Abstract
In less than five months, COVID-19 has spread from a small focus in Wuhan, China, to more than 5 million people in almost every country in the world, dominating the concern of most governments and public health systems. The social and political distresses caused by this epidemic will certainly impact our world for a long time to come. Here, we synthesize lessons from a range of scientific perspectives rooted in epidemiology, virology, genetics, ecology and evolutionary biology so as to provide perspective on how this pandemic started, how it is developing, and how best we can stop it.
Keywords: Coronavirus, Coevolution, Host susceptibility, Immune system, Pandemics, Phylodynamics
Highlights
-
•
A new coronavirus, SARS-CoV-2, emerged by the end of December 2019 in China.
-
•
In less than 3 months, it has spread worldwide causing a pandemic of COVID-19.
-
•
We review the genetic, virological, ecological, epidemiological and evolutionary factors of this viral pandemic.
1. Introduction
Pathogen X, the hypothetical unknown potentially devastating microorganism capable of causing a major pandemic (Friedrich, 2018), now has a name: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Despite warnings and preparedness efforts by WHO and other health agencies, the rapid spread of COVID-19, which in less than 4 months has moved from affecting a few persons in Wuhan (Hubei province, China) to more than 5 million people in almost every country in the world (Coronavirus Research Center, https://coronavirus.jhu.edu/map.html visited on May 21th, 2020) has caught by surprise most governments and public health systems. The result cannot be evaluated yet but the over 300,000 officially recognized deaths and the economic, social and political distresses caused by this epidemic will certainly impact our world in the coming months, probably years. In light of the wave of disinformation and mistrust by wide sectors in the public towards experts and scientists' views on how to stop this pandemic, we feel the need to bring a scientific perspective on the source, causes, uses, and possible ways to halt the spread of this new coronavirus from a basic science perspective, that marking the core nature of this journal, blending Genetics, Virology, Epidemiology, and Evolutionary Biology (Population genetics and population biology: what did they bring to the epidemiology of transmissible diseases? An E-debate, 2001).
2. The origin of SARS-CoV-2 and COVID-19
The novel human coronavirus (SARS-CoV-2), responsible for the current COVID-19 pandemic, was first identified in December 2019, in the Hubei province of China (Zhu et al., 2020). After SARS-CoV (severe acute respiratory syndrome coronavirus) and MERS-CoV (Middle East respiratory syndrome Coronavirus), SARS-CoV-2 is the third highly pathogenic coronavirus to emerge and spread in human populations. Phylogenetic analyses showed that, as SARS-CoV, SARS-CoV-2 is a member of the Sarbecovirus subgenus (genus Betacoronavirus) (Zhou et al., 2020b). Recently, the International Committee on Taxonomy of Viruses (ICTV) indicated that SARS-CoV-2 is to be classified within the species Severe acute respiratory syndrome-related coronavirus (Coronaviridae Study Group of the ICTV, 2020). MERS-CoV (subgenus Merbecovirus) is more distantly related to SARS-CoV-2 than SARS-CoV.
