Highlights
-
•
State-of-the-Art review on various Intelligent Computing based research for COVID-19
-
•
Analysing the limitations of prediction based models for COVID-19
-
•
Discussing the impact of clinical data and online data for COVID research
-
•
Focussing on Advance intelligent systems on symptom based identification of COVID-19
Keywords: COVID-19, SARS-CoV-2, Statistical methods, Machine learning, Deep learning
Abstract
The World Health Organization (WHO) declared novel coronavirus 2019 (COVID-19), an infectious epidemic caused by SARS-CoV-2, as Pandemic in March 2020. It has affected more than 40 million people in 216 countries. Almost in all the affected countries, the number of infected and deceased patients has been enhancing at a distressing rate. As the early prediction can reduce the spread of the virus, it is highly desirable to have intelligent prediction and diagnosis tools. The inculcation of efficient forecasting and prediction models may assist the government in implementing better design strategies to prevent the spread of virus. In this paper, a state-of-the-art analysis of the ongoing machine learning (ML) and deep learning (DL) methods in the diagnosis and prediction of COVID-19 has been done. Moreover, a comparative analysis on the impact of machine learning and other competitive approaches like mathematical and statistical models on COVID-19 problem has been conducted. In this study, some factors such as type of methods(machine learning, deep learning, statistical & mathematical) and the impact of COVID research on the nature of data used for the forecasting and prediction of pandemic using computing approaches has been presented. Finally some important research directions for further research on COVID-19 are highlighted which may facilitate the researchers and technocrats to develop competent intelligent models for the prediction and forecasting of COVID-19 real time data.
1. Introduction
Throughout history, several infectious diseases have alleged the lives of many people and induced critical situations that have taken a long time to overcome the situation. The terms epidemic and pandemic have been used to describe the disease that emerges over a definite period of time [1]. During a particular course of time, the existence of more cases of illness or other health situations than normal in a given area is defined as epidemics [2]. On the other hand, pandemics are outbreaks of the infectious disease that can enormously increase the morbidity and mortality over a vast geographical area. Due to the factors such as raise of worldwide travel, urbanization, changes in usage of land and misusing of the natural environment, the occurrence of the pandemics has increased from the past century [3]. In the past, the outbreak of smallpox has killed of nearly 500 million world population in the last 100 years of its survival [4]. Due to the outbreak of Spanish influenza in 1918, an estimate of 17 to 100 million deaths occurred [5]. From the last 20 years several pandemics have been reported such as acute respiratory syndrome coronavirus (SARS-CoV) in 2002 to 2003, H1N1 influenza in 2009 and the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2015. Since December 2019 the novel outbreak of coronavirus has infected more than thousand and killed above hundreds of individuals within the first few days in Wuhan City of Hubei Province in South China. In the 21st century, the pandemics such as SARS-CoV has infected 8096 individuals causing 774 deaths and MERS-CoV has infected 2494 individuals causing 858 deaths. While the SARS-CoV-2 has infected more than 3.48 million individuals causing 2,48,144 deaths across 213 countries as on May 3, 2020. These evidential facts state that, the transmission ratio of SARS-CoV-2 is greater than other pandemics. A list of some dangerous pandemics happened over time is listed in Table 1 .
Table 1.
List of Pandemics over time.
| Name | Time period | Death toll |
|---|---|---|
| Antonine Plague | 165-180 | 5M |
| Japanese smallpox epidemic | 735-737 | 1M |
| Prague of Justinian | 541-542 | 30-50M |
| Black death | 1347-1351 | 200M |
| New World Smallpox Outbreak | 1520-onwards | 56M |
| Great Plague of London | 1665 | 100 000 |
| Italian plague | 1629-1631 | 1M |
| Cholera Pandemics 1-6 | 1817-1923 | 1M+ |
| Third Plague | 1985 | 12M (China and India) |
| Yellow Fever | Late 1800s | 100 000-150 000 (US) |
| Russian Flu | 1889-1890 | 1M |
| Spanish Flu | 1918-1919 | 40-50M |
| Asian Flu | 1957-1958 | 1.1M |
| Hong Kong Flu | 1968-1970 | 1M |
| HIV/AIDS | 1981-Present | 25-35M |
| Swine Flu | 2009-2010 | 200,000 |
| SARS | 2002-2003 | 770 |
| Ebola | 2014-2016 | 11,000 |
| MERS | 2015-Present | 850 |
| COVID-19 | 2019-Present | 3.48 Million as on May 3, 2020 |
Due to the rapid increase of patients at the time of outbreak, it becomes extremely hard for the radiologist to complete the diagnostic process within constrained accessible time [6]. The analysis of medical images such as X-rays, Computer tomography and scanners plays a crucial role to overcome the limitations of diagnostic process within constrained accessible time. Now-a-days, machine learning and deep learning techniques helps the physicians in the accurate prediction of imaging modalities in pneumonia. ML is a wing of artificial intelligence that has the ability to acquire relationships from the data without defining them a priori [7]. Due to the availability of large number of intelligent tools for the analysis, collection and storage of large volume of data, machine learning techniques have been extensively utilized in the clinical diagnosis. Machine learning approaches can be efficiently used in applications of healthcare such as disease identification, diagnosis of disease, discovery and manufacturing of drug, analysis of medical images, collection of crowd sourced data, research and clinical trials, management of smart health records, prediction of outbreak etc. Some recent studies show the usage of machine learning techniques in the time series forecasting of Ebola outbreak [8]. In order to select the better performing classifier for forecasting Ebola casualties, experiments were conducted on ten different classifiers. Further, results demonstrate that the proposed model achieves 90.95 % accuracy, 5.39 % MAE and 42.41 % RMSE value. Even though machine learning approaches have rapidly used in the diagnosis of outbreaks, these approaches still have some limitations such as complete utilization of biomedical data, temporal dependency, owing to high-dimensionality, sparsity, heterogeneity and irregularity [9], [10], [11]. On the other hand, due to the deep architectural design, the deep learning models are the best accurate models for handling medical datasets such as classification of brain abnormalities, classification of different types of cancer, classification of pathogenic bacteria and segmentation of biomedical images [12], [13], [14], [15], [16]. Several studies show that DL models are adopted in the diagnosis and classification of pneumonia and other diseases on radiography. A deep learning model build on Mask-RCNN has been utilized for the detection and localization of pneumonia in chest X-ray images [17]. In order to perform pixel-wise segmentation, the model makes use of global and local features. The robustness of the system is achieved through the modification of training process and post processing step. Further, results show that the model outperforms in the identification of pneumonia in chest X-ray images. To improve the performance of prediction, a bioinspired meta-heuristic optimization algorithm has been presented by Martinez-Alvarez et al [18]. In this approach, to prevent the researches from initializing with arbitrary values, the input parameters are initialized with the disease statistics. Also this approach has the ability to stop after certain number of iterations without setting this value. Further, to explore wider search space in less number of iterations, a parallel multi-virus approach has been proposed. Finally, it has been integrated with DL models for finding the optimal parameters in the training phase. Deep learning prototypes have been widely used in the prediction and forecasting of outbreak over machine learning models because of its features such as greater performance, feature extraction without human intervention identification and not making the use of engineering advantage in training phase.
The major objective of this paper is to provide a review of different machine learning approaches used in the prediction, classification and forecasting of COVID-19. First, we describe the origin of SARS-CoV-2 virus, its transmission rate, comparison of SARS-CoV-2 with other pandemics in the history and its impact on the global health. Then the analysis is broaden by describing the advantages of computing approaches such as statistical and mathematical models, ML and DL approaches in the prediction of COVID-19 along with its applications. Further, an analysis of number of articles published in different computing approaches by different countries till date, impact of the nature of data in the prediction of COVID-19 have been presented. As the researchers and technocrats are the main targets of this review, we highlighted some of the challenges in the ongoing research of different computing approaches at the end of the paper. The remaining section of the paper is systematized as follows. Section 2 describes the origin of COVID-19 and its impact. The application of statistical, ML and DL models in the diagnosis and prognosis of COVID-19 has been portrayed in section 3. Section 4 illustrates the critical investigation on the analysed data types of COVID research, growth in publication of ML approaches for COVID, comparative analysis on the type of methods etc. Few challenges in ML related to covid-19 have been focused in Section 5. Section 6 outlines the conclusion with a brief discussion on covid-19 impact in real life and novel research directions.
2. Genesis of SARS-CoV-2 and its Impact on global health
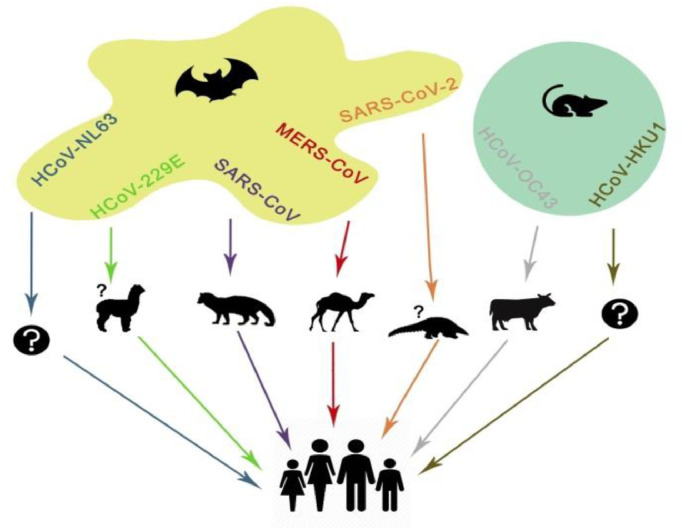
In the 21st century, Human corona viruses such as SARS-Coronavirus (SARS-CoV) and MERS-Coronavirus (MERS-CoV) that have emerged from the animal reservoirs caused global epidemic with distressing morbidity and mortality. These human corona viruses belong to the subfamily of Coronavirinae that is a part of Coronaviridae family. It was named as corona because of the presence of spike like structure on the outer surface of the virus under electron microscope. Its RNA is a single stranded with a diameter of 80-120 nm and nucleic material range varying from 26 to 32 kbs in length [19]. These are basically divided into four types of genera named as alpha (α), beta (β), gamma (γ) and delta (δ). α- and β-CoV usually infects mammals, while γ- and δ-CoV tends to affect birds. Among the six susceptible human viruses, HCoV-229E and HCoV-NL63 of α-CoVs and HCoV-HKU1 and HCoV-OC43 of β-CoVs shows low pathogenicity and moderate respiratory symptoms as common cold. The other two familiar β-CoVs such as SARS-CoV and MERS-CoV exhibit acute and malignant respiratory diseases [20]. The Fig. 1 shows the transmission process of corona viruses from animal sources to human.
Fig. 1.
Transmission of Corona viruses from animals to Human.
