Abstract
The pandemic of COVID-19 (Coronavirus Disease-2019) is an extremely contagious respiratory illness due to a novel coronavirus, SARS-CoV-2. Certain drugs have several protein targets and many illnesses share overlapping molecular paths. In such cases, reusing drugs for more than one objective and finding their novice uses can considerably decrease the time in finding new cures for unforeseen diseases. Remdesivir has been recently a strong candidate for the treatment of Covid-19. In this commentary, we have portrayed the structure of the coronavirus in a simple way as well as the site where remdesivir acts. We have also displayed the ongoing clinical trials, as well as a published study that was conducted on compassionate base. The covid-19 pandemic might wean down by the end of summer 2020, but the risk of seasonality exists. Therefore, future disposal of agents such as remdesivir might be crucial for ensuring an efficient treatment, decrease mortality and allow early discharge.
Communicated by Ramaswamy H. Sarma
Keywords: Remdesivir, Covid-19, treatment
Introduction
The pandemic of COVID-19 (Coronavirus Disease-2019) is an extremely contagious respiratory illness (Elmezayen et al., 2020; Sarma et al., 2020). Globally, scientists are attempting to study this novel virus, and to discover efficient management to control and prevent the illness (Enayatkhani et al., 2020). The common symptoms of Covid-19 symptoms are malaise, fever, shortness of breath, dry cough, malaise, while decreased or loss of taste and olfactory perceptions have been reported (Elfiky, 2020a).
The Covid-19 virus spreads mainly through saliva, droplets, or discharges from the nose of an infected individual after coughing or sneezing (Pant et al., 2020). The incubation period can range from two to fourteen days (Abdelli et al., 2020). Since reported in December 2019, it has infected 2, 160, 207 individuals, leading to 146,088 deaths globally (WHO, 2020a). The first reported death was on 11 January 2020 and the transmission from patients to healthcare workers was first documented on 20 January 2020 (Enmozhi et al., 2020). Moreover, the lives of millions of people have been affected due to isolations, required lockdowns, and quarantines. Thus the dreaded effect of the COVID-19 pandemic has imposed main challenges for worldwide well-being, economy and society (Joshi et al., 2020; Umesh et al., 2020).
Coronavirus is lavish in mammals and birds and institute a large family of non-segmented, enveloped, positive- sense, single-stranded RNA betacoronavirus of the family Coronaviridae (Schoeman & Fielding, 2019), and has a high frequency of genomic recombination and mutation (Khan, Zia, et al., 2020). This genetic modification of CoV can hinder the production of efficient vaccine (Hasan et al., 2020). Bats are the main harbor to the ultimate variety of genotypes with spread via an unknown intermediate mammal host to humans (Zhu et al., 2020). Overall, human and animal coronavirus comprises of four genera namely α, β, γ and δ coronavirus genus (Sinha et al., 2020). The β coronavirus includes the Middle East respiratory syndrome (MERS), severe acute respiratory syndrome (SARS), and SARS-CoV-2 (Khan, Jha, et al., 2020; Sahu et al., 2020).
Coronavirus (CoV) structure
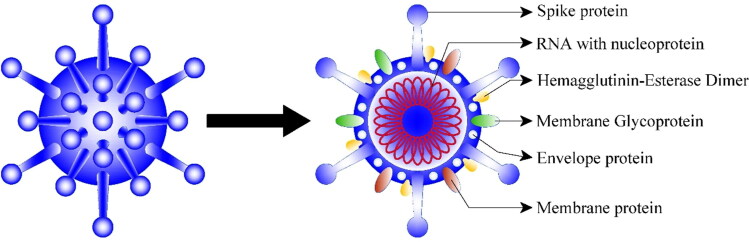
The CoV family has a sizable homogeneous “spike protein” (Figure 1). The role of the spike (S) protein, which is composed of 1300 amino acids (Elfiky, 2020b), is to interact with the host cells, such as the pulmonary and parabronchial epithelial cell, and assists the coronavirus to enter through the epithelial cell membrane (Boopathi et al., 2020; Xia et al., 2020).
Figure 1.
Structure of Covid-19.
