Abstract
Degree of genomic instability closely correlates with poor prognosis, drug resistance as well as poor survival across human cancer of different origins. This study assessed the relationship between DNA damage response (DDR) and chromosome instability in hepatocellular carcinoma (HCC). We investigated DDR signaling in HCC cells by analyzing DNA damage-dependent redistribution of major DDR proteins to damaged chromatin using immunofluorescence microscopy and Western blotting experimentations. We also performed gene conversion and metaphase analyses to address whether dysregulated DDR may bear any biological significance during hepatocarcinogenesis. Accordingly, we found that HCC cell lines suffered from elevated spontaneous DNA double-strand breaks (DSBs). In addition, analyses of HCC metaphases revealed marked aneuploidy and frequent sister chromatid exchanges when compared to immortalized hepatocytes, the latter of which were further induced following camptothecin-induced DSBs. We propose that genomic instability in HCC may be caused by erroneous DNA repair in a desperate attempt to mend DSBs for cell survival and that such preemptive measures inadvertently foster chromosome instability and thus complex genomic rearrangements.
Introduction
Liver cancer ranks among the top of all anatomical sites for cancer incidence and remains one of the major causes for cancer-related mortality worldwide [1,2]. Chronic liver conditions, including fatty liver disease, hepatitis infection, and inflammation, could lead to liver cirrhosis and subsequently liver cancer. Accordingly, the intrinsically high proliferative rate of hepatocytes makes them especially vulnerable to oxidative stress, replicative stress, and telomere exhaustion, where processing of collapsed replication forks or telomeric attrition may contribute to DNA double-strand break (DSB) formation [3,4].
The mammalian DNA damage response (DDR) is the frontline barrier against accumulation of spontaneous DNA damage from genotoxic stress conditions. The two cell fate decisions in the extremities are either the onset of programmed cell death or cellular transformation through the gain of prosurvival modifications on the genome. Cells that survive through these challenges share a characteristic of genomic instability, best illustrated by extensive chromosome instability and complex genomic rearrangements observed in cells derived from sporadic liver malignancies of different etiologies and clinical stages. The most prominent types of genetic aberration observed in precancerous lesions of hepatocellular carcinoma (HCC) include aneuploidy, chromosome fragility, microsatellite instability, as well as complex derivative chromosomes. Thus, isolating recurrent genetic loci that fuel tumorigenesis represents an important means to delineate the development of cancer [5].
With the advent in high-density array and sequencing technology, complex genomic rearrangements previously reported by conventional karyotyping techniques have been reassessed, and microrearrangements have been appended onto existing complex conditions. Indeed, an integrative analysis of genomic alterations derived from 18 HCC studies reported chromosomal gains and losses in more than 50% of the studies [6]. Computational study on somatic structural variations in human cancer genomes identified tandem duplications as major somatic genomic alterations in 16 HCC, followed by deletion and intra- and interchromosomal translocations [7]. Moreover, a high-resolution pan-cancer somatic copy number alteration study revealed that as high as 25% of the 140 peak alteration regions harbor oncogenes or tumor suppressor genes, including amplicons where the EGFR, MYC, and TERC genes resides, and deletions that cover the ATM and NOTCH1 gene loci [8]. However, little information is currently available to explain when and how these genetic aberrations have arisen. The fact that no known oncogene or tumor suppressor gene was found among the 75% peak alterations highlights the possibility that a large proportion of genomic alterations represent collateral events during carcinogenesis and that they do not impose survival advantage to malignant cells.
The two major mammalian DSB repair pathways are homology-directed repair (HDR) and nonhomologous end joining (NHEJ). Depending on the homology pairing mechanism, HDR can be further categorized into homologous recombination (HR), single-strand annealing, and break-induced replication (BIR). Ataxia telangiectasia mutated (ATM) and ATM and Rad 3-related (ATR) kinase signal activation underlie HR repair events that involve genomic mobility induced by uncapped telomeres and common fragile sites, respectively [9,10]. The ATM- and ATR-dependent DDR signaling cascades converge to HR activation, effected via the recombinase RAD51 that mends DNA breaks in a manner that entails use of homologous templates [11]. In addition, RAD51 also enforces stable chromatin association of fork proteins and replication fork restoration [12]. In contrast to HDR, NHEJ repair does not require homologous DNA as repair templates. DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is recruited to DSB termini by the Ku70/80 heterodimers [13], where its formation as homodimers serves as a molecular scaffold to bridge the DSB ends. DNA-PKcs homodimerization also allows for its autophosphorylation, which then signals the activation and/or recruitment of downstream repair proteins, for example, the Artemis and polymerases, to initiate broken DNA end processing [14]. Common to these protective strategies against genomic instability is the prerequisite of a highly cooperative DNA repair machinery to accommodate the massive DNA damage within cells.
Considering the need to balance HDR and NHEJ in genomic stability maintenance, especially in cancerous cells that experience overwhelming amounts of DNA damage, we hypothesize that hyperactive DSB repair may contribute to the manifestation of genomic instability in HCC. To address this idea, we charted DDRs in HCC with the goal to identify pathogenic DNA repair pathway(s) that may contribute to genomic instability during hepatocarcinogenesis.
Materials and Methods
Cell Culture
Human liver cancer cell lines HEP3B, PLC/PRF/5, HUH7, and HEPG2 and immortalized hepatocyte MIHA were maintained in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin in 37°C incubator with 5% CO2. HEP3B and PLC/PRF/5 are hepatocellular carcinoma HBV-related cell lines established from an 8- year-old male and a 24-year-old male, respectively. HUH7 is a non–HBV-/non–HCV-related hepatocellular carcinoma cell line derived from a 57- year-old male. HEPG2 is a non–HBV-/non–HCV-related hepatoblastoma cell line established from a 15- year-old male. MIHA is an immortalized nontumorigenic human hepatocyte cell line established from a 59- year-old male.