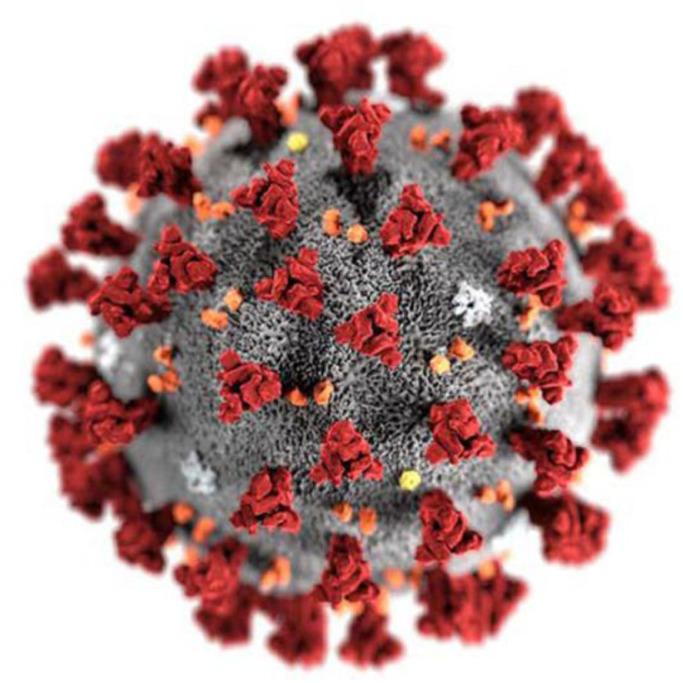
The people in Wuhan City of Hubei Province in south china were reported in local hospitals with an unidentified pneumonia on December, 2019 [21]. Initially, these cases were related to Huanan Seafood Wholesale Market which is famous for variety of live species. All these cases have clinical characteristics similar to those of viral pneumonia such as dry cough, fever, dyspnea and lung infiltrates on imaging. After the analysis of samples collected from the throat swab, the authorities from Centers for disease control (CDC) announced the unidentified pneumonia as novel coronavirus pneumonia (NCP) on 7th January, 2020 [22]. Later it was named as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses [23], [24] and the disease was renamed as COVID-19 by the WHO on 11th February 2020 [25]. The COVID-19 generated by SARS-CoV-2 is a β-coronavirus. The sequence analysis of SARS-CoV-2 matches with the typical structure of coronaviruses as depicted in Fig. 2 .
Fig. 2.
Interpretation of the SARS-CoV-2 virion [26].
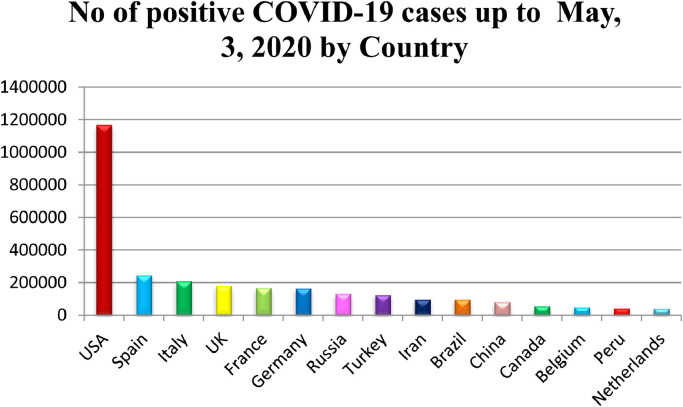
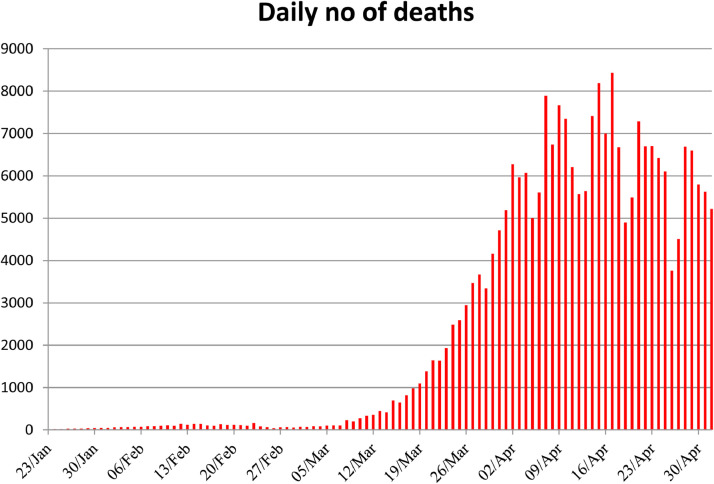
The structure of SAR-CoV consists of 14 binding remnants that collaborate precisely with human angiotensin-converting enzyme 2. As eight binding residues of the SARS-CoV has sustained in the SARS-CoV-2, the structure of SARS-CoV-2 genome contributes 79.5% similarity to SARS-CoV. [27]. In addition, both bat CoV and human SARS-CoV-2 share the identical ancestor as the genome sequencing of COVID-19 shows 96.2 % identity to Bat CoV RaTG13 [28]. Due to the genetic recombination occurrence at ‘S’ protein in the RBD area of SARS-CoV-2, the SARS-CoV-2 has greater transmission ability than SARS-CoV [29]. After the spread of COVID-19 to 18 countries through human-to human transmission, the WHO announced the epidemic as Public Health Emergency of International Concern (PHEIC) on 30th January, 2020. In addition, critical situation was created when the first case not imported from china was registered in the United States on 26th February, 2020. The WHO declared the COVID-19 as pandemic on 11th march 2020 when it imposes serious hazard to public health as the number of cases outside the china has raised 13 times and the number of countries distressed by COVID-19 has increased by three times. Since the last two months, the number of covid-19 cases registered has crossed all the previous records of the viral disease. Due to its rapid spread, it is considered to be the most dangerous disease till date. According to the WHO, the SARS-CoV-2 has infected about 3.48 million people and caused 2,48,144 deaths across 213 countries of the World as on May 3, 2020. Among the countries, the USA has reported about 1,188,122 positive cases and 68,595 deaths as on May 3, 2020 and stood in first place in both positive cases as well as in death rate. Similarly, other countries like Spain, Italy, UK, France, Germany, Russia, Turkey, Iran, Brazil, Canada, Belgium, Peru and Netherland are placed as top countries with more than 50,000 cases after the pandemic of COVID-19 outside the mainland of China. The number of deaths per day due to COVID1-9 pandemic is also gradually increasing from the starting day of transmission to till date. Fig. 3, Fig. 4 , predicts the top countries in the world having more than 50,000 and number of deaths per day as on May 3, 2020.
Fig. 3.
No of positive cases up to May, 3, 2020.
Fig. 4.
Daily no of deaths.
3. Developed Computing approaches in the classification, prediction and prevention of COVID-19
Due to the quick spread of the SAR-CoV-2, physicians are facing extreme difficulty in the diagnosis of COVID-19. Even though Reverse Transcription Polymerase Chain Reaction (RT-PCR) method is the standard method used in the diagnosis of Ncov-2019 [30], due to pandemic it suffers from limitation such as low sensitivity, requires more time, and short in supply. In recent years, analysis of medical images is one of the most promising rising research areas in the healthcare sector. Therefore the analysis of medical images such as X-rays, Computer tomography and scanners can overcome the limitations of RT-PCR. As digital technologies is also playing a crucial role in the prevention of disease, the worldwide health emergency is also seeking the support of digital technology such as the application of statistical, machine learning and deep learning models for efficient diagnosis of medical images to stop the spread of the disease.
3.1. Machine learning techniques
The fundamental capability of the machine learning is the derivation of predictive models without having any knowledge of the basic mechanism that are usually not known or insufficiently defined. These techniques are capable of developing complex patterns from huge, noisy or complex data. As the combination of predictive models with expert system reduces the subjectivity problem, these models offer outstanding support for the clinical diagnosis. Previously, machine learning techniques can be used to find the epidemic patterns as these algorithms are used in the analysis and forecasting of medical images [31]. Hence in the present pandemics, several researches have also used different approaches of machine learning such as Support Vector Machine, Regression, Random Forest, K-means and so on in the prediction and diagnosis of SARS-CoV-2.
3.1.1. Random Forest (RF)
Random forest algorithm (RF) is one of the most promising and recognized classifier that uses multiple trees to train and predict data samples. This approach has been extensively used in the fields of chemometrics and bioinformatics [32], [33]. Because of its praiseworthy characteristics, random forest has been used in resolving issues of the nCOVID-19 infection. For precise and rapid recognition of COVID-19, a tool based on random forest algorithm to extract 11 key blood indices from clinical available blood samples was suggested by Wu et al [34]. In this study, random forest algorithm is used as a discrimination tool to explore patients with COVID-19 symptoms. The proposed method achieved better outcome in the prediction of COVID-19 with accuracy of 0.9795 for the cross-validation set and 0.9697 for the test set. Further, the tools also achieved better performance in terms of sensitivity, specificity and overall accuracy of 0.9512, 0.9697, and 0.9595, respectively on an external validation set. Moreover, it achieved an accuracy of 0.9167 in a detailed clinical estimate of 24 samples collected from infected COVID-19 patients. After multiple verifications, the proposed approach has been emerged as a precise tool for the recognition of COVID-19 infection. To predict the hospital stay of patients infected with novel coronavirus, a model based on linear regression and random forest has been suggested by Qi et al [35]. The proposed model based on 6 second-order characteristics was refined on features obtained from pneumonia lesions in training and inter-validation datasets. Further, the predictive efficiency has been evaluated using lung-lobe and patients-level test dataset. From the conclusions, it is observed that model was efficient in segregating short and long-term stay of patients in hospital infected with coronavirus infection. Moreover, linear regression model exhibited a sensitivity and specificity of 1.0 and 0.89. While a sensitivity and specificity of 0.75 and 1.0 has been exhibited by the random forest model. The following Table 2 depicts the usage of random forest approach in the prognosis of SARS-CoV-2 infection.
Table 2.
Applicability of Random forest approach in the prognosis of SARS-CoV-2 infection.
| Author | Model Name | Problem category | Type of Data | Year | Ref |
|---|---|---|---|---|---|
| Cobb et al. | Random Forest | To examine the effect of social distancing on the cumulative spread of COVID-19 at country level | Text data | Apr,2020 | [36] |
| Shi et al. | Infection Size Aware Random Forest method | Classification of COVID-19 from CT images | Chest CT images | Mar,2020 | [37] |
| Tang et al. | Random Forest model | Severity assessment of COVID-19 patients | Chest CT images | Mar,2020 | [38] |
| Sarkar & Chakrabarti | Random Forest | To identify the important predictors and their effects on mortality of COVID-19 pateints. | Text data | Jan,2020 | [39] |
| Chen et al. | Random Forest | To identify factors influencing the viral clearance negatively. | Text data | Jan,2020 | [40] |
3.1.2. Support Vector Machine
As SVM is used as a powerful tool for data regression and classification, it has superior performance in many real world applications such as medical image analysis over other machine learning approaches. Because of its better performance, it has been used in the classification and analysis of COVID-19. To predict the threat of positive COVID-19 diagnosis, different machine learning approaches such as neural networks, random forests, gradient boosting trees, logistic regression and support vector machines for training the sample data was suggested by Batista et al [41]. The performance of the different machine learning approaches was trained on arbitrary sample data of 70% of patients and was tested on 30% of new not seen data. Form the results, it concludes that the support vector machine algorithm outperforms with AUC, sensitivity, specificity and Brier score of 0.85, 0.68, 0.85 and 0.16 respectively when compared with other machine learning algorithms. For the diagnosis of COVID-19 infected patients form the chest X-ray images, a machine learning model developed on multi-level thresholding and SVM has been suggested by Hassanien et al [42]. Furthermore, the results depict that the proposed model achieves better performance with an average sensitivity of 95.76%, specificity of 99.7%, and accuracy of 97.48%, respectively. Table 3 represents the usage of machine learning approaches in the recognition and diagnosis of COVID-19.
