Abstract
The proposed dataset provides annotations for the 552 cardiotocographic (CTG) recordings included in the publicly available “CTU-CHB intra-partum CTG database” from Physionet (https://physionet.org/content/ctu-uhb-ctgdb/1.0.0/). Each CTG recording is composed by two simultaneously acquired signals: i) the fetal heart rate (FHR) and ii) the maternal tocogram (representing uterine activity). Annotations consist in the detection of starting and ending points of specific CTG events on both FHR signal and maternal tocogram. Annotated events for the FHR signal are the bradycardia, tachycardia, acceleration and deceleration episodes. Annotated events for the maternal tocogram are the uterine contractions. The dataset also reports classification of each deceleration as early, late, variable or prolonged, in relation to the presence of a uterine contraction. Annotations were obtained by an expert gynecologist with the support of CTG Analyzer, a dedicated software application for automatic analysis of digital CTG recordings. These annotations can be useful in the development, testing and comparison of algorithms for the automatic analysis of digital CTG recordings, which can make CTG interpretation more objective and independent from clinician's experience.
Keywords: Cardiotocography, Electronic fetal monitoring, Fetal distress, Fetal heart rate, Uterine contractions, Labor, Childbirth
Specifications Table
| Subject | Biomedical Engineering |
| Specific subject area | Biomedical signal processing, specifically digital cardiotocographic (CTG) recordings |
| Type of data | MATLAB® data file (.mat) |
| How data were acquired | Visual inspection by an expert gynecologist, supported by a dedicated software application. Instruments: CTG Analyzer, dedicated software application for the automatic analysis of digital CTG recordings (https://dx.doi.org/10.1109/EMBC.2017.8037391). |
| Data format | Raw and derived |
| Parameters for data collection | Annotations refer to a total of 552 CTG recordings, each composed by two simultaneously acquired signals: the fetal heart rate and the maternal tocogram (uterine activity). Annotations are based on the analysis of both signals and indicate starting and ending points of specific events such as bradycardia, tachycardia, accelerations, decelerations and uterine contractions. |
| Description of data collection | This dataset contains the annotations of the digital CTG recordings constituting the “CTU-CHB intra-partum CTG database” available at Physionet (http://physionet.org). Annotations were visually identified by an expert gynecologist supported by CTG Analyzer, a dedicated software application (https://dx.doi.org/10.1109/EMBC.2017.8037391). |
| Data source location | Raw CTG data (digital CTG recordings) were obtained from Physionet (https://physionet.org/content/ctu-uhb-ctgdb/1.0.0/). Derived CTG annotation data were obtained and are stored at the Cardiovascular Bioengineering Lab, Department of Information Engineering, Università Politecnica delle Marche, Ancona, Italy. |
| Data accessibility | Raw CTG data are publicly available at the following Physionet link: https://physionet.org/content/ctu-uhb-ctgdb/1.0.0/. Derived CTG annotations are available with the article as supplementary material. |
| Related research article | S. Romagnoli, A. Sbrollini, L. Burattini, I. Marcantoni, M. Morettini, L. Burattini, Digital cardiotocography: What is the optimal sampling frequency?, Biomed. Signal Process. Control. 51 (2019) 210–215. https://doi.org/10.1016/j.bspc.2019.02.016[1] |
Value of the Data
-
•
This annotation dataset, together with the associated raw data (“CTU-CHB intra-partum CTG database”) may represent a gold standard useful for studies on digital CTG.
-
•
Beneficiaries of this dataset include biomedical engineers, who will have data to develop algorithms; clinicians, who will have computerized tools to support their diagnosis; pregnant women, who will receive objective diagnosis.
-
•
This annotation dataset is useful to support development, testing and comparison of algorithms for the automatic analysis of digital CTG recordings, possibly also obtained by scanning paper CTG reports.
-
•
The “CTU-CHB intra-partum CTG database” includes only tracings; by adding annotations, this dataset supports technical developments and thus passage from usual subjective CTG interpretation, mainly based on clinician's experience, to objective CTG interpretation.
1. Data Description
1.1. Raw CTG Data
The raw data consist of 552 digital CTG recordings constituting the “CTU-CHB intra-partum CTG database” [2,3] and are publicly available in Physionet at the link: https://physionet.org/content/ctu-uhb-ctgdb/1.0.0/. Each CTG recording is composed by two signals: i) the fetal heart rate (FHR, sometimes called tachogram); and ii) the maternal tocogram (representing the uterine activity).