Antibodies
ATM (Cell Signaling 2873s), ATM S1981 (Cell Signaling 4526L), ATR (N19) (Santa Cruz sc-1807), BRCA2 [15], DNA-PKcs S2056 (Abcam ab18192), gamma-H2AX [16], 53BP1 [17], RAD51 [15], Actin (Sigma A5441), GAPDH (Chemicon MAB374), H3 (Cell Signaling 9715s), CHK1 (Santa Cruz sc-8048), and mouse anti-BrdU (BD Biosciences 347-580 B44) were used at 1:500 dilution. All secondary antibodies were purchased from Jackson Immuno Research Laboratories and were used at 1:3000 dilution.
Immunofluorescence (IF) Microscopy
Cells seeded on coverslips were washed with PBS, fixed at room temperature with 3% paraformaldehyde for 15 minutes, permeabilized with 0.5% Triton for 15 minutes, blocked with 3% milk in 1× PBS-T for 20 minutes, and then immunostained with specified primary antibodies for 2 hours. After incubation with primary antibodies, cells were washed twice with PBS and were incubated with AlexaFluor 488–conjugated goat anti-mouse and AlexaFluor 596–conjugated goat anti-rabbit antibodies for 2 hours. Nuclei were counterstained with DAPI. Images were visualized and captured on Nikon Eclipse 800 microscope.
Cell Fractionation and Western Blot Analyses
Whole cell lysates (W) were prepared by lysing cells in NETN lysis buffer [20 mM Tris–HCL (pH 8.0), 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 0.5 mM NaF, 1 mM β-glycerophosphate] with Benzonase (Merck Millipore) and MgCl2 on ice for 30 minutes or until pellet dissolved. For soluble (S) and insoluble (I) cell fractions, cells were lysed with NETN buffer on ice for 30 minutes and then centrifuged at 13,000 rpm at 4°C. Supernatants were collected and labeled as soluble fractions. The remaining pellets enriched with chromatin were then washed three times with NETN. Subsequently, cell pellets were lysed with NETN with Benzonase until completely dissolved. The resultant cell fractions were labeled as insoluble fraction (I). Cell lysates were resolved on SDS-PAGE, transferred to PVDF membranes, and probed with specified primary and secondary antibodies. Chemiluminescence signals were captured by conventional x-ray film (Kodak) exposure.
Gene Conversion Assay
Gene conversion assay was performed with method described previously [15]. Briefly, cells were electroporated with DR-GFP plasmid and 8 μg pCBAs-I-SceI plasmid or pEGFP at 250 V, 975 μF using Bio-Rad gene pulsar II and then plated onto 10-cm dishes and incubated in culture media for 48 hours. Cells were then analyzed on Becton-Dickinson FACSCanto with green (FL1) versus orange (FL2) fluorescence plot. Results represent average of at least three independent experiments. Statistical analyses were performed in ImageJ.
Sister Chromatid Exchange (SCE) Assay
Cells seeded at 20% confluency were labeled with 40 μM BrdU (Sigma) for two doubling times and were treated with 0.03 μg/ml Colcemid (Thermofisher) for 3 hours prior to harvest. Cells were collected and incubated with KCl for 30 minutes and then fixed with 3:1 methanol:acetic acid. Metaphases were spread onto clean dry glass slides and aged at room temperature for 24 hours. To visualize SCE events, aged slides were treated with 2 N HCl for 30 minutes at 37°C and then neutralized with 0.1 M boric acid. Slides were then immunostained with mouse anti-BrdU antibody (BD Biosciences) and AlexaFluor 596–conjugated anti-mouse antibody. Images of metaphases were captured using a Carl Zeiss LSM880 and were processed with Airyscan processing in ZEN software. Chromosomes and SCE events were quantified using ImageJ.
Drug Sensitivity Assay
Cells were seeded onto 96-well plates in triplicates for 24 hours before treatment with different chemicals. After incubation for 72 hours with chemicals, cells were incubated in 20 μl 5 mg/ml thiazolyl blue tetrazolium blue (MTT, Sigma). Absorbance at 570 nm minus 690 nm was recorded by Victor plate reader. Statistical analyses and charts from two or more experiments were prepared with Prism 6.0.
Colony Formation Assay
HCC cells were seeded onto 60-mm dishes in triplicates for 24 hours before treatment with different chemicals. Cells were incubated until colonies (>50 cells) were formed. Colonies were then stained and fixed with Coomassie blue in methanol/acetic acid fixative for 20 minutes. Images were acquired using Biorad GelDoc, and colonies were counted using ImageJ. Statistical analyses and charts from two or more experiments were prepared with Prism 6.0.