Moreover, the alveolar epithelial cells have ample expression of angiotensin-converting enzyme 2 (ACE2), which is an aim by the virus. The detection of ACE2 by the S protein of the virus permits the invasion of the coronavirus into the human circulation system (Belouzard et al., 2009).
Single-strand RNA (22–26 kilobases) viruses such as the coronavirus family reproduce the virus genomes by capitalizing on host cells. For instance, after coronavirus comes near the ribosome of the epithelial cells or other host cells, it utilizes the ribosome of the host cell to replicate polyproteins. The replication and ensuing procedures of precursor polyproteins can arise in the epithelial cells (Hoffmann et al., 2020; Wahedi et al., 2020).
After the coronavirus polyproteins are exhibited, two enzymes, coronavirus main proteinase (3CLpro) and the papain-like protease (PLpro) are believed to be involved in cleaving the polyproteins into smaller products used for replicating new viruses. In order to produce the daughter RNA genome, the coronavirus exhibits an RNA-dependent RNA polymerase (RdRp), which is an important replicase that catalyzes the synthesis of a complementary RNA strand using the virus RNA (Wrapp et al., 2020). CoVs gather together near intracellular membranes within the Endoplasmic reticulum-Golgi intermediate compartment (or ERGIC) after infection. Here they bud within the lumen and eventually carried outside the cell through ‘exocytosis’ within vesicles (Gupta et al., 2020). Replication of SARS-CoV-2 depends on the viral RNA-dependent RNA polymerase (RdRp) (Elfiky & Azzam, 2020) which is the most probable target of the investigational nucleotide analogue remdesivir (RDV) (Agostini et al., 2018; Jordan et al., 2018; Siegel et al.,2017; Tchesnokov et al., 2019).
Search for treatment
After the outbreak, many kinds of medications alone or as adjuvant have been used in many countries (Islam et al., 2020). The usage of already known viral treatments have many advantages as the pharmacodynamics, pharmacokinetics, and safety profiles of these medications have already been recognized (Das et al., 2020). Certain drugs have several protein targets and many illnesses share overlapping molecular paths. In such cases, reusing drugs for more than one objective and finding their novice uses can considerably decrease the time in finding new cures for unforeseen diseases (Muralidharan et al., 2020).
Remdisivir (RDV)
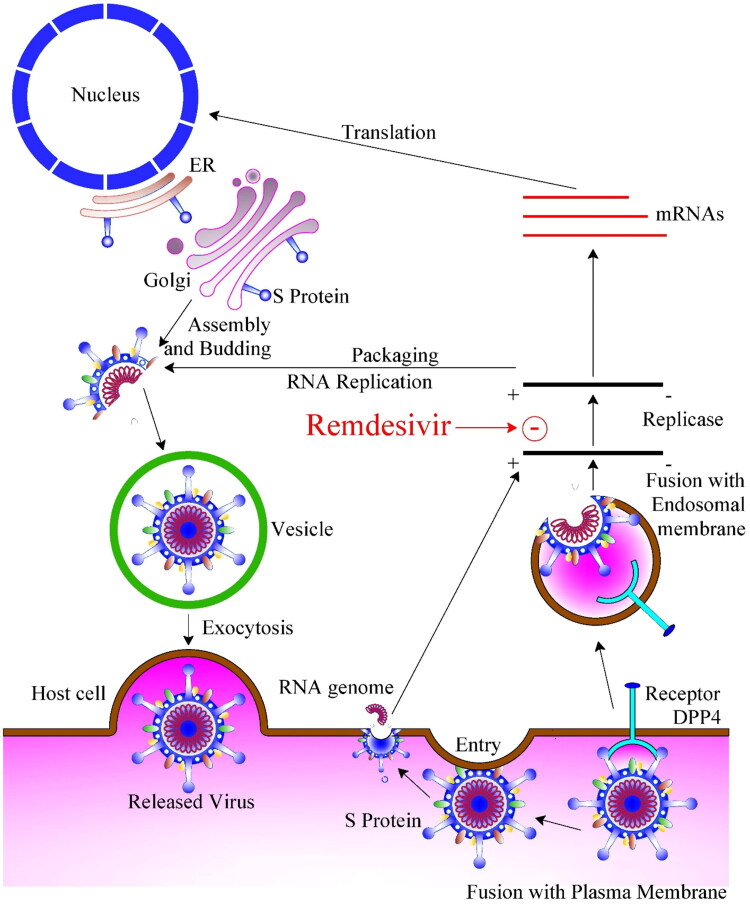
RDV exhibits broad-spectrum antiviral activity against RNA viruses, and former studies with RdRps from Ebola virus (EBOV) and Middle East respiratory syndrome coronavirus (MERS-CoV) have shown that delayed chain-termination is RDV’s conceivable mechanism of action (Figure 2) (Agostini et al., 2018; Jordan et al., 2018; Siegel et al.,2017; Tchesnokov et al., 2019).