1.2. Derived CTG Annotations
This dataset contains the annotations of the digital CTG recordings constituting the “CTU-CHB intra-partum CTG database” available at Physionet (http://physionet.org). Annotations were visually identified by an expert gynecologist supported by CTG Analyzer [4], a dedicated software application. They consist in the detection of starting and ending sample points of specific events on both FHR signal and maternal tocogram. Annotated events for the FHR signal are the bradycardia, tachycardia, acceleration and deceleration episodes. Annotated events for the maternal tocogram are the uterine contractions. Table 1 reports guidelines for identification of events on the CTG recording. The dataset also includes classification of each deceleration as early, late, variable or prolonged, in relation to the presence of a uterine contraction. Table 2 reports classification of the decelerations in relation to uterine contractions.
Table 1.
Guidelines for identification of events on the CTG recording.
| Bradycardia | FHR baseline below 110 bpm for longer than 10 minutes |
| Tachycardia | FHR baseline over 160 bpm for longer than 10 minutes |
| Acceleration | FHR increments above baseline of more than 15 bpm lasting longer than 15 s |
| Deceleration | FHR decrements below baseline of more than 15 bpm lasting longer than 15 s |
| Uterine contraction | Bell-shaped temporary increment of the amplitude of the uterine activity signal lasting 45−120 s |
Table 2.
Classification of the decelerations in relation to uterine contractions.
| Early | Deceleration onset synchronized with uterine-contraction onset; shallow short-lasting deceleration with normal variability within the deceleration |
| Late | Deceleration onset during the second half of uterine contraction; gradual onset and/or gradual return to the baseline and/or reduced variability within the deceleration |
| Variable | Deceleration uncorrelated to any uterine contraction; a rapid drop and good variability within the deceleration; variable in size and shape |
| Prolonged | Deceleration lasting more than 3 minutes |
The dataset includes a total of 552 MATLAB® data file; each data file is called annotation_i.mat where i corresponds to the patient identifier in the “CTU-CHB intra-partum CTG database” [2,3]. Each annotation_i.mat includes two variables called dataloss and ann.
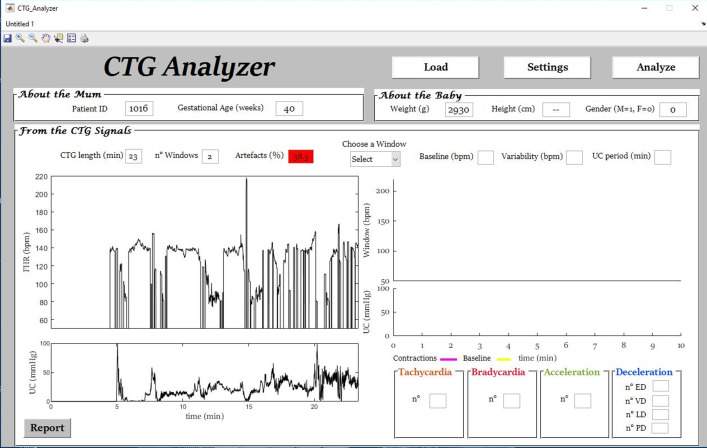
The dataloss variable (1 × 3 vector) contains information about the quality of the FHR signal divided in 3 consecutive segments of equal length (see Experimental Design, Materials, and Methods section, Signal-Quality Assessment paragraph). Percentage of data loss, contained in the dataloss variable and automatically determined by CTG Analyzer [4] (Fig. 1 to Fig. 4), ranges from 0% to 100%. FHR-signal quality decreases with increasing percentage [1].
Fig. 1.
Signal-quality assessment of a case (initial segment of signal ‘1016’) with data loss equal to 38.5% and for which automatic annotation by CTG Analyzer was not performed.
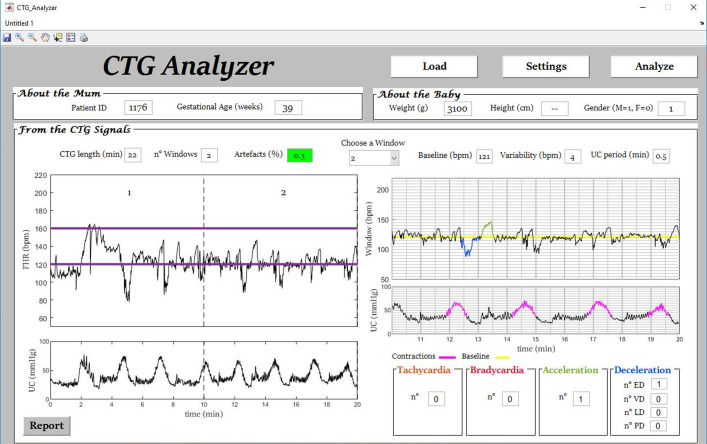
Fig. 4.
Example (initial segment of signal ‘1176’) of automatic identification of accelerations, decelerations and uterine contractions and classification of decelerations by CTG Analyzer.