Results
Elevated DNA Damage Response in HCC
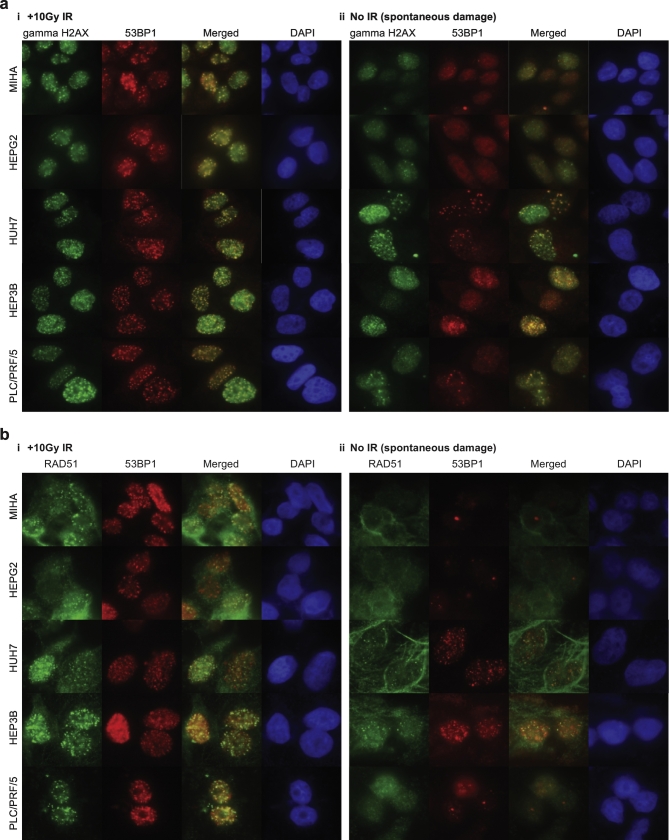
Upon genotoxic challenge, DNA damage signaling or repair proteins are rapidly recruited to sites of DNA breaks where they concentrate and form cytologically discernable punctate structures. Gamma-H2AX (Serine139 phosphorylation) plays an established role in initiating and orchestrating the local accumulation of DNA repair protein complexes at DNA damage sites [18]. We used a specific antibody that detects only gamma-H2AX as the surrogate marker of DNA breaks to estimate the level of DNA damage in HCC cell lines and the immortalized hepatocytes [16]. Foci formation IF experiments enabled the visualization of protein recruitment to the site of DNA breaks with or without exogenous DNA damage (Figure 1). Upon ionizing radiation [IR, 10 Gy (dose rate 10.85 Gy/min, recorded at 2004 July 06)], both HCC cell lines and immortalized hepatocytes demonstrated prominent H2AX serine139 phosphorylation as well as its focal concentration at site of DSBs, indicating that DNA damage sensing function is preserved (Figure 1Ai). The higher level of gamma-H2AX foci formation in cancerous cell lines compared to MIHA in the absence of exogenous DNA damage suggested a higher level of spontaneous DNA damage response in HCC cells (Figure 1Aii).
Figure 1.
Elevated endogenous DNA damage in HCC.
Immortalized hepatocyte MIHA and HCC cell lines were seeded onto 6-well culture plates with coverslips and treated (i) with or (ii) without IR (10Gy). After recovery for 6h, fixed and permeabilized cells were immunostained with (a) gamma-H2AX and 53BP1 and, (b) RAD51 and 53BP1 antibodies, followed by fluorescent-conjugated secondary antibodies. Nuclei were counterstained with DAPI. Images were captured on Nikon Eclipse 800 microscope.
HDR and NHEJ are competing DNA repair mechanisms. It has been demonstrated that 53BP1 loading at DSBs counters DNA end resection and suppresses RPA loading, and thus plays a role in determining the choice of DSB repair pathways [19]. In IF experiment in Figure 1A, we observed that, in both HCC cells and immortalized hepatocytes, 53BP1 colocalized with gamma-H2AX at sites of IR-induced DSBs, as demonstrated with the large overlapping foci (red) that colocalize with gamma-H2AX foci (green) in nuclei (Figure 1Ai). In addition, colocalization of 53BP1 and gamma-H2AX foci was also readily observed in nonirradiated cells, highlighting the presence of spontaneous DNA damage in liver cancer cells. Similar to gamma-H2AX, the number of 53BP1 foci formation was higher in cancerous cell lines compared to immortalized hepatocytes (Figure 1Aii). The presence of both gamma-H2AX and 53BP1 foci suggests that both HDR and NHEJ were functional and in force in both cancerous cell lines HEPG2, HUH7, HEP3B, and PLC/PRF/5 and immortalized hepatocytes for the maintenance of genome integrity against genotoxic stress.
RAD51 is a key component in the ATM-dependent DNA repair pathway and plays a role in the Fanconi anemia repair pathway [11]. RAD51 recruitment to chromatin is indicative of the processivity of the DNA broken ends by HDR as it displaces RPA complexes from single-strand DNA. Dysregulation in DDR signaling or replication will affect RAD51 redistribution upon DNA damage. We thus utilized RAD51 as readout to detect any altered DDR signaling and integrity of HDR in HCC. We assessed the functional concentration of RAD51 at DSBs in response to DNA damage (Figure 1B). Accordingly, the ability to support formation of ionizing radiation-induced foci of RAD51 suggests that HDR repair is functional in the HCC cells examined. Moreover, a higher level of spontaneous HDR was indicated by the higher level of RAD51 foci in nonirradiated HCC cells when compared to immortalized hepatocytes MIHA.
Apical DNA Damage and Repair Protein in HCC
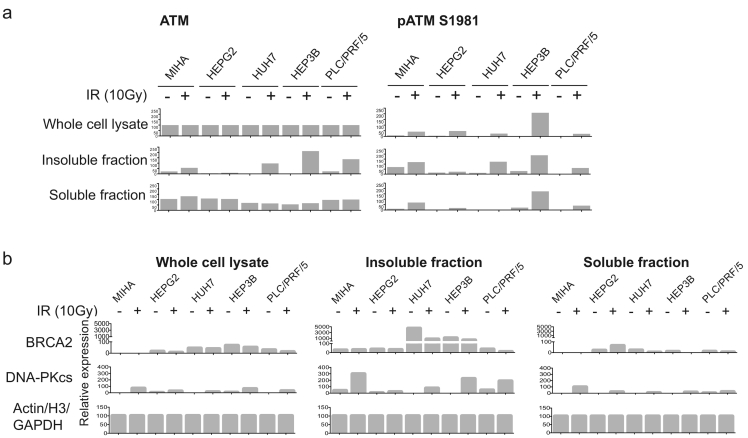
Upon detection of DSBs, DNA damage signaling and repair factors would accumulate in the chromatin surrounding DSBs as functional entities. Activated ATM triggers rapid amplification of downstream DDRs. BRCA2 is an essential recombination-mediating protein in HDR. On the other hand, DNA-PKcs is required for NHEJ repair [20]. We examined the expression level of the three proteins as well as their chromatin loading upon IR-induced DNA damage. Soluble and insoluble (chromatin) fractions as well as whole cell lysates were collected before and after ionizing irradiation damage (IR, 10 Gy) (Figure 2).