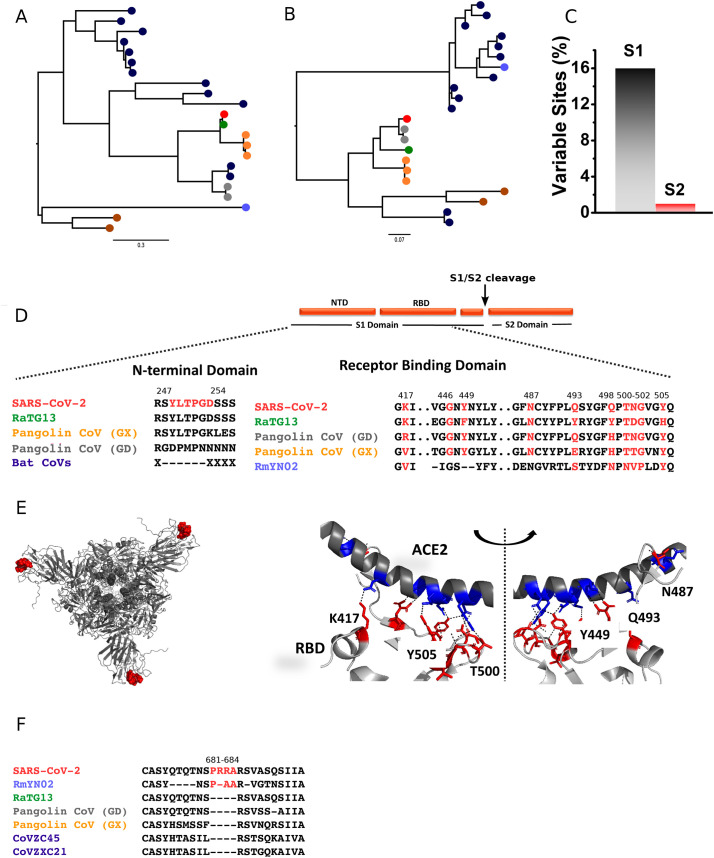
In the aftermath of the SARS and MERS epidemics, intense efforts were devoted to identify the animal reservoirs of these viruses and to reconstruct the chain of events that led to the human spillovers. It is now known that both viruses originated in bats and were transmitted to humans by intermediate hosts (Normile and Enserink, 2003; Killerby et al., 2020). In particular, the progenitor of SARS-CoV emerged through recombination among bat viruses and subsequently infected palm civets and other small carnivores, eventually spilling over to humans (Cui et al., 2019). MERS-CoV was most likely transmitted from bats to dromedary camels decades earlier than the first human cases were registered (Müller et al., 2014). MERS-CoV displays limited human-to-human spread, and most cases resulted from independent zoonotic transmission from camels (Cui et al., 2019). It is thus unsurprising that the closest known relatives of SARS-CoV-2 are two bat coronaviruses (BatCoV RmYN02 and BatCoV RaTG13) identified in horseshoe bats (Rhinolophus malyanus and R. affinis, respectively) (Zhou et al., 2020a, Zhou et al., 2020b). Both BatCoVs display an average nucleotide identity of ~95% with SARS-CoV-2, although with variations through the genome, and they were detected in bats sampled in Yunnan province, China, in 2019 and 2013, before the first detection of SARS-CoV-2 in humans (Zhou et al., 2020b). Both the place and timing of BatCoV RmYN02 and RaTG13 detection, as well as their levels of identity with SARS-CoV-2, indicate that these viruses are not direct progenitors of the SARS-CoV-2 pandemic strain, but clearly support the view that the latter had an ultimate bat origin (Zhou et al., 2020b) (Fig. 1A).
Fig. 1.
Comparative analysis of SARS-CoV-2 with other coronaviruses. Maximum-likelihood phylogenetic trees of the NTD (A) and RBD (B) regions of SARS-CoV-2 (red), RaTG13 (green), RmYN02 (light blue), Pangolin coronaviruses (Guangdong lineage, grey; Guangxi lineage, orange), and other Asian (dark blue) and non-Asian (brown) bat coronaviruses belonging to the Sarbecovirus subgenus. Trees are based on amino acid sequences and were built using PhyML (Guindon and Gascuel, 2003). Trees are mid-point rooted. (C) Combined variability in S1 (grey) and S2 (red) domains of SARS-CoV-2 when compared to RaTG13 and pangolin coronaviruses spike sequences. (D) Sequence alignments showing absence of the YLTPGD insert in bat sarbecoviruses, and the sequence of the RBD region involved in the interaction with ACE2. (E) The position of YLTPGD inserts forming conformational clusters (red spheres) at the NTD of SARS-CoV-2 spike protein is shown (left). The ribbon structure of the spike protein-ACE2 interaction surface is represented to show polar interactions (right). Polar interactions were analyzed using PyMol using PDB id: 6m0j (Lan et al., 2020). (F) Alignment of the region carrying the polybasic amino acid insertion (red) at the S1/S2 cleavage site. GenBank/GISAID accessions for the sequences included in trees are: NC_045512.2 (SARS-CoV-2), MN996532.1(RaTG13), EPI_ISL_412977 (RmYN02), MT084071.1 (MP789 or Guangdong 1), EPI_ISL_410544 (Guangdong P2S), MT040334.1 (GX-P1E),MT072865.1 (GX-P3B), MT040335.1 (GX-P5L), KY417148 (Rs4247), DQ071615.1 (Rp3), GQ153547.1 (HKU3–12), GQ153542 (HKU3–7), MK211378.1 (BtRs-BetaCoV/YN2018D), DQ648856.1 (BtCoV/273/2005), JX993987.1 (Rp/Shaanxi2011), KJ473816 (BtRs-BetaCoV/YN2013), MG772933 (CoVZC45), MG772934 (CoVZXC21), KY417151.1 (Rs7327), KF569996 (LYRa11), NC_014470.1 (BM48–31/BGR/2008), KY352407.1 (BtKY72). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In analogy to SARS-CoV and MERS-CoV, several lines of evidence suggest that an intermediate host was responsible for the cross-species transmission of SARS-CoV-2 to humans. First, most although not all, early COVID-19 detected cases were associated with the Huanan seafood and wildlife market in Wuhan city, where several mammalian species were traded (Huang et al., 2020). This is reminiscent of the circumstances associated with the initial phases of SARS-CoV spread, as palm civets were sold in wet markets and their meat consumed (Cui et al., 2019). Second, in vitro experiments have shown that, in addition to bats, SARS-CoV-2 can infect cells from small carnivores and pigs (Zhou et al., 2020b). Experimental in vivo infection and transmission in ferrets and cats was also reported (Kim et al., 2020; Shi et al., 2020a). Third, viruses very closely related (85.5% to 92.4% sequence similarity) to SARS-CoV-2 were very recently detected in Malayan or Sunda pangolins (Manis javanica) illegally imported in Southern China (Lam et al., 2020). The analysis of these viral genomes indicated that pangolins host at least two sub-lineages of sarbecoviruses, which are referred as the Guangdong and Guangxi lineages after the locations where the animals were sampled (Lam et al., 2020) (Fig. 1A and B). Viruses in the Guangdong lineage share high similarity with SARS-CoV-2 in the receptor-binding motif of the spike protein. This same region is instead the most divergent between BatCoV RaTG13 and SARS-CoV-2 (Lam et al., 2020; Zhou et al., 2020b). This observation is very relevant, as the binding affinity between the spike protein and the cognate cellular receptor (angiotensin-converting enzyme 2, ACE2, in the case of SARS-CoV and SARS-CoV-2) is a major determinant of coronavirus host range (Haijema et al., 2003;Kuo et al., 2000; McCray Jr et al., 2007; Moore et al., 2004; Schickli et al., 2004). Indeed, coronaviruses use a surface spike glycoprotein to attach to host receptors and gain entry into cells (Walls et al., 2020). It is a homo-trimeric protein formed by two subunits, S1 and S2. The S1 subunit contains an N-terminal domain connected by a linker of variable length to the receptor-binding domain (RBD). In various coronaviruses, the N-terminal domain (NTD) and RBD contribute to define host range (Lu et al., 2015). The S2 domain participates in membrane fusion (Duquerroy et al., 2005). Comparison of the complete spike protein, as well as of the NTD, indicated higher similarity of SARS-CoV-2 with RaTG13 than with pangolin coronaviruses (Fig. 1A). Domain based sequence analysis indicates that sequence variations are majorly confined to the S1-domain (~16% variable sites) than to the S2-domain (Fig. 1C), and mutations were observed in both NTD and RBD. Interestingly, an insertion (YLTPGD) is present only in the NTD of SARS-CoV-2, RaTG13, and Guangxi pangolin coronaviruses, but absent in related bat coronaviruses (in Guangdong pangolin viruses the region is not fully covered by sequencing) (Fig. 1D). This motif forms a conformational cluster at the exposed NTD regions of the spike trimer and may contribute to determine host range. However, as mentioned above, within the RBD, SARS-CoV-2 shows higher identity with pangolin viruses belonging to the Guangdong lineage than to RaTG13 (Fig. 1B and D). Structural analysis of the binding interface between SARS-CoV-2 RBD and human ACE2 shows a strong network of polar contacts (PDB id: 6m0j) (Lan et al., 2020). SARS-CoV-2 RBD mediates these polar interactions through Lys417, Gly446, Tyr449, Asn487, Gln493, Gln498, Thr500, Asn501, Gly502 and Tyr505 with ACE2 (Fig. 1E). These residues, which participate in polar interactions with the host protein, are almost completely conserved with pangolin coronaviruses of the Guangdong lineage (Fig. 1D).