Table 3.
Usage of Support Vector Machine in the prediction and diagnosis of COVID-19.
| Author | Model Name | Problem category | Type of Data | Year | Ref |
|---|---|---|---|---|---|
| Sonbhadra et al. | One-class SVM | To extract the activity and trends of corona-virus related research articles | —- | Apr,2020 | [43] |
| Zhang et al. | SVM | To detect severely ill COVID patients from mild symptom COVID patients | Text data | Apr,2020 | [44] |
| Hassanien et al. | SVM | To predict the recovery of COVID-19 patients | Text data | Apr,2020 | [45] |
| Barstugan et al. | SVM | For the classification of COVID-19 | CT images | Mar,2020 | [46] |
| Sethy et al. | SVM using deep features | To detect Coronavirus disease | X-ray image | Mar,2020 | [47] |
3.1.3. Other machine learning approaches
Besides random forest and support vector machine, other approaches of machine learning approaches such as linear and logistic regression, XGBoost, K-means, neural network have also been used in the clinical and public health approach. Many research works are being performed and Table 4 represents the usage levels of these approaches in solving some of the conflicts of Covid-19.
Table 4.
Other Machine learning approaches in the prediction and diagnosis of COVID-19.
| Author | Model Name | Problem category | Type of Data | Year | Ref |
|---|---|---|---|---|---|
| Pandey et al. | Linear Regression | For outbreak prediction of COVID-19 in India | Time series data | Apr, 2020 | [48] |
| Liu et al. | Machine Learning and mechanistic models | To predict COVID-19 outbreak in real time | Text data | Apr, 2020 | [49] |
| Dandekar et al. | Neural network augmented SIR quarantine model | To forecast the spread the of COVID-19 in Wuhan, China | Time series data | Mar,2020 | [50] |
| Carrillo-Larco et al. | K-Means | To categorize countries according to the number of confirmed COVID-19 cases | Text data | Mar,2020 | [51] |
| Magar et al. | Different Machine learning models | To predict the possible inhibitory synthetic antibodies for Corona virus | Text data | Mar,2020 | [52] |
| Yan et al. | XGBoost machine learning model | To foresee the survival ratio of severely ill Covid-19 patients | Time series data | Jan, 2020 | [53] |
| Chen et al. | Different Machine learning algorithms | Prediction of mortality risk in severe infected COVID-19 patients | Time series data | Jan, 2020 | [54] |
| Al-Karawi et al | FFT-Gabor | To distinguish between positive from negative cases using a dataset of COVID-19 | CT images | Jan,2020 | [55] |
| Metsky et al. | Multi Layered Perception (MLP), Adaptive Network-based Fuzzy Inference System (ANFIS) | CRISPR-based nucleic acid detection | Text data | Jan,2020 | [56] |
3.2. Deep learning techniques
Deep learning techniques are representation-learning algorithms that consist of simple but nonlinear modules which are used to alter the representation at one level into a presentation at slightly more intellectual levels [57]. The deep structural nature made the deep learning models capable of resolving the complex artificial intelligent tasks. Deep learning paradigms offer new opportunities in the field of biomedical informatics because of its features such as excellent performance, end-to-end learning scheme with combined feature learning, ability to handle complex and multi-modal data and so on. Deep learning techniques have also been used in the efficient classification and analysis of medical images of COVID-19 pandemic. Various deep learning approaches such as Convolutional Neural Network (CNN), Long Short-Term Memory (LSTM), Generative Adversarial Networks (GAN), Residual Neural network and Autoencoder have been attempted by many researchers in the classification and prediction of COVID-19 infection.
3.2.1. Convolutional Neural Network (CNN)
Convolutional Neural Network has proven to be one of the most successful algorithms in the analysis of medical image with high accuracy. Previously, the identification of the nature of pulmonary nodules in CT images, prediction of pneumonia in X-ray images, labeling of polyps automatically at the time of colonoscopic videos have been done using convolutional neural networks [58], [59], [60]. The authentication features for identifying COVID-19 in medical images are bilateral distribution of patchy shadows and ground glass opacity [61]. Abbas et al. [62] have developed a Decompose, Transfer, and Compose (DeTraC) model based on convolutional neural network to categorize the COVID-19 chest X-ray images. Using the class decomposition mechanism the class boundaries are investigated that helps the model to accord with any anomalies in the image dataset. Further, the results show that an accuracy of 95.12% was attained by DeTraC in the recognition of COVID-19 X-ray images from other normal and pneumonia cases. To recognize COVID-19 patient from chest X-ray images, distinct convolutional neural network frameworks namely ResNet50, InceptionV3 and Inception-ResNetV2 have been proposed by Narin et al. [63]. Further, insufficient data and training problem can be overwhelmed by applying deep transfer learning technique using ImageNet. From the results it can be observed that highest classification performance with 98% accuracy can be attained by the ResNet50 model over the other two models. To interpret and predict the number of positive cases, a COVID-19 forecasting model build on convolutional neural network (CNN) was suggested by the Huang et al [64]. The main focus of the study was to consider the cities with large number of positive cases in China. The overall competence of different algorithms was compared using mean absolute error (MAE) and root mean square error (RMSE). The outcomes indicate that CNN achieves greatest prediction efficacy when collated with other approaches of deep learning such as LSTM, GRU and MLP. Furthermore, the actual usage and feasibility of the proposed model in forecasting the total registered cases were also documented in their study. To automatically recognize the Ncov-2019 positive cases from chest X-ray images, Mukherjee et al. [65] have proposed tailored shallow convolutional neural network architecture. The architecture was designed with few parameters for validating 130 COVID-19 positive X-ray images and 5-fold cross validation was used to avoid possible bias in the experimental tests. Moreover, the proposed method achieved an accuracy, sensitivity and AUC of 96.92%, 0.942 and 0.9869 respectively, which is dominative over other compared methods. A multitask deep learning model has been proposed by Amyar et al. [66] to perform the automatic screening and segmentation of COVID-19 chest CT images. For reconstruction and segmentation, one encoder and two decoders along with multi-layer perceptron has been used in the architecture for the classification purpose. Then the model has been evaluated with a dataset of 1044 patients, which includes 449 patients suffering with COVID-19, 100 normal cases, 98 patients with lung cancer and 397 cases of different types of pathology. Moreover, results indicate that the model obtains better performance over the other image segmentation and classification techniques. The application of CNN in the classification and diagnosis of COVID-19 has been depicted in Table 5 .
Table 5.
Prediction and Diagnosis of COVID-19 using CNN.
| Author | Approach | Problem Type | Month&Year | Ref |
|---|---|---|---|---|
| Hammoudi et al. | Tailored CNN Models | To detect and evaluate COVID-19 patients using chest X-ray images | Apr, 2020 | [67] |
| Rajaraman et al. | Custom CNN and pre-trained CNN models | For detection of COVID-19 in chest X-ray images | Apr, 2020 | [68] |
| Hall et al. | Deep convolutional neural network ResNet50 | Detection of COVID-19 from chest X-ray | Apr, 2020 | [69] |
| Ozkaya et al. | CNN with Fusion and Ranking technique | Classification of COVID-19 images from CT images | Apr, 2020 | [70] |
| Zahangir et al. | Improved Inception Recurrent Residual Neural Network (IRRCNN) and NABLA-3 network models | For identification of COVID-19 patients from X-ray and CT images | Apr, 2020 | [71] |
| Rahimzadeh et al. | Modified CNN | Classification of X-ray images for identification of COVID-19 cases | Apr, 2020 | [72] |
| Zhang et al. | 18 layer residual CNN | For screening of Ncovid-19 from chest X-ray images | Mar, 2020 | [73] |
| Hemdan et al. | COVIDX-Net model based on CNN | To categorize COVID-19 in chest X-ray images. | Mar, 2020 | [74] |
| Wang et al. | DenseNet121-FPN and COVID-19Net | For COVID-19 Diagnostic and Prognostic Analysis of CT images | Jan, 2020 | [75] |
| Zheng et al. | 3D deep convolutional neural Network | For detection of COVID-19 from chest CT images | Jan, 2020 | [76] |
| Apostolopoulos et al. | Mobile Net based on Convolutional neural network | Classification of COVID-19 biomarkers from x-ray images | May, 2020 | [77] |
| Kumar et al. | ResNet152 model with SMOTE and Machine Learning Classifiers | To perform accurate recognition of COVID-19 from chest x-ray images | Jan, 2020 | [78] |
| Fu et al. | ResNet-50 based AI Framework | To classify CT images to distinguish COVID-19 from other known infectious diseases of the lung | Jan,2020 | [79] |
| Razzak et al. | Different convolutional Neural network architectures | Diagnosis of COVID-19 | Jan, 2020 | [80] |
3.2.2. Long Short-Term Memory (LSTM)
Long Short-Term Memory is a type of the Recurrent Neural Network that can store knowledge of previous states and can be trained for work that needs memory. LSTM is one of the efficient models for the prediction of time series sequential data [81]. As past data is retained in the hidden states, LSTM approach can perform more accurate predictions of the output. A novel multivariate spatiotemporal model has been proposed by Jana et al [82]. This model uses ensemble of convolutional LSTM to make accurate forecast of the dynamics of COVID-19 transmission in large geographic region. For converting the available spatial features into set of 2D images, a data preparation method is used. Further, the proposed model is trained using USA and Italy data. From the findings it can be observed that the model achieves 5.57% and 0.3% MAPE for USA and Italy respectively. To predict the number of COVID-19 cases in India, a data-driven estimation approach based on LSTM and curve fitting has been suggested by Tomar et al [83]. This approach was also used to estimate the effective of social distancing measures on the spread of the pandemic. Further, the findings show the accuracy of proposed approach in predicting the number of positive and recovered cases in India. Table 6 shows the applicability of LSTM in resolving the issues of COVID-19 pandemic.
Table 6.