Figure 2.
Homology-directed repair regulators were upregulated in HCC.
Immortalized hepatocyte MIHA and HCC cell lines were seeded onto 100mm culture dishes and treated with IR (10Gy) as indicated. After 6h recovery, whole cell lysates (W), soluble fractions (S) and insoluble fractions (I) were collected. (a) Activation of ATM autophosphorylation in HCC. Protein fractions were blotted with ATM and ATM Serine1981 phosphorylation (pATM S1981) antibodies. (b) Protein expression and subcellular distribution of BRCA2 and DNA-PKcs. Signal intensities were measured and normalized using ImageJ and plotted as bar chart using Prism 6.
Activation of ATM Autophosphorylation in HCC with Ionizing Radiation
ATM undergoes autophosphorylation at serine 1981 upon DSBs, causing homodimer dissociation and thus allowing phosphorylation of its downstream targets [21]. We examined if ATM protein kinase expressed in liver cancer cells and immortalized hepatocytes is catalytically active and responsive to DNA damage. Western blot results on whole cell lysates demonstrated that ATM was ubiquitously expressed in both liver cancer cells and immortalized hepatocytes (Figure 2A, Supplementary Figure 1A). Majority of ATM proteins were found in soluble fractions in cells without genotoxic stress. Notably, 6 hours after IR-induced DNA damage, ATM autophosphorylation at serine 1981 was observed in all cells with pATM serine 1981 phosphorylation antibody. Significant chromatin loading of the catalytically active pATM was also detected in insoluble fraction, suggesting functional activation of ATM-regulated DDR. Higher levels of activated pATM recruitment to chromatin in HCC cells were observed compared to nontumorigenic hepatocytes MIHA in response to IR.
Upregulated BRCA2-Chromatin Loading in HCC
BRCA2 harbors RAD51-binding BRC-repeats at its C terminus [22] and is required at DSBs to facilitate RPA displacement by RAD51. Absence of BRCA2 compromises RAD51 loading onto chromatin and in turn impairs HDR repair [23]. We asked if BRCA2 is functionally recruited to DSBs in HCC cells by analyzing its chromatin association. Western blot analyses using specific antibody against BRCA2 demonstrated that the overall expression of BRCA2 was higher in all cancer cells compared to MIHA (whole cell lysate, W) (Figure 2B, Supplementary Figure 1B). Constitutive BRCA2 chromatin loading was observed in all cells before and after exogenous genotoxic stress (IR 10 Gy) (insoluble fraction, I). More robust loading of BRCA2 onto chromatin was observed in insoluble protein fractions from HCC cells. These observations are in agreement with the higher number of RAD51 foci in HCC cells compared to MIHA in IF microscopy examination (Figure 1B), as BRCA2 is the major recruiting partner of RAD51 for chromatin recruitment. It also suggests that HDR repair is in effect in both liver cancer cells as well as in the immortalized hepatocyte MIHA. However, the higher level of total BRCA2 observed in malignant cells (both soluble and insoluble form) may allow a more rapid response to activate HDR repair and higher rate of repair upon sensing of DSBs.
Relatively Lower DNA-PKcs Chromatin Recruitment in HCC
DNA-PKcs expression was highly responsive to ionizing radiation (10 Gy IR) in MIHA (Figure 2B). While its expression was maintained at basal level in the absence of exogenous DNA damage, both protein expression and chromatin loading (insoluble fraction) of DNA-PKcs in MIHA were significantly induced upon IR. Comparable induction of DNA-PK expression and chromatin recruitment was not observed in HCC cells. HEPG2 and HUH7 demonstrated negligible induction of DNA-PKcs, while that in HEP3B and PLC/PRF/5 was suboptimal compared to MIHA. The disparity suggests that NHEJ repair pathway was downregulated in HCC and/or DNA repair might have been redirected to other repair mechanism(s).
Homology-Directed Recombination in HCC
Based on our findings on protein expression and chromatin recruitment of HDR proteins, we further examined if HCC cells may display upregulation in HDR for repairing DSBs.
Gene Conversion in HCC and Immortalized Noncancerous Hepatocytes
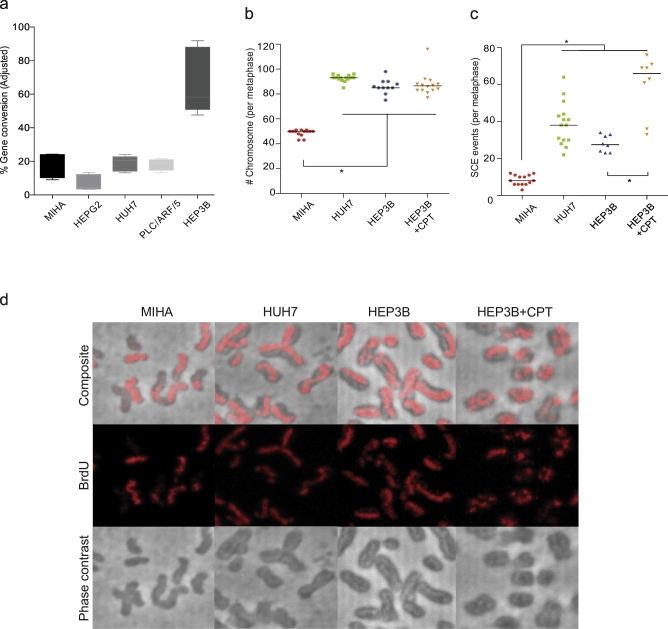
We employed the gene conversion functional assay to study HR capability of HCC cells in comparison to the noncancerous hepatocyte MIHA (Figure 3A, Supplementary Figure 2). All malignant HCC and noncancerous hepatocytes demonstrated capability in gene conversion to restore a complete GFP allele that gave green fluorescence signal in FACS analyses. The results indicate the existence of functional HR in these cells, albeit at different efficiencies. Highest gene conversion was observed in hepatitis B–carrier HEP3B, while the non–virus-related HEPG2 demonstrated the lowest level of gene conversion. However, statistical significance was not reached when we looked at HCC cell lines collectively against MIHA as a noncancerous control [Kruskal-Wallis test (P = .0013) with Dunn's multiple comparison post hoc test (P > .05)].