One possible explanation for this observation is that the RBDs of SARS-CoV-2 and Guangdong pangolin viruses have been progressively optimized through natural selection (convergent evolution) to bind ACE2 molecules from humans and pangolins (and possibly other non-bat mammalian species) (Lam et al., 2020). An alternative possibility is that recombination events among coronaviruses hosted by bats, pangolins, and possibly other mammals originated the progenitor of SARS-CoV-2 (Lam et al., 2020; Cagliani et al., 2020). It is presently impossible to disentangle these two alternative scenarios, and only the sequencing of additional related sarbecoviruses might eventually clarify the evolutionary history of SARS-CoV-2 RBD. It is also worth mentioning here that, as previously noted (Andersen et al., 2020), the similarity of the SARS-CoV-2 RBD with that of viruses only recently sequenced from pangolins can be regarded as a major evidence against the circulating theory that SARS-CoV-2 is the result of deliberate human manipulation.
In any case, these data do not necessarily imply that pangolins had a role in the emergence of SARS-CoV-2 and in its spread to humans, as these animals might have in turn contracted infection from a bat or other reservoir. Moreover, the SARS-CoV-2 spike protein displays a unique feature that is not shared with either BatCoV RaTG13 or the pangolin viruses, namely the presence of a furin cleavage site insertion (PRRA) at the S1-S2 junction (Walls et al., 2020) (Fig. 1D and F). This feature, also absent in SARS-CoV, was suggested to increase viral infectivity and/or pathogenicity (Walls et al., 2020; Andersen et al., 2020). It is presently unknown how and when SARS-CoV-2 acquired the furin cleavage site, but it is equally unexplored whether it affects any viral phenotype or if it contributed to adaptation to humans or other hosts. Importantly, though, the presence of a similar insertion in a virus isolated from wild bats is another strong indication in favor of a natural animal origin of SARS-CoV-2 (Zhou et al., 2020a).
3. Where did adaptation to humans occur?
Although, for the reasons mentioned above, an as-yet unidentified intermediate host is likely to have played a role in the zoonotic transmission of SARS-CoV-2 in the Wuhan market or elsewhere, the possibility that SARS-CoV-2 was directly transmitted from bats to humans cannot be discarded. Indeed, serological surveys on people living in proximity to bat colonies in Yunnan province indicated that direct bat-to-human transmission of SARS-CoV related coronaviruses might occur (Wang et al., 2018). In general, wild animal trade might reduce the ecological barriers separating humans from coronavirus hosts, including these bats or other mammals. However, the observation that some early COVID-19 cases had no apparent epidemiological link to the Huanan seafood and wildlife market (Huang et al., 2020) opens the possibility that the virus originated elsewhere and that the crowded market only contributed to the spreading of the epidemic. Clearly, wide sampling of coronavirus diversity in different mammals in China and neighboring countries will be necessary to track down the progenitor(s) of SARS-CoV-2.
The scenarios described above implicitly imply that, whatever the source of the zoonotic transmission, SARS-CoV-2 emerged and adapted in another host, eventually spilling over to humans (Andersen et al., 2020). It cannot however be excluded that, before being recognized in December 2019, SARS-CoV-2 had been circulating for a while in humans, maybe causing mild symptoms. The acquisition of some key viral mutations during unrecognized human transmission might have then fostered the current pandemic strain, characterized by sustained human-to-human transmissibility and virulence. In this case, the identification of such mutations and the assessment of the role of the above-mentioned furin cleavage site insertion would be of paramount importance. Nonetheless, it should be kept in mind that extreme caution should be exerted when ascribing phenotypic effect to mutations that arise during viral spreads (Grubaugh et al., 2020). Here, again, MERS-CoV and SARS-CoV have lessons to teach. Whereas in the case of SARS-CoV, changes in the spike protein RBD contributed to the adaptation of the virus to human cells (Wu et al., 2012; Qu et al., 2005), MERS-CoV adaptation to our species occurred with limited changes in the RBD (Cotten et al., 2014; Forni et al., 2015). However, during an outbreak of MERS-CoV in South Korea, viral strains carrying point mutations in the spike RBD emerged and spread (Kim et al., 2016). Notably, whereas these RBD mutations were found to decrease rather than increase binding to the cellular receptor, they facilitated viral escape from neutralizing antibodies (Kim et al., 2016; Kim et al., 2019; Rockx et al., 2010; Kleine-Weber et al., 2019). This suggests that changes in the spike protein do not necessarily arise as an adaptation to optimize receptor-binding affinity.