Application of LSTM in resolving issues of COVID-19 pandemic.
| Author | Approach | Problem Type | Month& Year | Ref |
|---|---|---|---|---|
| Jelodar et al. | NLP using LSTMRNN | Classification of novel COVID-19 | Apr,2020 | [84] |
| Ibrahim et al. | Variational-LSTM autoencoder | To forecast the spread of the COVID-19 | Apr,2020 | [85] |
| Patankar | Long Short Term Memory model | Drug Discovery | Mar,2020 | [86] |
| Yang et al. | Modified SEIR and LSTM | To predict the epidemic curve of COVID-19 | Mar,2020 | [87] |
| Kafieh et al. | M-LSTM | To predict the number of cases in Iran | Jan,2020 | [88] |
| Kolozsvari et al. | RNN-LSTM | To predict the epidemic of COVID-19 | Jan,2020 | [89] |
| Bandyopadhyay et. al. | LSTM-GRU based RNN | To predict the number of positive, negative and negative cases | Jan, 2020 | [90] |
| Ayyoubzadeh et al. | LSTM and Linear Regression | To predict the incidence of COVID-19 in Iran. | 2020 | [91] |
3.2.3. Other approaches of deep learning
In addition to CNN and LSTM approaches, several other deep learning approaches such as Generative Adversarial Networks and Autoencoder have also been used in the analysis and prediction of COVID-19 pandemic. Generative adversarial networks (GANs) are algorithmic architectures that consist of two neural networks called generator and discriminator to generate new, synthetic instances of data from real ones that have been never observed before. GANs have been widely used in image generation, video generation and voice generation. An autoencoder is a sort of artificial neural network used to grasp efficient data coding in an unsupervised way. The following Table 7 represents the usage of other deep learning approaches in the prediction and diagnosis of novel coronavirus disease.
Table 7.
Prediction and diagnosis of COVID-19 pandemic using other Deep learning approaches.
| Author | Approach | Problem Type | Month& Year | Ref |
|---|---|---|---|---|
| Liu et al. | GAN | Enhancing the quantification of COVID-19 using 3D Tomographic pattern synthesis | May,2020 | [92] |
| Khalifa et al. | GAN and fine tuned AlexNet, GoogleNet, SqueezeNet and ResNet18 | To identify COVID-19 cases from chest X-ray images | Apr, 2020 | [93] |
| Loey et al. | GAN | To detect coronavirus in chest X-ray images | Apr,2020 | [94] |
| Hartono et al. | Topological Autoencoder | To visualize the global trend of COVID-19 transmission | Apr,2020 | [95] |
| Shan et al. | DL based VB-Net | For automatic segmentation and on quantification of infectious region from CT images | Mar, 2020 | [96] |
| Hu et al. | Modified Autoencoder | To forecast and evaluate the spread of nCOVID-19 in world | Mar, 2020 | [97] |
| Zhavoronkov et al. | GAN | To generate novel drug compounds against nCoVID-19 | Feb,2020 | [98] |
3.3. Mathematical and Statistical methods
From the past few pandemics, mathematical and statistical models have been successfully used in the estimation of human loss and also in the prediction of total number of deaths until a specific period or end of the pandemic. As the mathematical and statistical approaches shows better performance, researchers have also used the same approaches in the estimation of spread rate and death count of the current pandemic. To foresee the window period for testing positive negative results as well as false negative result, a multivariate model has been developed by Xu et al [99]. The clinical characteristics that are responsible for false negative outcomes of SARS-CoV-2 nucleic acid identification are determined using these models. Zhong et al. [100] have used the simple mathematical models for the early prediction of novel coronavirus disease in china. The prediction models estimated that the total number of positive cases may reach to 76,000 to 230,000 in late February to early March. It is also estimated that from early May to late June, the number of registered cases will rapidly decrease. To predict the number of deaths in china, Boltzmann's function based analysis has been proposed by Gao et al [101]. The prediction model gives better prediction accuracy in the assessment of severity of situation. The impact of primary health conditions such as cardiac disease, diabetes and age of the patient in the prediction of death rate has been presented by Banerjee et al [102]. Azad et al [103] have used Holt's and autoregressive integrated moving average (ARIMA) model in the temporary forecasting of COVID-19 infected patients in India from 30 January to 21 April 2020. The following Table 8 shows some other analysis done in the prediction of COVID-19 using mathematical and statistical approaches.
Table 8.
Other reviews done on the forecasting of COVID-19 using mathematical and statistical approaches.
| Author | Type of COVID Analysis | Month & Year | Ref |
|---|---|---|---|
| Mohler et al. | Analysis of public health interventions on the transmission of COVID-19 | Apr, 2020 | [104] |
| Gupta et al. | Using fatality data in the estimation of total COVID-19 infections in Indian hot spots | Apr, 2020 | [105] |
| Dowd et al. | Impact of age in the dissemination and mortality rate of COVID-19 | Apr, 2020 | [106] |
| Nadim et al. | Analysis of prevention strategies in the prediction of COVID-19 pandemic | Mar, 2020 | [107] |
| Magal et al. | Predicting the cumulative number of positive cases | Mar, 2020 | [108] |
| Anzai et al. | Impact of reduced travel in the spread of COVID-19 in China | Feb,2020 | [109] |
| Kumar et al. | To estimate the total number of COVID-19 confirmed, death and recovery in India | Jan, 2020 | [110] |
| Oliveiros et al. | Impact of temperature and humidity on the dissemination of COVID-19 | Jan, 2020 | [111] |
| Lu et al. | To identify the number of positive cases | Jan, 2020 | [112] |
4. Critical Investigation
A systematic analysis of articles related to COVID-19 has been carried out in this study using various intelligent computing approaches. From the analysis, it is observed that the machine learning and deep learning approaches have been successfully utilized in the interpretation of medical images because of the facts such as extraction of rich features from multimodal clinical dataset, ability to distinguish between bacterial and viral pneumonia, capable of detecting various chest CT imaging features and early detection of epidemic patterns. Therefore, the data obtained using these approaches may help the physicians and researchers in understanding and prediction of COVID-19 infection at the early stage. In this section systematic analysis on the week wise publications of COVID-19 using ML, DL and other approaches, number of articles published journal wise, country wise analysis of the publications, number of articles published using different approaches, performance analysis of different approaches, the impact of natural data on the prediction of COVID-19 has been carried out.
4.1. Performance analysis of different intelligent computing approaches
From the systematic review, it has been observed different researchers have used different intelligent approaches in the prediction and diagnosis of COVID-19 infection.
4.1.1. Machine learning approaches
It has been evident from the system review that machine learning approaches also have been efficiently used in the prediction and diagnosis of COVID-19. Among the available machine learning approaches mostly support vector and random forest have been used in the recognition of novel COVID-19 outbreak. As support vector machine is one of the best classifier algorithms with minimum error rate and maximum accuracy, it gives better prediction results. A machine learning model suggested by Barstugan et al. [46], have been efficiently used in the classification of medical images. Using SVM for classification and Grey Level Co-occurrence Matrix (GLCM) for feature extraction, the proposed model achieves 99.68% classification accuracy. As random forest uses multiple tress to identify the samples and robust to noise, it has been extensively utilized in the classification of medical images. Moreover, random forest approach is suitable for multiclass problem, whereas SVM is suitable for two-class problem. A random forest model proposed by Tang et al. [38], have been successfully utilized in the severity assessment of coronavirus infected patients. Using 30 quantitative features, the proposed model achieves 0.933 true positive rate, 0.74 true negative rate and 0.875 accuracy. Table 9 represents the analysis of performance metrics of various machine learning algorithms in the diagnosis of COVID-19 infection. It has been evident from the systematic analysis that most of the machine learning approaches trained on small datasets. Moreover, the limitations specified in the Table provide scope for the researchers to develop more accurate prediction and forecasting models in the future.
Table 9.
Performance analysis of various machine learning approaches in forecasting and prediction of COVID-19 infection.
| Author | Month & Year | Method | Classes | Accuracy | Specificity | Sensitivity | AUC | Limitation | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Hassanien et al. | Jan, 2020 | Multi level thresholding of SVM | 2 | 97.48% | 97.48% | 95.76% | —- | Not trained on large dataset | [42] |
| Wu et al. | Jan, 2020 | Random forest algorithm | 4 | 95.95% | 96.97% | 95.12% | —- | Not considered atypical symptoms | [34] |
| Al-Karawi et al. | Jan, 2020 | FFT-Gabor scheme | 2 | 95.37% | 94.76% | 95.99% | — | —– | [55] |
| de Moraes et al. | Jan, 2020 | Support vector Machine | - | —- | 85% | 68% | 0.85 | Not consideredTime interval from first symptom, analyzed small sample dataset | [41] |
| Shi et al. | Mar, 2020 | Infection size aware Random forest method | 2 | 87.9 | 83.3 | 90.7 | —- | Not included findings from follow-up CT scans and not considered pneumonia related clinical characteristics | [37] |
4.1.2. Deep learning approaches
Due to the advantages of deep learning approaches over machine learning approaches such as excellent performance, feature extraction without human intervention and handling complex and multimodal, more number of researches has been carried in the diagnosis of COVID-19 infection using deep learning approaches. From the systematic review, it is observed that CNN is one of the most commonly used deep learning approaches for the prediction of pandemic from the medical Images over other approaches due to its ability to extract features automatically. A deep learning algorithm proposed by Wang et al. [113], have achieved an overall accuracy of 73.1% on external validation. This is due to the limitations such as large number of variable objects and presence of only one radiologist in outlining the ROI area. It has been also observed that the implementation of exact architecture may not yield in better solutions. A new modified CNN for categorizing X-ray images proposed by Rahimzadeh et al. [72], improves the overall performance by extracting multiple features using Xception and ResNet50V2 networks. The model has been applied on 11302 images and conclusions show that the proposed method achieves an overall average accuracy of 99.56% for COVID-19 cases. An iteratively pruned model proposed by Rajaraman et al. [68], improves the classification performance by combining distinct ensemble schemes. The empirical results show that the proposed model outperforms by achieving 99.01% accuracy and 0.9972 of area under curve value. A ResNet based framework proposed by Farooq et al [114], have achieved an overall accuracy of 96.2% with augmentation. Hence, the performance can be enhanced by considering some aspects such as presence of expert knowledge about the task to be solved, additional augmentation and preprocessing steps, optimization of hyperparameters and so on in the implementation. The following Table 10 depicts the performance analysis of some deep learning approaches that may enable the researchers to select an appropriate deep learning approach and architecture for resolving conflicts in COVID-19 pandemic as well as further scope to improve the overall performance of different approaches.
Table 10.