Figure 3.
Gene conversion and SCE in HCC and immortalized hepatocytes.
(a) Unique DSBs were introduced into cells by electroporation of DR-GFP substrate and pCBAs-I-SceI plasmids. Relative gene conversion efficiency was expressed as a function of green fluorescent signal from the repair of DSBs on DR-GFP by gene conversion. (b) Number of chromosomes and (c) SCE events by metaphase analyses in HCC or immortalized hepatocytes were assessed using ImageJ and analyzed by Prism 6. Asterisks (*) indicates significant data point (p<0.005) by Kruskal-Wallis test. d) Representative metaphases demonstrating “Harlequin” chromosomes in HCC but MIHA. Number of Harlequin chromosomes increased with camptothecin treatment.
Prominent Hyperploidy and SCEs in HCC
We also set up metaphase spreading and SCE assays to determine both chromosomal abnormality and SCE incidences. In analyses on metaphases collected from HEP3B, HUH7, and MIHA, a significant difference was observed in the number of chromosomes per metaphase (Kruskal-Wallis test, P < .0001) (Figure 3B, Supplementary Figure 3). Chromosome number in HCCs (mostly tetraploid, 4N) deviated significantly from the noncancerous diploid (2N) MIHA.
SCE frequency is reflective of HDR capacity. Cells with hyperactive HDR would show notable increase in SCE frequency and vice versa (Figure 3C). By analyzing BrdU signals, we observed an average of 8.29 ± 2.76 SCE events per metaphase in MIHA, which is comparable to that of the reported spontaneous SCE per metaphase in normal cycling cells [24]. By contrast, both non–virus-related HUH7 and HBV-related HEP3B demonstrated frequent spontaneous recombination incidence between sister chromatids, with an average number of SCE events per metaphase of 38.8 ± 11.59 events and 28.00 ± 4.53 events, respectively. The frequent SCE in HCCs led to the formation of Harlequin chromosome; examples are shown in Figure 3E. Statistical analyses of SCE events per metaphase from HCCs and MIHA by Kruskal-Wallis test indicated that HCC demonstrated a significantly more frequent SCE than MIHA (P < .0001) (Figure 3C).
We examined whether HDR could be further elevated by the introduction of replication-dependent spontaneous DSBs, thus leading to more severe SCE phenotype in HCC. HEP3B was treated with low-dose 1 nM camptothecin (CPT) in the second BrdU-labeling cell cycle before metaphases were collected. CPT is a topoisomerase-I inhibitor that would induce replication-associated DSBs by stalling TOPOI on replication forks [25]. Chromosome numbers and ploidy status in CPT-treated HEP3B metaphase remained comparable to untreated control (Figure 3B, D). Nonetheless, both the number of “Harlequin” chromosomes and that of SCE in each metaphase with CPT treatment were increased significantly (P < .001) (Figure 3B, E).
Testing Knowledge-Based Chemotherapy Design for HCC
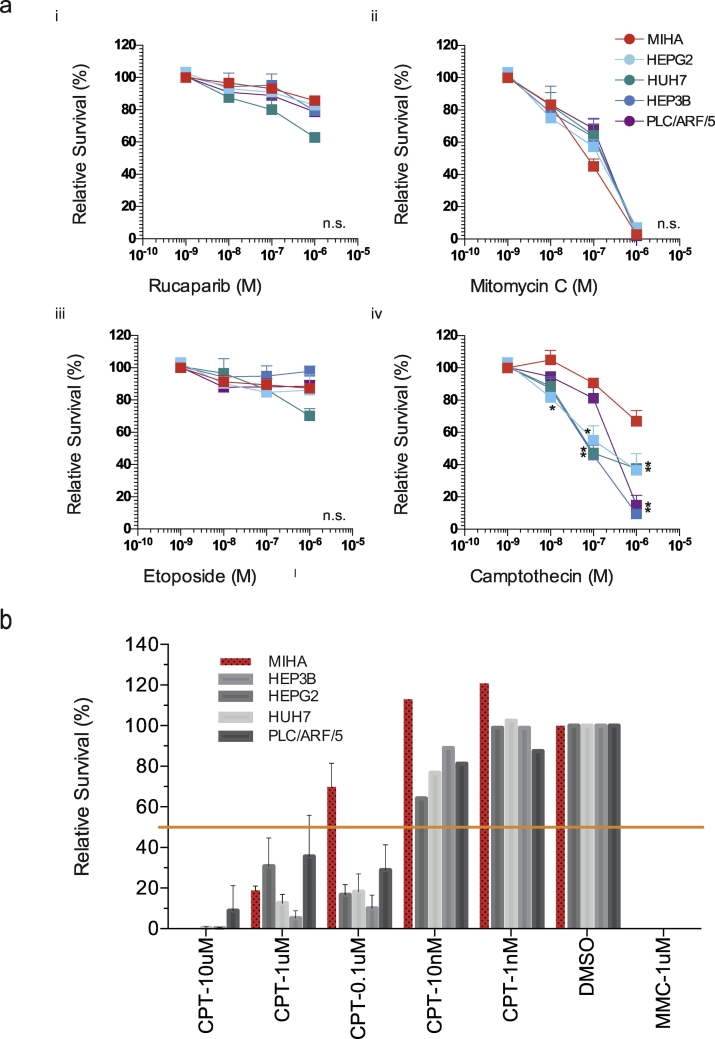
We performed follow-up study to test if the disparity in HDR capability and capacity in HCC could serve as predictive marker for targeted chemotherapy. We tested if CPT would have selective cytotoxic activity towards HCC cells. While treatment with rucaparib (PARP inhibitor), mitomycin C (DNA cross-linker), and etoposide (topoisomerase II inhibitor) demonstrated no differential cytotoxicity to HCC, the MTT assay demonstrated a significant selective anticancer effect of CPT towards HCC at concentration 1 × 10−8 to 1 × 10−6 M (P < .05-.0001) (Figure 4A). We further tested the cytotoxicity of CPT on HCC using long-term colony formation assay. Agreeing to our finding in MTT tests, CPT demonstrated specific cytotoxicity towards HCC at 1 × 10−7 M concentration (Figure 4B).