Even more emblematic is the case of SARS-CoV ORF8, encoding an accessory viral protein. In the early stages of the epidemic, SARS-CoV strains acquired a 29-nucleotide deletion in ORF8 (Chinese SARS Molecular Epidemiology Consortium, 2004). Together with the observation that the encoded protein is fast evolving in SARS-CoV strains, this finding was taken to imply that deletions in ORF8 were driven by natural selection and favored human infection (Lau et al., 2015). The evidence for adaptation was subsequently not confirmed, and recent data indicate that the 29-nucleotide deletion most likely represents a founder effect causing fitness loss in bat and human cells (Forni et al., 2017; Muth et al., 2018). This observation clearly indicates that a mutation sweeping at high frequency in the viral population does not necessarily represent a selectively advantageous change.
Clearly, huge gaps remain in our understanding of SARS-CoV emergence and adaptation to our species. Most likely, acquisition of inserts in the spike protein and remodeling of the RBD did not occur in a single animal-to-human jump event. The majority of relevant changes might have been present in a reservoir/intermediate species and only minor changes may then have been required to gain full transmissibility in humans.
Alternatively, the progressive optimization from multiple zoonotic events may have followed short events of human-to-human transmission over an extended period of time. This has been earlier observed during MERS transmission, entailing repeated jumps of MERS-CoV from camels (Cui et al., 2019). Accumulation of mutations in NTD and RBD during global and local transmissions further strengthens the likelihood that the progenitor virus may have been transmitted among humans over time, adapting to become more efficiently transmitted in people.
In a nutshell, the missing knowledge about the reservoir/intermediate host of SARS-CoV-2, as well as about the extent of its distribution in wild and domestic animals, raises concerns about zoonotic transmissions. If pre-adaptation routinely occurs as these viruses incubate in animal reservoirs, there is a high probability for new pandemics to recur. Thorough surveillance of animal populations and early diagnosis in people will be necessary to prevent future COVID-19 like pandemics.
4. Ecological factors
Zoonotic spillover of infectious pathogens is threatening socioeconomic development and public health worldwide (Jones et al., 2008). The emergence and rapid worldwide propagation of the SARS-CoV-2 coronavirus has led to the COVID-19 pandemic. The chain of events that facilitated the zoonotic spillover likely required the alignment of ecological, epidemiological and behavioral determinants that allowed a precursor lineage of a bat virus to trespass a series of barriers, possibly after establishing infections in intermediate hosts, before evolving to become capable of efficient human-to-human transmission. The mechanisms involved in the spillover and evolution of this coronavirus as a human pathogen remain unclear, including contrasting information regarding the immediate reservoir or other hosts, including horseshoe bats (Rhinolophus affinis) or the ant and termite-feeders, critically endangered pangolin species (Manis javanica). The latter is the most trafficked animal worldwide and illegally imported from Malaysia into China for trade in markets (Chan et al., 2020; Huang et al., 2020; Xu, 2020; Zhou et al., 2020b). Two lineages related to that of SARS-CoV-2 were identified in Malayan pangolins trafficked into China, showing that these animals can potentially be involved in the zoonotic transfer to humans (Lam et al., 2020).
Two scenarios have been proposed to explain the cross-species transfer and evolution of the new betacoronavirus to a human-to-human transmission. The precursor of the SARS-CoV-2, cycling in an animal host before the zoonotic transfer, might have evolved the bind to an ACE2 receptor similar to that of humans (Andersen et al., 2020). For this to have occurred, a high host population density would have been needed to achieve efficient natural selection for this important phenotype. The second scenario proposed by Andersen and colleagues hypothesizes that natural selection for this trait occurred in humans after the virus established itself in human beings, continuously adapting before transmission caught the attention of the medical and public health communities. In either scenario, the new coronavirus emergence required a combination of successful mutations that enabled the virus to cause infection in humans, and ecological and behavior determinants that enabled the virus to breach host species barriers and then spread in large and dense human populations before being recognized as the agent of a potential pandemic. Such an alignment of genetic, ecological, epidemiological, and behavioral factors was proposed by Geoghegan and Holmes (2017) and Plowright et al. (2017) to explain virus disease emergence at the human-animal interface.