Figure 2.
Mode of infection of Covid-19 and Remdasivir mechanism of action. ER: Endoplasmic Reticulum; DPP4: Dipeptidyl peptidase-4.
Recently, Gordon, Tchesnokov, Woolner, et al. (2020) proved that RdRp effectively incorporates the active triphosphate form of RDV (RDV-TP) into RNA, causing termination of RNA synthesis.
The investigators obtained almost identical results with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRps and concluded that the distinguished property of RDV-TP is its high discernment over incorporation of its natural nucleotide counterpart ATP.
RDV, formerly known as GS-5734, is a monophosphoramidate prodrug of an adenosine analog that was developed in response to the Ebola outbreak in West Africa from 2014.
RDV binds to RdRp and acts as RNA chain terminator. It exhibits effective in vitro activity against SARS-CoV-2 with an EC50 at 48 h of 0.77 µM in Vero E6 cells (Wang et al., 2020). Comparable activity has been shown against other zoonotic coronaviruses with EC50 values of 0.07 µM demonstrated for both SARS-CoV-1 and MERS-CoV (Gordon, Tchesnokov, Feng, et al., 2020; Sheahan et al., 2017, 2020; Wang et al., 2020).
RDV is very discerning for viral polymerases, hence a low propensity to cause human toxicity. In addition, it has shown to have a wide therapeutic index in a human airway epithelial cell model (Sheahan et al., 2020). The drug also exhibits a high genetic hurdle to resistance in coronaviruses and has an extended intracellular half-life that permits for once-daily dosing (Agostini et al., 2018; Sheahan et al., 2020).
The safety and pharmacokinetics of RDV were assessed in single- and multiple-dose phase Intravenous infusions between 3 mg and 225 mg and were well-tolerated without any evidence of kidney or liver toxicity. RDV showed linear pharmacokinetics within this dose range and an intracellular half-life of more than 35 h. Ensuing multiple-dose infusions, reversible aspartate aminotransferase and alanine transaminase elevations ensued (WHO, 2020b).
The dose under investigation for treatment of COVID-19 is 200 mg intravenously (IV) on day 1 followed by 100 mg IV daily for up to 10 days, infused over 30–60 min. The initial clinical use of RDV was conducted by Jacob et al in 2016 to treat Ebola (Jacobs et al., 2016), followed by case series (Holshue et al., 2020; Kujawski et al., 2020).
Clinical trials
As of April 19, 2020, the following studies are being conducted in the United States using RDV to treat covid-19 (ClinicalTrials.gov, 2020): NCT04302766, NCT04292899, NCT04292730, NCT04257656, NCT04252664, NCT04280705.
In a recent study published by the New England Journal of Medicine, investigators have used RDV on a compassionate-use basis to patients hospitalized with Covid-19. Participants were patients with confirmed SARS-CoV-2 infection and who had an oxygen saturation of 94% or less in room air or who were receiving oxygen support. Patients received a 10-day course of RDV, comprising of 200 mg administered intravenously on day 1, ensued by 100 mg daily for the remaining 9 days of management. Overall, 61 patients were recruited but only data from 53 patients were analyzed. Participants were from the United States, Europe, Canada, and Japan. More than 50% of patients (n = 30) were receiving mechanical ventilation and almost 10 % (n = 4) were receiving extracorporeal membrane oxygenation. On follow-up (median =18 days), 36 patients (68%) had an amelioration in oxygen-support, including 17 of 30 patients (57%) getting mechanical ventilation who were extubated. Moreover, 25 patients (47%) were discharged, and 7 patients (13%) expired. The authors concluded that use RDV, led to clinical improvement in 36 of 53 patients (68%) infected with Covid-19.