The ann variable (5 × L cell array, where L is the length of the CTG recording in samples) contains the annotations (i.e. strings indicating start and ending points; see Experimental Design, Materials, and Methods section, Annotations of CTG events and Decelerations Classification paragraph) for the 5 considered events: 1-bradycardia, 2-tachycardia, 3-acceleration, 4-deceleration and 5-uterine contraction. CTG annotations, contained in ann variable, were visually identified by an expert gynecologist supported by CTG Analyzer [4] (Fig. 2 to Fig. 4). Table 3 reports the symbols used in the proposed dataset for the annotations of the CTG events.
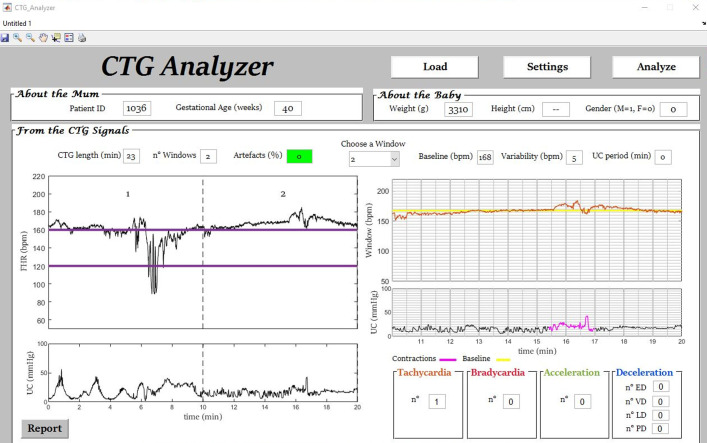
Fig. 3.
Example (initial segment of signal ‘1036’) of automatic tachycardia identification by CTG Analyzer.
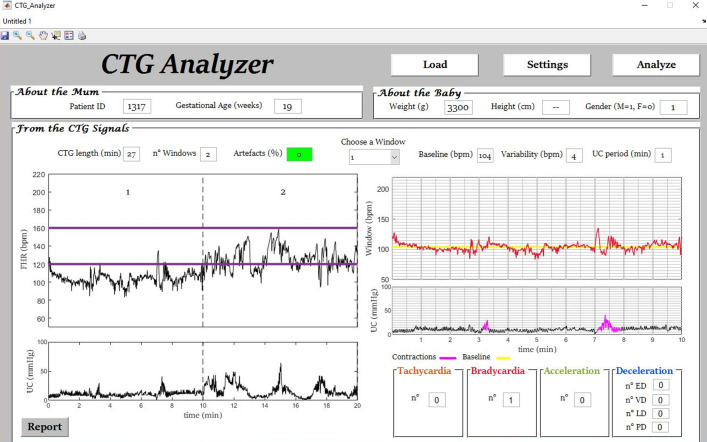
Fig. 2.
Example (initial segment of signal ‘1317’) of automatic bradycardia identification by CTG Analyzer.
Table 3.
Symbols used for the annotations of the CTG events.
| Bradycardia | Starting point of the kth bradycardia event | (BCk |
| Ending point of the kth bradycardia event | )BCk | |
| Tachycardia | Starting point of the kth tachycardia event | (TCk |
| Ending point of the kth tachycardia event | )TCk | |
| Acceleration | Starting point of the kth acceleration event | (ACCk |
| Ending point of the kth acceleration event | )ACCk | |
| Deceleration | Starting point of the kth deceleration event, classification in relation to uterine contraction | (DECkE (DECkL (DECkV (DECkP |
| Ending point of the kth deceleration event, classification in relation to uterine contraction | )DECkE )DECkL )DECkV )DECkP |
|
| Uterine contraction | Starting point of the kth uterine contraction | (UCk |
| Ending point of the kth uterine contraction | )UCk |
2. Experimental Design, Materials, and Methods
2.1. Data Acquisition
The raw CTG recordings constituting the “CTU-CHB intra-partum CTG database” were acquired from pregnant women during labor at the Czech Technical University in Prague (Czech Republic) and at the University Hospital in Brno (Czech Republic) using, as electronic fetal monitors [3], the STAN S21 and S31 (Neoventa Medical, Mölndal, Sweden) and the Avalon FM40 and FM50 (Philips Healthcare, Andover, MA). CTG recordings have a duration ranging from 30 minutes to 90 minutes. Both FHR signal and maternal tocogram were sampled at 4 Hz [1,3]. These data, as all Physionet data, were fully anonymized and may be used without further approval of the Institutional Review Board.
2.2. Data Annotation
2.2.1. CTG Analyzer
CTG Analyzer [4] is a dedicated software application developed under MATLAB® GUI (Graphical User Interface) for the automatic analysis of digital CTG recordings obtained by digital recorders or by scanning paper CTG reports [5,6]. Details of CTG Analyzer can be found in [4]. Briefly, it provides a measure of the FHR signal quality and quantitative characterization of FHR signal and maternal tocogram according to FIGO (International Federation of Gynecology and Obstetrics) guidelines [7].