Zoonotic transfer of pathogens to humans has been attributed to changes in natural environments for expanded global food production (Rohr et al., 2019; Wolfe et al., 2007), climate change (Zinsstag et al., 2018), habitat degradation (Allen et al., 2017), biodiversity loss (Keesing et al., 2010; Keesing et al., 2006), wildlife trade (Smith et al., 2009), and changes in the distribution and prevalence of host reservoirs and their parasites (Vanwambeke et al., 2019). In fragmented forest landscapes, generalist reservoir species are capable of adapting to new ecological conditions, becoming more abundant, expanding their distribution range, and accumulating parasites and pathogens. In this scenario, the host species richness and high population density increase the human-animal reservoir contact rate and the exposure to parasites (Borremans et al., 2019; Johnson et al., 2020). In addition, the close proximity of people to forest ecosystems, increased human population density, poor living conditions, low socioeconomic condition and human mobility facilitate human exposure to wild animals and the probability of spillover events (Geoghegan and Holmes, 2017; Wilkinson et al., 2018).
The probability of a spillover event establishing new human infection is influenced by (1) the pathogen pressure (that is the amount of pathogen available at a given time and space), (2) human and reservoir host behavior (mediating the risk of human exposure to the pathogen), and (3) factors linked to human susceptibility to infection, together with parasite dose and route of exposure (Plowright et al., 2017).
Decline in wildlife populations caused by predatory hunting activities, decreased habitat quality and habitat loss driven by extensive deforestation has been linked to increased probability of zoonotic virus spillover to humans at the human-animal interface (Geoghegan and Holmes, 2017). In these human-dominated landscapes, primates and bats are reservoir hosts of more viruses than other mammal species, increasing the potential for new infections in humans to become established (Johnson et al., 2020). The potential risk of SARS-CoV spillover from horseshoe bat population to humans was demonstrated by Menachery et al. (2015) using a reverse genetics system entailing a chimeric virus expressing the bat SHCO14 in a mouse-adapted backbone. This study illustrated scenarios for the emergence of bat SARS-CoV in humans: infection of an intermediate nonhuman host might be followed by human infection. Direct bat-human transmission could be followed by selection in the human population (distinct from closely-related viruses circulating in the source host). In a third scenario, the circulation of quasi-species pools in the animal reservoir might maintain multiple virus strains, some of which capable of causing infection in humans without the need for additional mutations. Alphacoronaviruses and betacoronavirues were identified in free-ranging bats from Myanmar, showing the potential for zoonotic virus emergence in humans in close contact with sylvatic animals in forest areas disturbed by ongoing process of changes in land use (Valitutto et al., 2020).
4.1. Climate variability anomalies
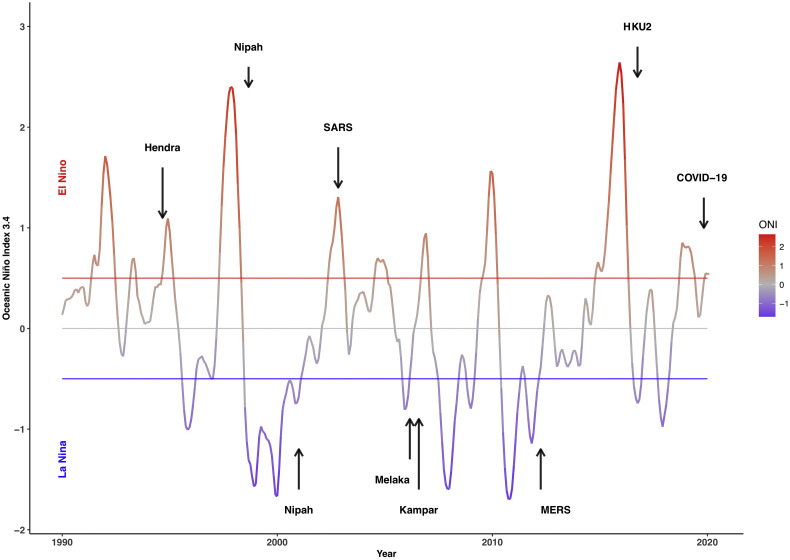
Climate variability is known to affect the outbreaks of many infectious diseases (Morand et al., 2013). Vector-borne diseases such as Murray Valley encephalitis, Rift Valley fever, Ross River virus disease and dengue, among many others, have all been linked with anomalies in climate El Niño Southern Oscillation (ENSO) (Anyamba et al., 2019; Nicholls, 1993). Two emerging viral diseases transmitted by bats have been found associated with El Niño events: Hendra virus in Australia (McFarlane et al., 2011; Giles et al., 2018) and Nipah virus in Malaysia (Daszak et al., 2013). The recent emergence of SARS-CoV-2 has also followed an important El Niño event (NOAA 2019, https://www.climate.gov/news-features/blogs/enso/august-2019-el-ni%C3%B1o-update-stick-fork-it), which has particularly affected China.