Figure 4.
Cytotoxic effect of camptothecin on HCC.
(a) Drug sensitivity of HCC upon treatment with i. PARP inhibitor Rucaparib, ii. DNA crosslinker mitomycin C, iii. Topoisomerase II inhibitor etoposide and iv. Topoisomerase I inhibitor camptothecin was assessed. Two-way ANOVA and Dunnett’s posthoc test were employed for statistical significance. Asterisks (*) indicates significant data points. N.s.: no statistical significance was observed. (b) Long term cytotoxic effect of CPT was assessed using colony formation assay on HCC and MIHA.
Discussion
In this study, we report elevated DNA damage and HDR in HCC cells. The excessive DNA damage foci observed in HCC cells without exogenous genotoxic stress indicated that the level of spontaneous DNA damage is higher among HCC cell lines. Theoretically, the number of DNA lesions that resulted from difference sources of endogenous DNA damage, including oxidation (400-1500), depurination (104), cytosine deamination (100-500), and SAM-induced methylation (10 – 4 × 103) per cell per day, could have activated programmed cell death [26] before cells acquire adequate prosurvival oncogenic genetic modification or inactivation in the apoptotic signaling pathway(s). To survive from all the genotoxic stresses during the course of hepatocarcinogenesis, the highly proliferative hepatocytes would need to tailor DDRs to accommodate the excessive DNA damage in the cell.
Our study showed that HDR is functional and is augmented for DSB repair in HCC. The observed upregulated phosphorylation of ATM in HCC (Figure 2a) suggests that ATM activation and the downstream ATM-licensed molecular pathways might be essential in the initiation and/or progression of hepatocarcinogenesis. Indeed, hepatocarcinogenesis was abrogated in ATM null mouse model [27]. On the contrary, diethylnitrosamine treatment in knockout mouse model lacking Ku70, an essential protein for Ku70/Ku80 heterodimer that recruits DNA-PK for NHEJ repair initiation, suggests that NHEJ may be dispensable for hepatocarcinogenesis [28].
It was demonstrated in Bloom (Blm tumor suppressor)-deficient mouse model that elevated mitotic recombination is a major mechanism for the loss of heterozygosity (LOH) and also increased tumor susceptibility [29]. The elevated HDR observed in HCC could be the primary driving mechanism for LOH. In fact, frequent LOH and allelic losses in primary HCC have been repeatedly reported [[30], [31], [32]]. The fact that there were no significant differences in the frequency of allelic losses with different HCC etiologies suggests that HDR and LOH might be fundamental to hepatocarcinogenesis.
With the highly rearranged genome and frequent LOH at a variety of loci observed in HCC, it is possible that hyperactive HDR in HCC might take advantages of nonallelic homologous templates for recombination processes. The availability of homologous template is one of the rate-determining steps for HDR [33]. Heightened replicative stress and increased DNA damage during cell proliferation might prompt HCC cells to switch to the use of nonallelic homologous recombination (NAHR) so as to speed up the rate of DNA repair and avoid the onset of cell death. NAHR recognizes only small stretches of homologous regions for speedy template matching. The utility of nonallelic repair template compromises the fidelity of HDR repair processes and thus leads to allelic imbalances and LOH. It is also reported that telomere attrition, i.e., single-sided DSB lesions at eroded telomeric ends, could induce NAHR between chromosomes, which in turn causes massive GCRs and genomic imbalances through “breakage-fusion-bridge” cycles [34].
Under physiologic condition, replication stress is a major source of spontaneous DNA damage as a result of vigorous cell proliferation. Most of the recurrent chromosome aberrations occur at fragile sites, where nucleotide content and microhomology make the locations difficult for replication. Problematic DNA replication may lead to replication fork collapse, subsequent premature chromosome condensation, and the formation of DSBs. Results from our metaphase experiments confirmed the higher genomic instability in HCC cells. The significantly higher exchange events between sister chromatids directly reflected a hyperrecombination phenotype in HCC. Results from CPT treatment in HEP3B also demonstrated that acute replication-dependent DNA damage can induce genomic instability through HDR (Figure 3, C-D).
According to a high-coverage whole genomic sequence analysis on somatic structural variation in various human cancers, the major structural aberration observed in HCC patient samples was tandem duplications [7]. The major DNA repair mechanisms that could lead to tandem duplications are SCE by HR and the formation of palindromic sequence that causes tandem repeat inversion duplications in inverted orientations [35]. The frequent tandem duplications observed by high-coverage whole genomic sequence analysis could be the resultant phenotype for elevated HDR observed in HCC.
The effect of CPT on the elevated frequencies of SCE and Harlequin chromosomes in HCC highlights the role of HDR in the restoration of collapsed replication fork and genomic instability. It was demonstrated that CPT-induced DSBs require replication. It is possible that BIR, one of the alternative recombination mechanisms, participates in the repairing of collapsed forks that arise from CPT treatment. BIR would cause chain reaction of leading strand invasion to dsDNA with short homology, thus resulting in massive nonrecurrent rearrangements [36]. BIR might be responsible for the ongoing chromosome instability observed during HCC progression.
