattass M. Lung nodule classification using artificial crawlers, directional texture and support vector machine. Expert Systems with Applications. 2016;69(12):176–188. [Google Scholar]
- 121.Han H, Li L, Han F, Song B, Moore W, Liang Z. Fast and adaptive detection of pulmonary nodules in thoracic CT images using a hierarchical vector quantization scheme. IEEE Journal of Biomedical and Health Informatics. 2014;19(2):648–659. doi: 10.1109/JBHI.2014.2328870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yuan D, Du W, Duan X, Wang J, Ma Y, Zhang, H: Detection of slices including a ground-glass opacity nodule in CT volume data with semi-supervised learning. In: IEEE Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/ Distributed Computing (SNPD), pp 557–561, 2017. 10.1109/SNPD.2017.8022778
- 123.Taşci E, Uğur A: Shape and texture based novel features for automated Juxtapleural nodule detection in lung CTs. Journal of Medical Systems 39–46, 2015 [DOI] [PubMed]
- 124.Wang Q, Kang W, Wu C, Wang B. Computer-aided detection of lung nodules by SVM based on 3D matrix patterns. Clinical Imaging. 2013;37(1):62–69. doi: 10.1016/j.clinimag.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 125.Retico A, Fantacci ME, Gori I, Kasae P, Golosio B, Piccioli A, Cerello P, De Nunzio G, Tangaro S. Pleural nodule identification in low-dose and thin-slice lung computed tomography. Computers in Biology and Medicine. 2009;39(12):1137–1144. doi: 10.1016/j.compbiomed.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 126.Sahoo PK, Soltani S, Wong AKC. A survey of thresholding techniques. Computer Vision, Graphics, and Image Processing. 2004;41(2):233–260. [Google Scholar]
- 127.Freixenet J., Muñoz X., Raba D., Martí J., Cufí X. Computer Vision — ECCV 2002. Berlin, Heidelberg: Springer Berlin Heidelberg; 2002. Yet Another Survey on Image Segmentation: Region and Boundary Information Integration; pp. 408–422. [Google Scholar]
- 128.McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Medical Image Analysis. 2002;1(2):91–108. doi: 10.1016/s1361-8415(96)80007-7. [DOI] [PubMed] [Google Scholar]
- 129.Yokota K, Maeda S, Kim H, Tan JK, Ishikawa S, Tachibana R, Hirano Y, Kido S: Automatic detection of GGO regions on CT images in LIDC dataset based on statistical features. In: IEEE Conference on Soft Computing and Intelligent Systems (SCIS) and 15th International Symposium on Advanced Intelligent Systems (ISIS), pp 1374–1377, 2015. 10.1109/SCIS-ISIS.2014.7044692
- 130.Wang H, Guo XH, Jia ZW, Li HK, Liang ZG, Li KC, He Q. Multilevel binomial logistic prediction model for malignant pulmonary nodules based on texture features of CT image. European Journal of Radiology. 2009;74(1):124–129. doi: 10.1016/j.ejrad.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 131.Yu SJ, Wantroba JS, Raicu DS, Furst JD, Channin DS, Armato III SG: A study on the effect of CT imaging acquisition parameters on lung nodule image interpretation. In: SPIE Proceeding of Medical Imaging, 7263, 1R-1–1R-8, 2009. 10.1117/12.813695
- 132.Dhara AK, Mukhopadhyay S, Khandelwal N: 3D texture analysis of solitary pulmonary nodules using co-occurrence matrix from volumetric lung CT images. In: SPIE Proceedings of Medical Imaging, 8670, pp 39-1–39-6, 2013. 10.1117/12.2007016
- 133.Ciompi F, Jacobs C, Scholten ET, Wille MM, De Jong PA, Prokop M, Van Ginneken B. Bag-of-frequencies: a descriptor of pulmonary nodules in computed tomography images. IEEE Journal of Biomedical and Health Informatics. 2014;34(4):962–973. doi: 10.1109/TMI.2014.2371821. [DOI] [PubMed] [Google Scholar]
- 134.Vapnik VN. Statistical Learning Theory. USA: Wiley-Interscience; 1998. [Google Scholar]
- 135.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van der Laak Jeroen AWM, Ginneken BV, Sanchez CI. A survey on deep learning in medical image analysis. Medical Image Analysis. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 136.Wong KKL, Wang L, Wang D. Recent developments in machine learning for medical imaging applications. Computerized Medical Imaging and Graphics. 2017;57:1–3. doi: 10.1016/j.compmedimag.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 137.Bruijne M. Machine learning approaches in medical image analysis: from detection to diagnosis. Medical Image Analysis. 2016;33:94–97. doi: 10.1016/j.media.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 138.Suzuki K. Overview of deep learning in medical imaging. Radiological Physics and Technology. 2017;10(3):257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 139.Tajbakhsh N, Suzuki K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. Pattern Recognition. 2016;63:476–486. [Google Scholar]