Performance analysis of various deep learning approaches in diagnosis of COVID-19.
| Author | Month & Year | Method | Classes | Accuracy | Specificity | Sensitivity | AUC | Ref |
|---|---|---|---|---|---|---|---|---|
| Chen et al. | Jan, 2020 | UNet++ (Retrospective testing per patient) |
2 | 95.24% | 93.55% | 100% | — | [115] |
| Wang et al. | Jan, 2020 | DenseNet121-FPN, COVID19-Net | 2 | 85% | 71.43% | 79.35% | [75] | |
| Li et al. | Mar, 2020 | COVNet | 3 | —- | 96% | 90% | 0.96 | [116] |
| Asnaoui et al. | Mar, 2020 | MobileNet_V2 | 2 | 96.27% | 94.61% | 98.02% | —- | [117] |
| Inception_Resnet_V2 | 96.09% | 93.88% | 98.53% | |||||
| Resnet50 | 96.61% | 94.92 | 98.43% | |||||
| Abbas et al. | Mar, 2020 | DeTrac-ResNet-18 | 3 | 95.12% | 91.87%, | 97.91%, | —- | [62] |
| Ozkaya et al. | Mar, 2020 | CNN using Deep Features Fusion and Ranking Technique | — | 98.27% | 97.60% | 98.93% | —- | [70] |
| Wang et al. | Mar, 2020 | Tailored CNN | 3 | 92.3% | — | —- | —- | [118] |
| Chowdhury et al. | Mar, 2020 | Sgdm-SqueezeNet | 3 | 98.3% | 99.0% | — | — | [119] |
| Apostolopoulos et al. | Apr, 2020 | CNN with transfer learning | 3 | 96.78% | 98.66% | 96.46% | — | [120] |
| Afshar et al. | Apr, 2020 | Capsule Network | 4 | 95.7% | 95.8% | 90% | 0.97 | [121] |
| Butt et al. | Apr, 2020 | Multiple CNN's | 3 | —- | 92.2% | 98.2% | 0.99 | [122] |
| Li et al. | Apr, 2020 | DenseNet | 3 | 88.9% | — | —- | —- | [123] |
| Ucar et al. | Apr, 2020 | Bayes-SqueezeNet | 3 | 98.3% | 99.1% | —- | —- | [124] |
4.2. Efficiency of COVID-19 Prediction model
Since the start of outbreak of COVID-19 in December 2019, several researchers and modeling groups around the world have developed abundant number of prediction models [125], [126], [127], [128] using mathematical and intelligent computing approaches to predict the trends of COVID-19 in different regions of the world. The list of COVID-19 forecasting attempts all over the world using various statistical models is publicly accessible1 . The modeling results have forecasted information about trends in COVID-19 around the world such as infection cases, future deaths, recovered cases, hospitality needs, impact of social distancing, travelling restrictions and so on. These models have shown a vast range of variations in predictions due to uncertainty of data. It has been found that the design issues have also been observed in the most cited forecasting technique from the IMHE [129], [130]. Though the issues are resolved in later revised model, the prediction errors still persist high. One of the reason for these variations is only small amount of information is available at the beginning of the outbreak and lack of reliable data due to frequent segregation of data over different geographical regions. Another reason is most of the prediction models have forecasted the future results by considering data of confirmed cases of those who got infected with symptoms and tested at the hospitals. These predictions have not taken into consideration the data of asymptomatic patients. Furthermore, the factors affecting the positive and death rate such as age, gender, hypertension, chronic diseases etc., have not considered in some prediction models. The selection of suitable model for performing epidemic study also influences the predictions of the model. A simple model is not realistic as it does not include more epidemiological information, while the complex model is biologically authentic. Though the complex model includes more biological information, it requires more parameters when compared to simple model. The complex model also leads to larger degree of uncertainty, if the increased parameters are unknown. Therefore, to make more accurate predictions in the future more research has to be carried on improving the tools and models of the prediction on large biological information.
4.3. Growth in publication of COVID-19 research using ML and other methods
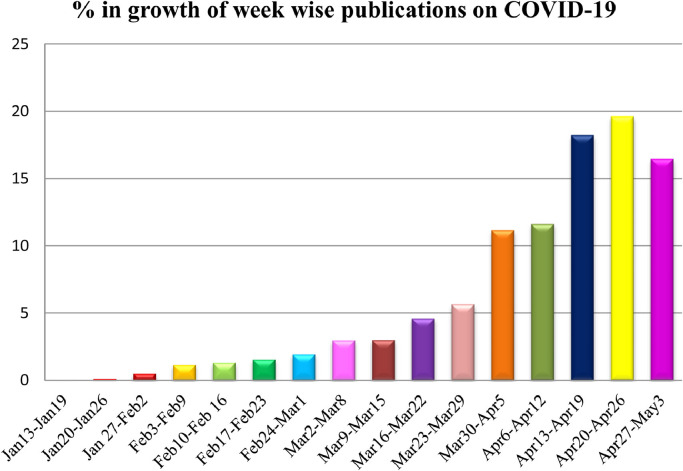
Despite having lower fatality rate, SARS-CoV-2 caused thrice the total of deaths when compared to the combined statistics of deaths caused by both MERS and SARS-CoV. As the symptoms of COVID-19 are similar to common influenza, it becomes difficult to detect the infection. In addition, COVID-19 is much more contagious than influenza, due to asymptomatic condition. Further, the shortage of medical supply, rapid spread and the non-availability of a vaccine or drug for treating COVID-19 are the major reasons that attracted most of the researchers to carry vast research on COVID-19 when compared to other pandemics. Fig. 5 represents the articles contributed for the analysis from Jan 13, 2020 to May 3, 2020. In the initial stage of disease, less than 1% of articles have been published in the month of January. Around 6% of articles issued in the month of February, 16% of articles reported in the month of March and 77% of the articles are published in the month of April. So it can be concluded that up to 3rd May 2020, majority of the articles have been published in the month of April as the number of infected COVID-19 cases increased world wise. It is worthy to note that, with the increase of COVID-19 cases throughout the world, researchers have shown incredible interest to decipher the problem of various aspects on COVID-19 through intelligent ML and other computing approaches.
Fig. 5.
Week wise Analysis of publications on COVID-19.
4.4. Distribution of articles on COVID-19 by Machine Learning, Deep Learning and statistical and mathematical techniques
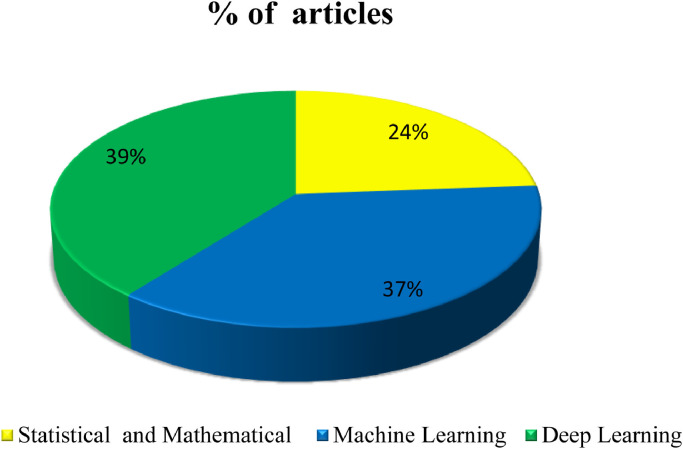
It is evident from the Fig. 6 , that majority of work on COVID-19 prediction/diagnosis has been conducted using deep learning approaches (39%). Next, 37% of research has been experimented using the machine learning approaches. Only 24% of the work has been done using mathematical and statistical models in the prediction of COVID-19. From the figure, it can be concluded that more appropriate prediction and diagnosis of COVID-19 diseases can be performed using deep learning approaches. More work has been contributed on deep learning approaches when compared to other approaches because of the features such as excellent performance, ability to handle complex and multi-modal data, feature extraction without human intervention and absence of engineering advantage in training phase.
Fig. 6.
Articles published on COVID-19 using different approaches.
4.5. Number of articles distributed by Journals
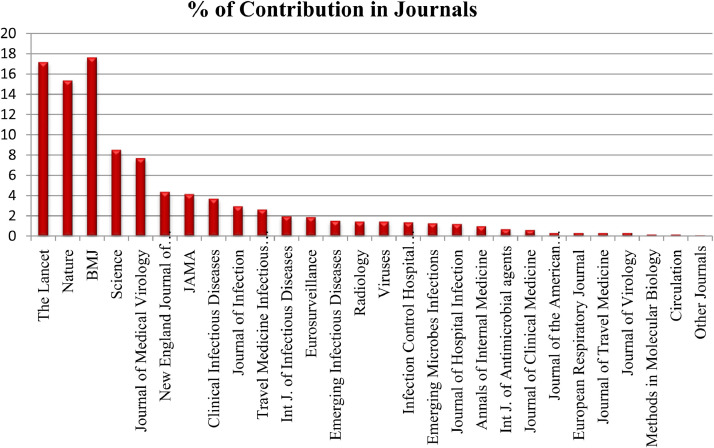
Articles from different sources such as Lancet, JAMA, NEJM, Elsevier, Oxford, Wiley, Nature, BMJ, Science and medRxiv have been considered for the systematic review. It is evident from the Fig. 7 that majority of the articles have been published in BMJ i.e., 17.6%. Next 17.1% articles have been published in Lancet. The articles from Nature have contributed 15.3 %. The science has contributed 8.5% of the studies. 7.6% of the studies have been contributed by Journal of Medical virology. NEJM, JAMA, Clinical Infectious diseases, Journal of Infection, Travel Medicine Infectious Diseases, International Journal of Infectious Diseases, Eurosurveillance, Emerging Infectious Diseases, Radiology, Viruses, Infection Control Hospital Epidemiology, Emerging Microbes Infection, Journal of Hospital Infection have been contributed between 2 to 4% of the studies. Remaining journals like Annals of Internal Medicine, Journal of the American Academy of Dermatology International Journal of Antimicrobial agents, Journal of Clinical Medicine, Journal of Travel Medicine, Journal of Virology, Methods in Molecular Biology have contributed less than 1% of the studies.
Fig. 7.
Distribution of articles by Journals.
4.6. Distribution of articles by Country on the diagnosis and Prognosis of COVID-19 using different approaches
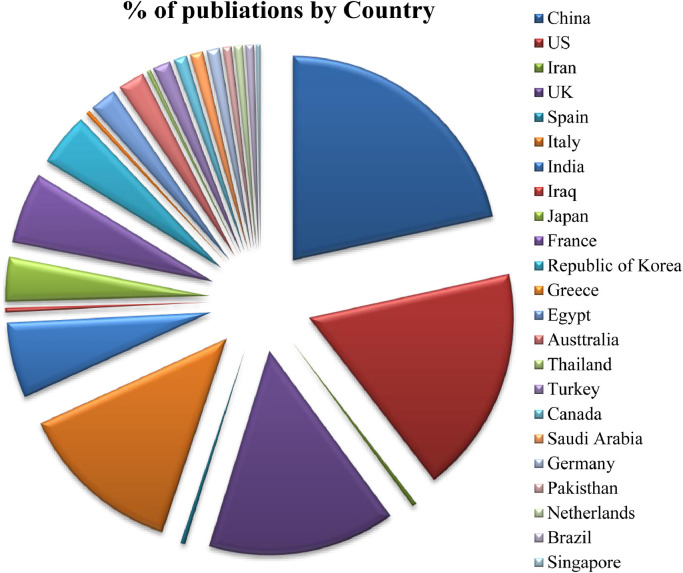
The Fig. 8 displays the number of articles published country wise on the diagnosis and prognosis of COVID-19 using statistical. Machine learning and Deep learning approaches. From the figure. it can observed that majority of the articles i.e., 21.7% have been contributed by the researchers of China. 18.1% articles have been contributed from the US researchers. The researchers of UK have contributed 14.9% of the articles in the study. 13.16% of articles have been contributed by Italy. The countries India, France, Republic of Korea and Japan have contributed 6.04%, 5.6%, 4.2% and 3.5% of the studies respectively. 2.13% have been contributed by the researchers of the countries like Egypt and Australia. The contributions from the researchers of Turkey are 1.4% and nearly 1.06% of total research has been contributed by the researchers of Canada, Germany and Saudi Arabia. However, the researchers of Pakistan, Netherlands and Brazil have contributed the 0.711% of the studies. Next, the researchers in the countries like Iran, Spain, Iraq, Greece, Thailand, and Singapore have contributed 0.35% of the studies.