In terms of safety, 32 patients (60%) reported adverse events during follow-up. The most common side events were renal impairment, rash, diarrhea, increased hepatic enzymes, and hypotension. Overall, side effects were more common in patients on invasive ventilation. A total of 12 patients (23%) had grave adverse events, most commonly septic shock, multiple-organ-dysfunction syndrome, hypotension, and acute kidney injury. Four patients (8%) terminated RDV treatment prematurely: one because of deteriorating of preexisting renal failure, one because of multiple organ failure, and two because of transaminitis, including one patient with a maculopapular rash (Grein et al., 2020).
On April 29, 2020, the National Institute of Allergy and Infectious Diseases (NIAID), published preliminary results on the effect RVD on covid-19 illness. The randomized, controlled trial that was initiated on February 21, 2020 involved 1063 patients. The mortality rate for the group of individuals who received RVD was 8 % as compared to 11.6% in the placebo group (p = 0.059). For patients who survived the illness, the median time to recovery was 31% quicker for patients who received RVD compared with those who received placebo (11 vs 15 days) (p < 0.001) (NIH, 2020).
On April 29, 2020, Gilead revealed results from the open-label, Phase 3 SIMPLE trial evaluating 5-day and 10-day dosing durations of the investigational antiviral remdesivir in hospitalized patients with severe manifestations of COVID-19 disease. Inclusion criteria was pneumonia and reduced oxygen levels that did not require mechanical ventilation at the time of study. The study showed that patients who received a 10-day treatment course of remdesivir attained comparable improvement in clinical status compared with those taking a 5-day treatment course (Odds Ratio: 0.75 [95% CI 0.51 – 1.12] on Day 14).
The time to clinical improvement for 50 percent of patients was 10 days in the 5-day treatment group and 11 days in the 10-day treatment group. More than 50% of patients in both treatment groups were discharged from the hospital by Day 14 (5-day: 60.0%, n = 120/200 vs.10-day: 52.3% n = 103/197; p = 0.14). At Day 14, 64.5 percent (n = 129/200) of patients in the 5-day treatment group and 53.8 percent (n = 106/197) of patients in the 10-day treatment group achieved clinical recovery. The overall mortality rate at Day 14 was 7 percent (n = 23/320) across both treatment groups, with 64 percent (n = 205/320) of patients showing clinical improvement at Day 14 and 61 percent (n = 196/320) of patients discharged from the hospital.
No unforeseen side effects were detected with the use of RDV across either treatment group (Gilead, 2020).
On May 1, 2020, the Food and Drug Administration (FDA) issued an Emergency Use Authorization (EUA) for emergency use of RDV for the treatment of hospitalized 2019 coronavirus disease (COVID-19) patients based on review of the topline data from the Gilead-sponsored open-label trial that evaluated different durations of RDV (NCT04292899), and from the randomized, double-blinded, placebo-controlled trial conducted by NIAID (NCT04280705) (FDA, 2020).
Conclusion
Remdesivir might be crucial for ensuring an efficient treatment, decrease mortality and allow early discharge in relation to Covid-19. Ongoing randomized, placebo-controlled trials are critical in delineating its efficacy.
Acknowledgements
Special appreciation for architect Riad Younes for developing the figures.
Disclosure statement
The authors have no potential conflicts of interest relevant to this article to disclose.
References
- Abdelli I., Hassani F., Brikci S. B., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siege l D., Mackman R. L., Clarke M. O., Baric R. S., & Denison M. R. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio, 9(2), 1–15. 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V. C., & Whittaker G. R. (2009). Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5871–5876. 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov 2020. Retrieved April 19, 2020, from https://clinicaltrials.gov/
- Das S., Sarmah S., Lyndem S., & Roy A. S. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (FDA ). (2020). Retrieved May 3, 2020, from https://www.fda.gov/media/137564/download.