- 140.Hasegawa A, Benedict Lo SC, Freedman MT, Mun SK: Convolution neural-network-based detection of lung structures. In: SPIE Proceedings of Medical Imaging, 2167, pp 654–662, 1994. 10.1117/12.175101
- 141.Lin JS, Ligomenides PA, Lo SCB, Hasegawa A, Freedman MT, Mun SK: An application of convolution neural networks: reducing false-positives in lung nodule detection. In: IEEE Proceedings of 1994 IEEE Nuclear Science Symposium - NSS'94, pp 1842–1846, 1994. 10.1109/NSSMIC.1994.474706
- 142.Lo SB, Lou SA, Lin JS, Freedman MT, Chien MV, Mun SK. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Transactions on Medical Imaging. 1995;14(4):711–718. doi: 10.1109/42.476112. [DOI] [PubMed] [Google Scholar]
- 143.Dumoulin V, Visin F: A guide to Convolution Arithmetic for Deep Learning, 2018. https://arxiv.org/pdf/1603.07285.pdf/. Accessed 8 Feb 2018
- 144.http://cs231n.stanford.edu/index.html. Accessed 9 Feb 2018
- 145.http://ufldl.stanford.edu/tutorial/supervised/ConvolutionalNeuralNetwork/. Accessed 9 Feb 2018
- 146.Jiang F, Grigorev A, Rho S, Tian Z, Fu YS, Jifara W, Adil K, Liu S. Medical image semantic segmentation based on deep learning. Neural Computing and Applications. 2017;29(5):1257–1265. [Google Scholar]
- 147.Yan Xingjian, Pang Jianing, Qi Hang, Zhu Yixin, Bai Chunxue, Geng Xin, Liu Mina, Terzopoulos Demetri, Ding Xiaowei. Computer Vision – ACCV 2016 Workshops. Cham: Springer International Publishing; 2017. Classification of Lung Nodule Malignancy Risk on Computed Tomography Images Using Convolutional Neural Network: A Comparison Between 2D and 3D Strategies; pp. 91–101. [Google Scholar]
- 148.Tan Jiaxing, Huo Yumei, Liang Zhengrong, Li Lihong. Smart Health. Cham: Springer International Publishing; 2017. Apply Convolutional Neural Network to Lung Nodule Detection: Recent Progress and Challenges; pp. 214–222. [Google Scholar]
- 149.Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J. Multi- crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognition. 2016;61:663–673. [Google Scholar]
- 150.Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, Van Riel SJ, Wille MMW, Naqibullah M, Sánchez CI, Van Ginneken B. Pulmonary nodule detection in CT images: f alse positive reduction using multi-view convolutional networks. IEEE Transactions on Medical Imaging. 2016;35(5):1160–1169. doi: 10.1109/TMI.2016.2536809. [DOI] [PubMed] [Google Scholar]
- 151.Ciompi F, De Hoop B, Van Riel SJ, Chung K, Scholten ET, Oudkerk M, De Jong PA, Prokop M, Van Ginneken B. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Medical Image Analysis. 2015;26(1):195–202. doi: 10.1016/j.media.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 152.Dou Q, Chen H, Yu L, Qin J, Heng PA. Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Transactions on Biomedical Engineering. 2016;64(7):1558–1567. doi: 10.1109/TBME.2016.2613502. [DOI] [PubMed] [Google Scholar]
- 153.Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y, Dong D, Gevaert O, Tian J. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Medical Image Analysis. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Da Silva GLF, Da Silve Neto OP, Silva AC, De Paiva AC, Gattass M. Lung nodules diagnosis based on evolutionary convolutional neural network. Multimedia Tools and Applications. 2017;76(18):19039–19055. [Google Scholar]
- 155.Yuan J, Liu X, Hou F, Qin H, Hao A. Hybrid-feature-guided lung nodule type classification on CT images. Computers & Graphics. 2017;70:288–299. [Google Scholar]
- 156.Xie Y, Zhang J, Xia Y, Michael F, Zhang Y. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Information Fusion. 2017;42:102–110. [Google Scholar]
- 157.Liu X, Hou F, Qin H, Hao A. Multi-view multi-scale CNNs for lung nodule type classification from CT images. Pattern Recognition. 2018;77:262–275. [Google Scholar]