Fig. 8.
Country wise distribution of articles on diagnosis and prognosis of COVID-19.
4.7. Type of data used in COVID research
The following are the different types of data that have been used in the prediction, classification and forecasting of COVID-19 by the researchers and technocrats.
4.7.1. Usage of Clinical data
The forecasting of pandemics can be done with the information of registered number of COVID-19 cases along with their geographical locations. The most used dataset for the forecasting and prediction of COVID-19 is collated by John Hopkins University [131]. This dataset contains information such as the daily positive cases, total patients recovered per day and the death rates at a country as well as state level. Another data source Kaggle also contains the daily number of COVID-19 cases [132]. This dataset is annotated with attributes such as patient demographics, case reporting date and location. When working with real datasets, most of the researchers face class imbalance distribution issues. Another limitation is the variations in interventions, population densities and demographics have a major impact on the prediction. However, many good researchers have suggested for the use of real clinical data under the supervision of doctors for further diagnosis of COVID-19 other than online data. The main problem with online data is the presence of large extent of missing values, which may affect the proper analysis using any intelligent based methods.
4.7.2. Usage of Online data
The growth, nature and spread of COVID-19 can be predicted using the rich textual data available from various online sources. The interpretation of the epidemic is quite complicated as most of the studies have taken into account only a few determinants. Assuming the affect of virus in terms of positive and death case everywhere in the globe could produce inaccurate predictions. To make accurate predictions, it is necessary for the researches to understand the spread at the local, regional national as well as international levels. To be accurate, analysts would also need data from the medical authorities that distinguish between deaths caused by the coronavirus and deaths that would have happened due to anonymous disease. In most of the studies this data is not known, or not available. The range to which people pursue the local government's quarantine policies or measures to prevent the spread of infection is also difficult to find. These factors need to be considered to make accurate prediction of COVID-19 using online data while experimenting with any ML based techniques.
4.7.3. Usage of Biomedical data
The screening of medical images such as chest CT images or X-ray images is considered as an alternative solution to overcome the shortage of RT-PCR supply. Now-a-days, most of the research for the classification and prediction of COVID-19 has been carried on medical image data set, as medical image analysis helps the physicians in the accurate prediction of imaging modalities in pneumonia. From the literature review it has been observed, there are still some limitations in the usage of medical image datasets. The vast challenge in the medical diagnosis is the classification of medical images due to the limited accessibility of medical images. The process of automated distinction is difficult in CT images, as those share some common imagery characteristics among novel COVID and other forms of pneumonia. None of the studies reviewed by the authors accurately reported the high specificity of CT in distinguishing COVID-19 from other pneumonia with identical CT findings, thus restricting the usage of CT as a confirmatory diagnostic test. The presence of mild or no CT findings in many early cases of infection highlights the difficulties of early detection. Moreover, the CT scan tools are expensive and patients are exposed to more radiation. Even though chest x-ray images are cheaper and expose the patient to less radiation, these images have higher false diagnosis rate.
4.8. Distribution of articles by Prediction, Classification and Forecasting of COVID-19
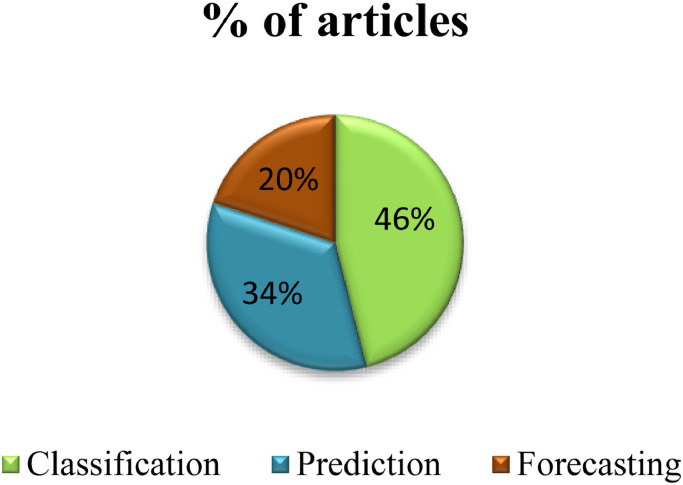
From the Fig. 9 it has been observed that majority of work has been done on classification of COVID-19 (46%). Next, 36% of work has been done on the prediction of COVID-19. Only 20% of the work has been done on the forecasting of COVID-19.From the Figure., it can be concluded that less work has been done on prediction and forecasting due to the lack of real world datasets and availability of less number of training medical images.
Fig. 9.
Articles published on COVID-19 Prediction, Classification and Forecasting.
5. Challenges
This section focuses on some of the challenges that raised while implementing the intelligent diagnostic tools in the prediction of COVID-19.
5.1. Limitation of data
The implementation of predictive tools using deep learning and machine learning requires huge volume of data. Even though few datasets for medical images and textual analysis are publicly available, these datasets are small when compared to the needs of the deep learning approaches. The main reason for the scarcity of measured data is the frequent segregation of data over different geographical regions. Therefore, aligning of the data sources is one of the key issues that need to be solved. Another limitation arises in the development of quality datasets, as real time datasets contain poorly quantified biases. As results of this, poor outcomes will be produced if models are trained on unrepresentative data. Although transfer learning allow models to be specific with regional characteristics, it is difficult to perform model selection due to the fast moving nature of the data. Therefore, designing an analytical approach to overcome these limitations is one of the key challenges that need to be addressed. Most of the researchers and technocrats are also facing the problem of lack of real data. This issue can be accomplished by creating more real world datasets with updated COVID-19 data. Another issue that needs to be observed is the less involvement of medical community. In most of the studies, either few or no physician has involved in the assessment of medical images such as X-rays and CT images.
5.2. Accuracy of prediction
There exists a hidden risk in all scientific work as most of the methods in the study are based on statistical learning on quickly produced datasets. The outcome of the research may have biases that may impact the policies taken by the government in controlling the spread of disease. Therefore the challenge is to find the uncertainty of conclusions produced in this research. The correctness of the data can be ensured by providing reproducible conclusions. This, in turn creates the challenge of balancing the requirements against the urgency.
5.3. Usage of advance approaches
Some data science approaches such as ultrasound scans and magnetic resonance imaging (MRI) have limited exposure in combating COVID-19. Even though ultrasound scans have shown good performance as that of chest CT scans, no studies have explored the usage of ultrasound scans in the prediction of COVID-19. Though some studies [133], have shown the efficient usage of MRI in predicting COVID-19 infections, still the approach remained unexplored due to the scarcity of adequate training data. Therefore, the challenge is to develop well-annotated dataset to make potential usage of new approaches in the prediction of COVID-19 infection.
5.4. Providing feasible solutions to developing countries
The usage of predictive tools in diagnosis of COVID-19 imposes a problem in developing countries that have limited access to healthcare facilities. Therefore, a key challenge is the development of tools that should be capable of deployment in economically underprivileged regions. For example, the development of mobile app for contact tracing should consider factors such as low cost, limited resources, accessibility to illiterates or disability people and support of multiple languages, so that it can be effectively deployed in economically deprived regions.
5.5. Necessity of advance intelligent systems on symptom based identification of COVID-19
Most of the studies have carried out only by considering the characteristics of COVID-19 and other pneumonia. The outcome of these studies may not produce accurate results as they have not considered the impact of other factors such as age, gender, diabetes, hypertension, chronic liver and kidney disease and so on. Therefore, to perform accurate predictions more research has to be carried on symptom based identification of COVID-19. Moreover, apart from prediction and forecasting based models, future research requires more attention on the classification problems on COVID-19 through various symptoms for easy and quick diagnosis. Also, most of the research is dedicated to top affected countries with this pandemic disease and further research may be enhanced for remaining mostly affected countries around the globe. Further, accurate prediction in the number of deaths and infections with advance machine learning approach is of utmost importance in the present scenario. As most of the machine learning models are highly accurate with large amount of data, so it may be worthy to note that with the increasing no of data and datasets of majorly COVID affected countries, many highly accurate models will be developed as a leading solution to this outbreak.
6. Conclusion
The researchers are always active in addressing the emerging challenges that arises in different application domains. In recent days, COVID-19 an infection caused by SARS-CoV-2 is one of the most emerging research areas as it affected more than 3 million people in 213 countries within a short span of time. Therefore, to empower the government and healthcare sector, it is necessary to analyze various forecasting and prediction tools. In this paper, an overall comprehensive summary of ongoing work in the prediction of COVID-19 infection using various intelligent approaches has been presented. Initially, the origin, dissemination and the affect of COVID-19 on the public health has been discussed. The major contribution of the study is the analysis of various prediction and forecasting models such as statistical, machine learning and deep learning approaches and their applications in the control of the pandemic. Following this, the analysis is broadened by making a critical investigation on the growth of studies carried on COVID-19 in various journals by country and the performance analysis of the statistical, machine learning and deep learning approaches. Finally, at the end of the paper some of the challenges observed as a part of systematic review are highlighted that may further help the researchers and technocrats to develop more accurate prediction models in the prediction of COVID-19.
The SARS-CoV-2 has infected about 3.48 million people and caused 2,48,144 deaths across 213 countries of the World as on May 3, 2020 according to the WHO. As the total of covid-19 cases registered has crossed all the previous records of the viral disease since last moths, it is considered to be the most dangerous disease till date. The whole world political, social, economic and financial structures have been disturbed because of the outbreak of pandemic. The economies of the world's topmost countries such as the US, China, UK, Germany, France, Italy, Japan and many others are at the edge of destruction. As 162 countries have moved into the lockdown to prevent the transmission of pandemic, the business across the world is operating in fear of an impending collapse of global financial markets. The sectors such as supply chains, trade, transport, tourism, and the hotel industry have been damaged extremely because of the pandemic. Other sectors like Apparel & textile, Building and Construction sectors have been affected adversely due to the lack of labour supply and availability of raw material. The other sector that is badly affected is the aviation sector as both international and domestic flights cancelled due to the implementation of lockdown in many countries. Even though the effect of lock down is less on essential goods retailer, other retailers such as shops and malls have been highly impacted by the pandemic. Due to the pandemic, the educational institutions have also been seriously affected and led to the shutdown of institutions which caused an interruption in the students learning activity as well as in internal and external assessment necessary for the qualification of the student. Even though several sectors have been affected, there are some sectors such as Digital and Internet Economy, Food based retail, Chemicals and Pharma sectors that have seen growth during the pandemic lockdown. The deep learning and machine learning approaches are useful in forecasting the impact of COVID-19 on different sectors which may help the government in implementing proper policies to overcome the economic crisis. From the systematic analysis, it is evident that computing intelligent approaches such as ML, DL, mathematical and statistical approaches have been profitably used in the prediction and screening of COVID-19 pandemic. It is observed that SVM, RF,K-means and linear regression of ML approaches have been mostly used for solving issues of COVID-19. While in case of DL, CNN and its variants are mostly utilized for predicting the pandemic. Even though computing approaches have been successfully used in the prediction and forecasting of COVID-19 pandemic, still there exist certain limitations such as limited availability of annotated medical images, not taking into account predictive end events such as mortality or admission into critical care unit while forecasting, not considering some features of medical images such as GGO, crazy-paving pattern, and bilateral involvement which are prerequisite in the diagnosis of COVID-19, training on small datasets and not coping with data irregularities need urgent focus to develop more accurate models. It is also observed that every researchers and modeling groups all over the world are presently facing the issue of scarcity of data. Therefore, real-world datasets with more epidemiological data need to be created for the development of more accurate prediction models. Moreover, the accuracy of prediction tools can be enhanced by the usage of advanced computing intelligent approaches such as ensemble method like bagging, stacking etc., application of optimization techniques, usage of artificial neural networks and higher order neural networks in the screening and prediction of COVID-19 which is considered as further scope of research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare that this manuscript has no conflict of interest with any other published source and has not been published previously (partly or in full). No data have been fabricated or manipulated to support our conclusions.
