Figure 4.
Identification of the BMAL1 Nucleolar Interactome
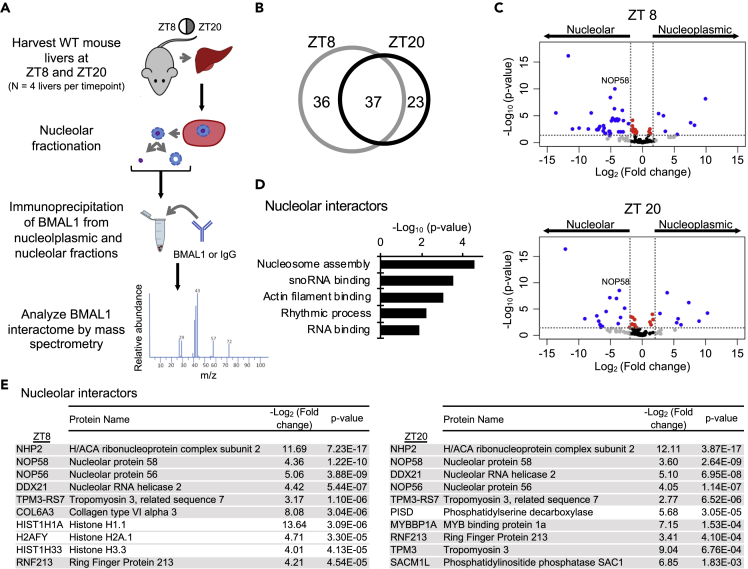
(A) Schematic representation of the experimental design for the nucleolar BMAL1 interactome analysis. Mass spectrometric analysis was performed on N = 4 biological replicates/time point.
(B) Venn diagram of the number of total significant BMAL1-interacting proteins at ZT8 and ZT20 compared with IgG control. Significant interactors were identified when having 2-fold differences between nucleoplasmic and IgG or nucleolar and IgG, respectively, with p < 0.05 as determined by two-way ANOVA adjusted for multiple comparisons.
(C) Volcano plots of nucleoplasmic versus nucleolar BMAL1 interactors at ZT8 and ZT20. Significant interactors were identified when having 4-fold differences (Log2 (Fold change) nucleoplasmic/nucleolar > |2| and p < 0.05 (blue) as determined by two-way ANOVA adjusted for multiple comparisons).
(D) Gene Ontology (GO) analysis of the significant nucleolar BMAL1-interacting proteins (Fisher exact p value denotes annotation significance). (E) Top 10 significant nucleolar BMAL1 interactors listed with p value and -Log2 (Fold change) of the comparison between nucleoplasmic/nucleolar. Proteins highlighted in gray appear at both time points.