- Gilead 2020. Gilead announces results from phase 3 trial of investigational antiviral remdesivir in patients with severe COVID-19. Gilead Sciences Retrieved April 29, 2020, from https://www.gilead.com/news-and-press/press-room/press-releases/2020/4/gilead-announces-results-from-phase-3-trial-of-investigational-antiviral-remdesivir-in-patients-with-severe-covid-19.
- Gordon C. J., Tchesnokov E. P., Woolner E., Perry J. K., Feng J. Y., Porter D. P., & Gotte M. (2020). Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. Journal of Biological Chemistry, 295(15), 4773–4779. 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J., Tchesnokov E. P., Feng J. Y., Porter D. P., & Gotte M. (2020). The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. Journal of Biological Chemistry, 295(15), 4773–4779. 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., & Feldt T. (2020). Compassionate use of remdesivir for patients with severe Covid-19. New England Journal of Medicine, 1–10. 10.1056/NEJMoa2007016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., & Pöhlmann S. (2020). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv, 1–23. 10.1101/2020.01.31.929042 [DOI] [Google Scholar]
- Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., … Pillai S. K. (2020). First case of 2019 novel coronavirus in the United States. The New England Journal of Medicine, 382(10), 929–936. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M., Rodger A., Bell D. J., Bhagani S., Cropley I., Filipe A., Gifford R. J., Hopkins S., Hughes J., Jabeen F., Johannessen I., Karageorgopoulos D., Lackenby A., Lester R., Liu R. S. N., MacConnachie A., Mahungu T., Martin D., Marshall N., … Thomson E. C. (2016). Late Ebola virus relapse causing meningoencephalitis: A case report. Lancet (London, England), 388(10043), 498–503. 10.1016/S0140-6736(16)30386-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. C., Liu C., Raynaud P., Lo M. K., Spiropoulou C. F., Symons J. A., Beigelman L., & Deval J. (2018). Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathogens, 14(2), e1006889 10.1371/journal.ppat.1006889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease$. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawski S. A., Wong K., Collins J. P., Epstein L., Killerby M. E., & Midgley C. M. (2020). First 12 patients with coronavirus disease 2019 (COVID-19) in the United States. medRxiv, 1–21. 10.1101/2020.03.09.20032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- NIH (2020). Clinical trial shows remdesivir accelerates recovery from advanced COVID-19 Retrieved April 30, 2020, from https://www.niaid.nih.gov/news-events/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu K. K., Mishra A. K., & Lal A. (2020). COVID-2019: Update on epidemiology, disease spread and management. Monaldi Archives for Chest Disease, 90(1), 1–9. 10.4081/monaldi.2020.1292 [DOI] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., & Fielding B. C. (2019). Coronavirus envelope protein: Current knowledge. Virology Journal, 16(1), 69 10.1186/s12985-019-1182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P., Sims A. C., Leist S. R., Schäfer A., Won J., Brown A. J., Montgomery S. A., Hogg A., Babusis D., Clarke M. O., Spahn J. E., Bauer L., Sellers S., Porter D., Feng J. Y., Cihlar T., Jordan R., Denison M. R., & Baric R. S. (2020). Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nature Communications, 11(1), 222 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P., Sims A. C., Graham R. L., Menachery V. D., Gralinski L. E., & Case J. B. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Science Translational Medicine, 9, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H. C., Doerffler E., Clarke M. O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K. M., Barauskas O., … Mackman R. L. (2017). Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. Journal of Medicinal Chemistry, 60(5), 1648–1661. 10.1021/acs.jmedchem.6b01594 [DOI] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E. P., Feng J. Y., Porter D. P., & Gotte M. (2019). Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses, 11(4), 326 10.3390/v11040326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh D. K., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., & Xiao G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO ). (2020. a). Coronavirus disease 2019 (COVID-19) Situation Report – 89 Retrieved April 19, 2020, from https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200418-sitrep-89-covid-19.pdf?sfvrsn=3643dd38_2
- World Health Organization (WHO ). (2020. b). WHO R&D blueprint: Ad-hoc expert consultation on clinical trials for Ebola therapeutics Published October 2018. Retrieved April 19, 2020, from https://www.who.int/ebola/drc-2018/summaries-of-evidence-experimental-therapeutics.pdf
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., & Lu L. (2020). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular and Molecular Immunology, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]