Footnotes
https://ddi.sutd.edu.sg/
References
- 1.Orbann Carolyn. Defining epidemics in computer simulation models: How do definitions influence conclusions? Epidemics. 2017;19:24–32. doi: 10.1016/j.epidem.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. "Principles of epidemiology in public health practice: an introduction to applied epidemiology and biostatistics." (2006).
- 3.Morse Stephen S. Plagues and politics. Palgrave Macmillan; London: 2001. Factors in the emergence of infectious diseases; pp. 8–26. [Google Scholar]
- 4.Henderson Donald Ainslie. Vol. 237. Prometheus Books; Amherst, NY: 2009. (Smallpox: the death of a disease). [Google Scholar]
- 5.Spreeuwenberg Peter, Kroneman Madelon, Paget John. "Reassessing the global mortality burden of the 1918 influenza pandemic. Am J Epidemiol. 2018;187(12):2561–2567. doi: 10.1093/aje/kwy191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Gensheng. Proceedings of the 2011 ACM Symposium on Research in Applied Computation. 2011. A review of breast tissue classification in mammograms. [Google Scholar]
- 7.Bishop Christopher M. springer; 2006. Pattern recognition and machine learning. [Google Scholar]
- 8.Pandey Manish Kumar, Karthikeyan S. Performance analysis of time series forecasting of ebola casualties using machine learning algorithm. Proceedings ITISE. 2011:201. [Google Scholar]
- 9.Hripcsak George, Albers David J. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen Peter B., Jensen Lars J., Brunak Søren. "Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 11.Luo Jake. Big data application in biomedical research and health care: a literature review. Biomedical Informatics Insights. 2016;8 doi: 10.4137/BII.S31559. BII-S31559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talo Muhammed. Application of deep transfer learning for automated brain abnormality classification using MR images. Cognitive Systems Res. 2019;54:176–188. [Google Scholar]
- 13.Ismael Sarah Ali Abdelaziz, Mohammed Ammar, Hefny Hesham. "An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artificial Intel Med. 2020;102 doi: 10.1016/j.artmed.2019.101779. [DOI] [PubMed] [Google Scholar]
- 14.Khalifa Nour Eldeen M. Artificial Intelligence Technique for gene expression by tumor RNA-seq data: a novel optimized deep learning approach. IEEE Access. 2020;8:22874–22883. [Google Scholar]
- 15.Ho Chi-Sing. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat Commun. 2019;10(1):1–8. doi: 10.1038/s41467-019-12898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque Intisar Rizwan I., Neubert Jeremiah. "Deep learning approaches to biomedical image segmentation. Informatics in Medicine Unlocked. 2020 [Google Scholar]
- 17.Jaiswal Amit Kumar. Identifying pneumonia in chest X-rays: A deep learning approach. Measurement. 2019;145:511–518. [Google Scholar]
- 18.Martínez-Álvarez, F., et al. "Coronavirus optimization algorithm: a bioinspired metaheuristic based on the COVID-19 propagation model." arXiv preprint arXiv:2003.13633 (2020). [DOI] [PubMed]
- 19.Zhu Na. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Yudong, Wunderink Richard G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(2020). Wuhan Municipal Health Commission Infection Data. [Online] Available: https://wjw.wuhan.gov.cn/front/web/list2nd/no/710
- 22.Lu, Hongzhou, Charles W. Stratton, and Yi‐Wei Tang. "Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle." J Med Virol. [DOI] [PMC free article] [PubMed]
- 23.Huang Chaolin. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet North Am Ed. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai Chih-Cheng. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Organization WH. World HealthOrganization; 2020. Laboratory testing for coronavirus disease 2019 (COVID-19)in suspected human cases: interim guidance, 2 March 2020. [Google Scholar]
- 26.The Harvard Gazette, Coronavirus Cases Hit 17,400 and Are Likely to Surge, (2020) https://news.harvard.edu/gazette/story/2020/02/as-confirmed-cases-ofcoronavirus-surge-path-grows-uncertain/
- 27.Fehr Anthony R., Perlman Stanley. Coronaviruses. Humana Press; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Peng. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shereen Muhammad Adnan. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, Xiaowei, et al. "Deep learning system to screen coronavirus disease 2019 pneumonia." arXiv preprint arXiv:2002.09334 (2020).
- 31.Singh Rameshwer, Singh Rajeshwar, Bhatia Ajay. Sentiment analysis using machine learning technique to predict outbreaks and epidemics. Int J Adv Sci Res. 2018;3(2):19–24. [Google Scholar]
- 32.Breiman Leo. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 33.Petralia Francesca. Integrative random forest for gene regulatory network inference. Bioinformatics. 2015;31(12):i197–i205. doi: 10.1093/bioinformatics/btv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, Jiangpeng, et al. "Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results." medRxiv (2020).
- 35.Qi, Xiaolong, et al. "Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 36.Cobb J.S., Seale M.A. Examining the Effect of Social Distancing on the Compound Growth Rate of SARS-CoV-2 at the County Level (United States) Using Statistical Analyses and a Random Forest Machine Learning Model. Public Health. 2020 doi: 10.1016/j.puhe.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, Feng, et al. "Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification." arXiv preprint arXiv:2003.09860 (2020). [DOI] [PubMed]
- 38.Tang, Zhenyu, et al. "Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images." arXiv preprint arXiv:2003.11988 (2020).
- 39.Sarkar, Jit, and Partha Chakrabarti. "A Machine Learning Model Reveals Older Age and Delayed Hospitalization as Predictors of Mortality in Patients with COVID-19." medRxiv (2020).
- 40.Chen, Xiaoping, et al. "Hypertension and diabetes delay the viral clearance in COVID-19 patients." medRxiv (2020).
- 41.de Moraes Batista, Andre Filipe, et al. "COVID-19 diagnosis prediction in emergency care patients: a machine learning approach." medRxiv (2020).
- 42.Hassanien, Aboul Ella, et al. "Automatic X-ray COVID-19 Lung Image Classification System based on Multi-Level Thresholding and Support Vector Machine." medRxiv (2020).
- 43.Sonbhadra, Sanjay Kumar, Sonali Agarwal, and P. Nagabhushan. "Target specific mining of COVID-19 scholarly articles using one-class approach." arXiv preprint arXiv:2004.11706 (2020). [DOI] [PMC free article] [PubMed]
- 44.Zhang, Nan, et al. "Severity Detection For the Coronavirus Disease 2019 (COVID-19) Patients Using a Machine Learning Model Based on the Blood and Urine Tests." (2020). [DOI] [PMC free article] [PubMed]
- 45.Hassanien, Aboul Ella, Aya Salama, and Ashraf Darwsih. Artificial intelligence approach to predict the COVID-19 patient's recovery. No. 3223. EasyChair, 2020.
- 46.Barstugan, Mucahid, Umut Ozkaya, and Saban Ozturk. "Coronavirus (covid-19) classification using ct images by machine learning methods." arXiv preprint arXiv:2003.09424 (2020).
- 47.Sethy, Prabira Kumar, and Santi Kumari Behera. "Detection of coronavirus disease (covid-19) based on deep features." Preprints 2020030300 (2020): 2020.
- 48.Pandey, Gaurav, et al. "SEIR and Regression Model based COVID-19 outbreak predictions in India." arXiv preprint arXiv:2004.00958 (2020).
- 49.Liu, Dianbo, et al. "A machine learning methodology for real-time forecasting of the 2019-2020 COVID-19 outbreak using Internet searches, news alerts, and estimates from mechanistic models." arXiv preprint arXiv:2004.04019 (2020).
- 50.Dandekar, Raj, and George Barbastathis. "Neural Network aided quarantine control model estimation of COVID spread in Wuhan, China." arXiv preprint arXiv:2003.09403 (2020).
- 51.Carrillo-Larco Rodrigo M., Castillo-Cara Manuel. Using country-level variables to classify countries according to the number of confirmed COVID-19 cases: An unsupervised machine learning approach. Wellcome Open Res. 2020;5(56):56. doi: 10.12688/wellcomeopenres.15819.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magar, Rishikesh, Prakarsh Yadav, and Amir Barati Farimani. "Potential neutralizing antibodies discovered for novel corona virus using machine learning." arXiv preprint arXiv:2003.08447 (2020). [DOI] [PMC free article] [PubMed]
- 53.Yan, Li, et al. "Prediction of survival for severe Covid-19 patients with three clinical features: development of a machine learning-based prognostic model with clinical data in Wuhan." medRxiv (2020).
- 54.Chen, Xingdong, and Zhenqiu Liu. "Early prediction of mortality risk among severe COVID-19 patients using machine learning." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 55.Al-Karawi, Dhurgham, et al. "Machine learning analysis of chest ct scan images as a complementary digital test of coronavirus (COVID-19) Patients." medRxiv (2020).
- 56.Metsky, Hayden C., et al. "CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach." bioRxiv (2020).
- 57.LeCun, Yann, Yoshua Bengio, and Geoffrey Hinton. "Deep learning. nature521." (2015): 530-531. [DOI] [PubMed]
- 58.Choe Jooae. Deep Learning–based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292(2):365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 59.Kermany Daniel S. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 60.Wang Pu. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nature Biomed Eng. 2018;2(10):741–748. doi: 10.1038/s41551-018-0301-3. [DOI] [PubMed] [Google Scholar]
- 61.Wang Dawei. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abbas, Asmaa, Mohammed M. Abdelsamea, and Mohamed Medhat Gaber. "Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network." arXiv preprint arXiv:2003.13815 (2020). [DOI] [PMC free article] [PubMed]
- 63.Narin, Ali, Ceren Kaya, and Ziynet Pamuk. "Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks." arXiv preprint arXiv:2003.10849 (2020). [DOI] [PMC free article] [PubMed]
- 64.Huang Chiou-Jye. Multiple-Input Deep Convolutional neural network model for COVID-19 forecasting in China. medRxiv. 2020 [Google Scholar]
- 65.Mukherjee, Himadri, et al. "Shallow convolutional neural network for COVID-19 outbreak screening using chest X-rays." (2020). [DOI] [PMC free article] [PubMed]
- 66.Amyar, Amine, Romain Modzelewski, and Su Ruan. "Multi-task deep learning based ct imaging analysis for COVID-19: classification and segmentation." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 67.Hammoudi, Karim, et al. "Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of COVID-19." arXiv preprint arXiv:2004.03399 (2020). [DOI] [PMC free article] [PubMed]
- 68.Rajaraman, Sivaramakrishnan, et al. "Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays." arXiv preprint arXiv:2004.08379 (2020). [DOI] [PMC free article] [PubMed]
- 69.Hall, Lawrence O., et al. "Finding COVID-19 from chest x-rays using deep learning on a small dataset." arXiv preprint arXiv:2004.02060 (2020).