Inducing Synthetic Lethality in HCC
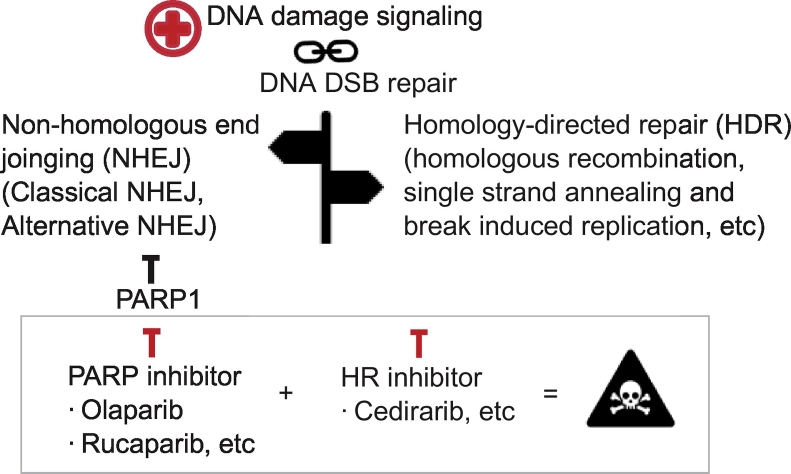
In order to induce synthetic lethality, the lack of HDR where DSBs would be directed to NHEJ repair is a prerequisite to the action of PARP inhibitor. PARP inhibitors would deregulate DNA-PK and thus the error-prone NHEJ repair [37]. Sensitivity towards PARP inhibitors was observed in other cancer types that demonstrated defective HR, such as familial breast and ovarian cancer with mutated BRCA1, BRCA2, or RAD51 [38]. However, our DNA repair functional assays revealed that HDR repair is augmented in liver cancer cells. In this scenario, DSBs will be repaired by HDR, which leaves the NHEJ pathway redundant, and thus, inhibition on NHEJ with PARP inhibitor, e.g., rucaparib, gave no lethal phenotype. Nonetheless, the recently identified novel action of vascular endothelial growth factor receptor inhibitor cediranib might serve as adjunct targeted therapy for liver cancer [39].
It was demonstrated that cediranib can downregulate expression of BRCA proteins and RAD51, thus suppressing HR in cancer cells [40]. A clinical trial on ovarian cancer patients demonstrated that combining the inhibiting action of cediranib on HR proteins with olaparib significantly improved progression-free and overall survival in patients without BRCA mutations [41,42]. Elaborating on the scenario and applying to the equation for synthetic lethality in liver cancer where HDR serves as the major repair pathway, the use of cedirarib alone or in combination with PARP inhibitors and/or DNA damaging agents would induce therapy sensitivity in HCC (Figure 5). However, this therapeutic approach should be adopted with care as simultaneous blockade of both HDR and NHEJ repair pathways could lead to the loss of therapeutic specificity towards cancer cells and be detrimental to adjacent noncancerous tissues. Albeit targeting both HDR and NHEJ repair pathways could be toxic to noncancerous tissues, it also could allow for lower doses of each drug to be used in combination which could also reduce overall toxicity to adjacent tissue.
Figure 5.
Inducing synthetic lethality in HCC by suppressing DSB repair.
Mechanistic diagram on adjunct therapy using PARP inhibitors and HDR inhibitors in HCC. Blocking both NHEJ and HDR DSB repair would lead to accumulation of DNA damages in HDR-capable HCC cells and thus induce cell death.
With our new findings describing augmented HDR repair in HCC, HDR could be useful targets for the development of prognostic biomarkers for predicative prescription in HCC therapy. Information on DDR molecular signature of HCC might be a useful tool for clinical decision on radiation and chemotherapies. HDR biomarker(s) could aid the identification of HCC patients who are suitable for treatment with therapeutic reagent(s) that targets the DDR pathway(s). Owing to the use of only cell line models in the current study, it would be best to extend the study to human primary HCC cells for consolidation on the role of HDR in hepatocarcinogenesis.
Acknowledgments
Acknowledgements
We thank Drs. Stephanie Ma and Wilson Ching (The University of Hong Kong) for sharing hepatocellular carcinoma and immortalized hepatocyte cell lines. We also thank the Core facilities of the Li Ka Shing Faculty of Medicine and Centre for Genomic Sciences, HKU, for their technical assistance.
Declaration of Interest
All authors declare no conflict of interest.
Financial support
This work is supported by the Health and Medical Research Fund, Hong Kong (Grant number: 14131132).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100796.
Appendix A. Supplementary data
Supplementary Figure 1 Western Blot images for protein expression and subcellular distribution of a) ATM and pATM S1981 and b) BRCA2 and DNA-PKcs in HCC and immortalized hepatocytes.
Supplementary Figure 2 Flow cytometry analyses on gene conversion in HCC and immortalized hepatocytes. Green fluorescence positive cells are indicative of successful gene conversion on GFP coding region by HR. Red square highlights the green fluorescent positive cell population. A separate electroporation of pEGFP was performed for adjustment of plasmid take-up efficiency in individual cell line.
Supplementary Figure 3 Representative metaphases of HCC and immortalized hepatocytes. Phase contrast and BrdU/Alexa fluor 594 images of metaphases were captured using Carl Zeiss LSM880 with Airyscan processing using ZEN software.