2.2.2. Signal-Quality Assessment
FHR signals are often affected by artifacts due to data loss represented by samples the value of which is set to zero. Since data loss may occur locally, FHR signals were divided into 3 consecutive segments of equal length (initial, middle and last segment) and data loss correction was performed in each segment independently by using the CTG Analyzer [1,4].
Quality of each FHR segment was assessed by computing the percentage of data loss, computed as 100 by the number of corrected samples over the total number of samples in the FHR segment. If FHR-segment quality was greater than 10%, the FHR segment was considered of bad quality [4,8] and no further automatic analysis by CTG Analyzer was performed; however, where possible, annotations were still obtained by visual inspection. Fig. 1 reports signal-quality assessment of a case with data loss equal to 38.5% and for which automatic annotation by CTG Analyzer was not performed. If FHR-segment quality was no more than 10%, linear interpolation was performed to compensate the data loss before automatic CTG events identification [4].
2.2.3. Annotations of CTG events and Decelerations Classification
Each CTG recording was annotated by an expert gynecologist by visual inspection and with the support of CTG Analyzer. All CTG events, that are bradycardia, tachycardia, acceleration, deceleration and uterine contraction, were identified according to their definitions provided by FIGO guidelines as reported in Table 1 [7,8] and indicated as BC, TC, ACC, DEC and UC, respectively. Classification of decelerations in early, late, variable and prolonged, was also performed according to the definitions of FIGO guidelines as reported in Table 2 [7] and deceleration class was indicated as E, L, V and P, respectively. Fig. 2 to Fig. 4 report examples of automatic CTG events identification and classification by CTG Analyzer. The starting sample point of an event is indicated as a string composed by an opening bracket ‘(’, the acronym of the event, a progressive number and, if a deceleration, the deceleration class as reported in Table 3. Analogously, the ending sample point of an event is indicated as a string composed by a closing bracket ‘)’, the acronym of the event, a progressive number and, if a deceleration, the deceleration class as reported in Table 3. When a sample does not represent a starting/ending point of an event, the cell has been left empty.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105690.
Appendix. Supplementary materials
References
- 1.Romagnoli S., Sbrollini A., Burattini L., Marcantoni I., Morettini M., Burattini L. Digital cardiotocography: What is the optimal sampling frequency? Biomed. Signal Process. Control. 2019;51:210–215. doi: 10.1016/j.bspc.2019.02.016. [DOI] [Google Scholar]
- 2.Goldberger A.L., Amaral L.A., Glass L., Hausdorff J.M., Ivanov P.C., Mark R.G., Mietus J.E., J.E. G.B.Moody, Peng C.K., Stanley H.E., PhysioBank PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. doi: 10.1161/01.CIR.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 3.Chudáček V., Spilka J., Burša M., Janků P., Hruban L., Huptych M., Lhotská L. Open access intrapartum CTG database. BMC Pregnancy Childbirth. 2014;14:16. doi: 10.1186/1471-2393-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sbrollini A., Agostinelli A., Burattini L., Morettini M., Di Nardo F., Fioretti S., Burattini L. CTG Analyzer: A graphical user interface for cardiotocography. Conf. IEEE Eng. Med. Biol. Soc. 2017:2606–2609. doi: 10.1109/EMBC.2017.8037391. [DOI] [PubMed] [Google Scholar]
- 5.Sbrollini A., Agostinelli A., Marcantoni I., Morettini M., Burattini L., Di Nardo F., Fioretti S., Burattini L. eCTG: an automatic procedure to extract digital cardiotocographic signals from digital images. Comput. Methods Programs Biomed. 2018;156:133–139. doi: 10.1016/j.cmpb.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 6.Cömert Z., Şengür A., Akbulut Y., Budak Ü., Kocamaz A.F., Bajaj V. Efficient approach for digitization of the cardiotocography signals. Physica A: Statistical Mechanics and its Applications. 2020;537 doi: 10.1016/j.physa.2019.122725. [DOI] [Google Scholar]
- 7.Ayres-De-Campos D., Spong C.Y., Chandraharan E. FIGO consensus guidelines on intrapartum fetal monitoring: Cardiotocography. Int. J. Gynecol. Obstet. 2015;131:13–24. doi: 10.1016/j.ijgo.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Agostinelli A., Braccili E., Marchegiani E., Rosati R., Sbrollini A., Burattini L., Morettini M., Di Nardo F., Fioretti S., Burattini L. Statistical baseline assessment in cardiotocography. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2017:3166–3169. doi: 10.1109/EMBC.2017.8037529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.