- 70.Ozkaya, Umut, Saban Ozturk, and Mucahid Barstugan. "Coronavirus (COVID-19) classification using deep features fusion and ranking technique." arXiv preprint arXiv:2004.03698 (2020).
- 71.Zahangir Alom, Md, et al. "COVID_MTNet: COVID-19 detection with multi-task deep learning approaches." arXiv (2020): arXiv-2004.
- 72.Rahimzadeh, Mohammad, and Abolfazl Attar. "A New Modified deep convolutional neural network for detecting COVID-19 from X-ray Images." arXiv preprint arXiv:2004.08052 (2020). [DOI] [PMC free article] [PubMed]
- 73.Zhang, Jianpeng, et al. "Covid-19 screening on chest x-ray images using deep learning based anomaly detection." arXiv preprint arXiv:2003.12338 (2020).
- 74.Hemdan, Ezz El-Din, Marwa A. Shouman, and Mohamed Esmail Karar. "Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images." arXiv preprint arXiv:2003.11055 (2020).
- 75.Wang, Shuo, et al. "A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 76.Zheng, Chuansheng, et al. "Deep learning-based detection for COVID-19 from chest CT using weak label." medRxiv (2020).
- 77.Apostolopoulos, Ioannis, Sokratis Aznaouridis, and Mpesiana Tzani. "Extracting possibly representative COVID-19 Biomarkers from X-Ray images with Deep Learning approach and image data related to Pulmonary Diseases." arXiv preprint arXiv:2004.00338 (2020). [DOI] [PMC free article] [PubMed]
- 78.Kumar, Rahul, et al. "Accurate prediction of COVID-19 using chest x-ray images through deep feature learning model with smote and machine learning classifiers." medRxiv (2020).
- 79.Fu, Min, et al. "Deep learning-based recognizing COVID-19 and other common infectious diseases of the lung by chest ct scan images." medRxiv (2020).
- 80.Razzak, Imran, et al. "Improving Coronavirus (COVID-19) diagnosis using deep transfer learning." medRxiv (2020).
- 81.Sherstinsky Alex. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network. Physica D. 2020;404 [Google Scholar]
- 82.Jana, Saikat, and Parama Bhaumik. "A multivariate spatiotemporal spread model of COVID-19 using ensemble of ConvLSTM networks." medRxiv (2020).
- 83.Tomar Anuradha, Gupta Neeraj. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jelodar, Hamed, et al. "Deep Sentiment Classification and Topic discovery on novel Coronavirus or COVID-19 online discussions: NLP using LSTM Recurrent Neural Network Approach." arXiv preprint arXiv:2004.11695 (2020). [DOI] [PubMed]
- 85.Ibrahim, Mohamed R., et al. "Variational-LSTM autoencoder to forecast the spread of coronavirus across the globe." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 86.Patankar, Sayalee. "Deep learning-based computational drug discovery to inhibit the RNA Dependent RNA Polymerase: application to SARS-CoV and COVID-19." (2020).
- 87.Yang Zifeng. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thoracic Dis. 2020;12(3):165. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kafieh, Rahele, et al. "COVID-19 in Iran: a deeper look Into the future." medRxiv (2020).
- 89.Kolozsvari, Laszlo Robert, et al. "Predicting the epidemic curve of the coronavirus (SARS-CoV-2) disease (COVID-19) using artificial intelligence." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 90.Bandyopadhyay, Samir Kumar, and Shawni Dutta. "Machine learning approach for confirmation of COVID-19 cases: positive, negative, death and release." medRxiv (2020).
- 91.Ayyoubzadeh Seyed Mohammad. Predicting COVID-19 incidence through analysis of google trends data in Iran: data mining and deep learning pilot study. JMIR Public Health and Surveillance. 2020;6(2):e18828. doi: 10.2196/18828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu, Siqi, et al. "3D tomographic pattern synthesis for enhancing the quantification of COVID-19." arXiv preprint arXiv:2005.01903 (2020)
- 93.Khalifa, Nour Eldeen M., et al. "Detection of Coronavirus (COVID-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset." arXiv preprint arXiv:2004.01184 (2020).
- 94.Loey, Mohamed, Florentin Smarandache, and Nour Eldeen M. Khalifa. "Within the lack of COVID-19 benchmark dataset: a novel gan with deep transfer learning for corona-virus detection in chest x-ray images." (2020).
- 95.Hartono, Pitoyo. "Generating similarity map for COVID-19 transmission dynamics with topological autoencoder." arXiv preprint arXiv:2004.01481 (2020).
- 96.Shan+, Fei, et al. "Lung infection quantification of covid-19 in ct images with deep learning." arXiv preprint arXiv:2003.04655 (2020).
- 97.Hu, Zixin, et al. "Forecasting and evaluating intervention of Covid-19 in the World." arXiv preprint arXiv:2003.09800 (2020).
- 98.Zhavoronkov Alex. Potential COVID-2019 3c-like protease inhibitors designed using generative deep learning approaches. Insilico Medicine Hong Kong Ltd A. 2020;307:E1. [Google Scholar]
- 99.Xu, Hui, et al. "Analysis and prediction of false negative results for SARS-CoV-2 detection with pharyngeal swab specimen in COVID-19 patients: a retrospective study." MedRxiv (2020).
- 100.Zhong Linhao. Early Prediction of the 2019 novel coronavirus outbreak in the mainland China based on simple mathematical model. IEEE Access. 2020;8:51761–51769. doi: 10.1109/ACCESS.2020.2979599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao Yuanyuan. Forecasting the cumulative number of COVID-19 deaths in China: a Boltzmann function-based modeling study. Infection Control & Hospital Epidemiology. 2020:1–16. doi: 10.1017/ice.2020.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Banerjee, Amitava, et al. "Estimating excess 1-year mortality from COVID-19 according to underlying conditions and age in England: a rapid analysis using NHS health records in 3.8 million adults." medRxiv (2020).
- 103.Azad, Sarita, and Neeraj Poonia. "Short-Term Forecasts of COVID-19 Spread Across Indian States Until 1May 2020." (2020).
- 104.Mohler, George, et al. "Analyzing the World-Wide Impact of Public Health Interventions on the Transmission Dynamics of COVID-19." arXiv preprint arXiv:2004.01714 (2020).
- 105.Gupta, Sourendu, and R. Shankar. "Estimating the number of COVID-19 infections in Indian hot-spots using fatality data." arXiv preprint arXiv:2004.04025 (2020).
- 106.Dowd Jennifer Beam. "Demographic science aids in understanding the spread and fatality rates of COVID-19. Proc Natl Acad Sci. 2020 doi: 10.1073/pnas.2004911117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nadim, Sk Shahid, Indrajit Ghosh, and Joydev Chattopadhyay. "Short-term predictions and prevention strategies for COVID-2019: A model based study." arXiv preprint arXiv:2003.08150 (2020). [DOI] [PMC free article] [PubMed]
- 108.Magal Pierre, Webb Glenn. "Predicting the number of reported and unreported cases for the COVID-19 epidemic in South Korea, Italy, France and Germany. Italy, France and Germany. 2020 doi: 10.1016/j.jtbi.2020.110501. March 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anzai Asami. Assessing the impact of reduced travel on exportation dynamics of novel coronavirus infection (COVID-19) J Clin Med. 2020;9(2):601. doi: 10.3390/jcm9020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar, Pavan, et al. "Forecasting COVID-19 impact in India using pandemic waves Nonlinear Growth Models." medRxiv (2020).
- 111.Oliveiros, Barbara, et al. "Role of temperature and humidity in the modulation of the doubling time of COVID-19 cases." medRxiv (2020).
- 112.Lu, Jian. "A new, simple projection model for covid-19 pandemic." medrxiv (2020).
- 113.Wang, Shuai, et al. "A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19)." MedRxiv (2020). [DOI] [PMC free article] [PubMed]
- 114.Farooq, Muhammad, and Abdul Hafeez. "Covid-resnet: A deep learning framework for screening of covid19 from radiographs." arXiv preprint arXiv:2003.14395 (2020).
- 115.Chen, Jun, et al. "Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study." medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 116.Li Lin. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Asnaoui, Khalid El, Youness Chawki, and Ali Idri. "Automated methods for detection and classification pneumonia based on x-ray images using deep learning." arXiv preprint arXiv:2003.14363 (2020).
- 118.Wang, Linda, and Alexander Wong. "COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images." arXiv preprint arXiv:2003.09871 (2020). [DOI] [PMC free article] [PubMed]
- 119.Chowdhury, Muhammad EH, et al. "Can AI help in screening Viral and COVID-19 pneumonia?." arXiv preprint arXiv:2003.13145 (2020).
- 120.Apostolopoulos Ioannis D., Mpesiana Tzani A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Afshar, Parnian, et al. "Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images." arXiv preprint arXiv:2004.02696 (2020). [DOI] [PMC free article] [PubMed]
- 122.Butt Charmaine. Deep learning system to screen corona virus disease 2019 pneumonia. Applied Intelligence. 2020:1. doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li, Xin, and Dongxiao Zhu. "Covid-xpert: An ai powered population screening of covid-19 cases using chest radiography images." arXiv preprint arXiv:2004.03042 (2020).
- 124.Ucar Ferhat, Korkmaz Deniz. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based Diagnostic of the Coronavirus Disease 2019 (COVID-19) from X-Ray Images. Med Hypotheses. 2020 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fanelli Duccio, Piazza Francesco. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos, Solitons Fractals. 2020;134 doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boccaletti Stefano. Modeling and forecasting of epidemic spreading: The case of Covid-19 and beyond. Chaos, Solitons, and Fractals. 2020 doi: 10.1016/j.chaos.2020.109794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ndairou Faical. Mathematical modeling of covid-19 transmission dynamics with a case study of wuhan. Chaos, Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Xiaolei, Ma Renjun, Wang Lin. Predicting turning point, duration and attack rate of COVID-19 outbreaks in major Western countries. Chaos, Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.109829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Woody, Spencer, et al. "Projections for first-wave COVID-19 deaths across the US using social-distancing measures derived from mobile phones." medRxiv (2020).
- 130.Jewell Nicholas P., Lewnard Joseph A., Jewell Britta L. Caution warranted: using the Institute for Health Metrics and Evaluation Model for predicting the course of the COVID-19 pandemic. Ann Intern Med. 2020 doi: 10.7326/M20-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.CSSEGIS and Data, “CSSEGIS and Data/COVID-19,” Mar 2020. [Online]. Available:https://github.com/CSSEGISandData/COVID-19
- 132.Sudalairaj kumar Data, “Novel corona-virus dataset,” Mar 2020. [Online]. Available:https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset
- 133.Poyiadji Neo. COVID-19–associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features. Radiology. 2020 doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]