References
- 1.E.M., Ferlay J., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. International Agency for Research on Cancer; Lyon, France: 2018. Global cancer observatory: cancer today. [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Lee H.W., Blasco M.A., Gottlieb G.J., Horner J.W., II+, Greider C.W., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 4.Neelsen K.J., Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 6.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 7.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H., Zhang C., Ren X., Protopopov A., Chin L., Kucherlapati R., Lee C., Park P.J. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhsng C.Z., Wala J., Mermel C.H., Sougnez C., Gabriel S.B., Hernandez B., Shen H., Laird P.W., Getz G., Meyerson M., Beroukhim R. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denchi E.L., de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 10.Casper A.M., Nghiem P., Arlt M.F., Glover T.W. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 11.Livingston D.M. Cancer. Complicated supercomplexes. Science. 2009;324:602–603. doi: 10.1126/science.1174839. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto Y., Puddu F., Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol. 2011;19:17–24. doi: 10.1038/nsmb.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sibanda B.L., Chirgadze D.Y., Ascher D.B., Blundell T.L. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science. 2017;355:520–524. doi: 10.1126/science.aak9654. [DOI] [PubMed] [Google Scholar]
- 14.van der Burg M., Ijspeert H., Verkaik N.S., Turul T., Wiegant W.W., Morotomi-Yano K., Mari P.O., Tezcan I., Chen D.J., Zdzienicka M.Z., van Dongen J.J., van Gent D.C. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sy S.M., Huen M.S., Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X., Fu S., Lai M., Baer R., Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappold I., Iwabuchi K., Date T., Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 19.Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C.X., Finkel T., Nussenzweig M., Stark J.M., Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith G.C., Jackson S.P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 21.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 22.Davies O.R., Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connor F., Bertwistle D., Mee P.J., Ross G.M., Swift S., Grigorieva E., Tybulewicz V.L., Ashworth A. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 24.Crossen P.E., Drets M.E., Arrighi F.E., Johnston D.A. Aanlysis of the frequency and distribution of sister chromatid exchanges in cultured human lymphocytes. Hum. Genet. 1977;35:345–352. doi: 10.1007/BF00446625. [DOI] [PubMed] [Google Scholar]
- 25.Saleh-Gohari N., Bryant H.E., Schultz N., Parker K.M., Cassel T.N., Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teoh N., Pyakurel P., Dan Y.Y., Swisshelm K., Hou J., Mitchell C., Fausto N., Gu Y., Farrell G. Induction of p53 renders ATM-deficient mice refractory to hepatocarcinogenesis. Gastroenterology. 2010;138:1155–1165. doi: 10.1053/j.gastro.2009.11.008. e1151–1152. [DOI] [PubMed] [Google Scholar]
- 28.Teoh N.C., Dan Y.Y., Swisshelm K., Lehman S., Wright J.H., Haque J., Gu Y., Fausto N. Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology. 2008;47:2078–2088. doi: 10.1002/hep.22194. [DOI] [PubMed] [Google Scholar]
- 29.Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 30.Walker G.J., Hayward N.K., Falvey S., Cooksley W.G. Loss of somatic heterozygosity in hepatocellular carcinoma. Cancer Res. 1991;51:4367–4370. [PubMed] [Google Scholar]
- 31.Buetow K.H., Murray J.C., Israel J.L., London W.T., Smith M., Kew M., Blanquet V., Brechot C., Redeker A., Govindarajah S. Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8852–8856. doi: 10.1073/pnas.86.22.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong C.M., Lee J.M., Lau T.C., Fan S.T., Ng I.O. Clinicopathological significance of loss of heterozygosity on chromosome 13q in hepatocellular carcinoma. Clin. Cancer Res. 2002;8:2266–2272. [PubMed] [Google Scholar]
- 33.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. elife. 2014;3 doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazzerini-Denchi E., Sfeir A. Stop pulling my strings — what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol. 2016;17:364–378. doi: 10.1038/nrm.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reams A.B., Roth J.R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 2015;7:a016592. doi: 10.1101/cshperspect.a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 37.Patel A.G., Sarkaria J.N., Kaufmann S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 39.Wedge S.R., Kendrew J., Hennequin L.F., Valentine P.J., Barry S.T., Brave S.R., Smith N.R., James N.H., Dukes M., Curwen J.O., Chester R., Jackson J.A., Boffey S.J., Kilburn L.L., Barnett S., Richmond G.H., Wadsworth P.F., Walker M., Bigley A.L., Taylor S.T., Cooper L., Beck S., Jurgensmeier J.M., Ogilvie D.J. AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res. 2005;65:4389–4400. doi: 10.1158/0008-5472.CAN-04-4409. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan A.R., Gueble S.E., Liu Y., Oeck S., Kim H., Yun Z., Glazer P.M. Cediranib suppresses homology-directed DNA repair through down-regulation of BRCA1/2 and RAD51. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J.F., Barry W.T., Birrer M., Lee J.M., Buckanovich R.J., Fleming G.F., Rimel B.J., Buss M.K., Nattam S.R., Hurteau J., Luo W., Curtis J., Whalen C., Kohn E.C., Ivy S.P., Matulonis U.A. Overall survival and updated progression-free survival outcomes in a randomized phase II study of combination cediranib and olaparib versus olaparib in relapsed platinum-sensitive ovarian cancer. Ann. Oncol. 2019;30:551–557. doi: 10.1093/annonc/mdz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J.F., Barry W.T., Birrer M., Lee J.M., Buckanovich R.J., Fleming G.F., Rimel B., Buss M.K., Nattam S., Hurteau J., Luo W., Quy P., Whalen C., Obermayer L., Lee H., Winer E.P., Kohn E.C., Ivy S.P., Matulonis U.A. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol. 2014;15:1207–1214. doi: 10.1016/S1470-2045(14)70391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Western Blot images for protein expression and subcellular distribution of a) ATM and pATM S1981 and b) BRCA2 and DNA-PKcs in HCC and immortalized hepatocytes.
Supplementary Figure 2 Flow cytometry analyses on gene conversion in HCC and immortalized hepatocytes. Green fluorescence positive cells are indicative of successful gene conversion on GFP coding region by HR. Red square highlights the green fluorescent positive cell population. A separate electroporation of pEGFP was performed for adjustment of plasmid take-up efficiency in individual cell line.
Supplementary Figure 3 Representative metaphases of HCC and immortalized hepatocytes. Phase contrast and BrdU/Alexa fluor 594 images of metaphases were captured using Carl Zeiss LSM880 with Airyscan processing using ZEN software.