In Asia and the Pacific, eight new viruses transmitted by bats have emerged in humans and livestock since the 1990s (Table 1 ). Eight of these nine outbreaks of newly emerging bat-borne diseases appear associated with El Niño - La Niña events using values of ‘NINO 3.4’ index (Fig. 2 ) retrieved from the National Oceanic and Atmospheric Administration (NOAA, https://www.noaa.gov). Four bat-borne viruses have emerged during an El Niño phase and four during a La Niña phase according to the monthly classification of the ENSO provided by NOAA. Only Kampar virus has emerged during a neutral phase, although following closely a La Niña event.
Table 1.
Emergence of bat-borne viral diseases in Asia (Middle East, China, South Asia, Southeast Asia) and Australia in relation to El Niño Southern Oscillation (ENSO)-driven climate anomalies. Note that the existence of lag time between the index ENSO 3.4 and its effects on a country or region may vary from less than one month (Australia) to two months (Southeast Asia), three months (South Asia, Middle East) and up to four-six months (China).
| Emergence / outbreaks | Intermediate host | Date, location | ENSO | Reference |
|---|---|---|---|---|
| Hendra | Horse | Sep 1994, Australia | El Niño | (Selvey et al., 1995) |
| Nipah | Swine | Sep 1998, Malaysia | La Niña | (Lam and Chua, 2002) |
| Nipah | Unknown | Jan 2001, India | La Niña | (Chadha et al., 2006) |
| SARS | Civet cat | Nov 2002, China | El Niño | (Liang et al., 2003) |
| Melaka | Unknown | Mar 2006, Malaysia | La Niña | (Chua et al., 2007) |
| Kampar | Unknown | Aug 2006, Malaysia | Neutral Phase | (Chua et al., 2008) |
| MERS | Camel | Apr 2012, Middle East | La Niña | (Zaki et al., 2012) |
| HKU2 | Swine | Oct 2016, China | La Niña | (Gong et al., 2017) |
| COVID-19 | Unknown | Dec 2019, China | El Niño | (Zhu et al., 2020) |
Fig. 2.
Onset of bat-borne viral diseases in Asia and the Pacific since 1990 (see Table 1) in relation to El Niño Southern Oscillation (ENSO) driven climate anomalies using ‘NINO 3.4’ index retrieved from the National Oceanic and Atmospheric Administration (NOAA, https://www.noaa.gov).
This observation, although needing more in depth analysis, suggests that viral diseases transmitted by bats seem likely driven by ENSO climatic anomalies. Abnormal rainfall, temperature, and vegetation development, whether above or below normal condition during El Niño - La Niña events, are known to create appropriate ecological conditions for pathogens, their reservoirs and vectors that may enhance transmission, risk of spill-over, emergence, and propagation of disease clusters (Anyamba et al., 2019). Stresses induced by climate variability may have a profound effect on disease dynamics in wild animal populations, mostly in relation to immune or behavioral changes (Subudhi et al., 2019).
Then, climate anomalies by their effects on food shortage, behavioral mobility, and modulation of the immune system of bats are likely to increase the risks of disease emergence by putting them in contact with other animals, wild or livestock, and favoring viral spillover. A condition that has prevailed in 2019 for SARS-CoV2.
5. Lessons from molecular epidemiology
We know by now that SARS-CoV2 transmission has occurred by droplets through human to human contact similar to SARS and MERS (Han et al., 2020). Spread by aerosols is also possible but it is still under investigation. Some studies have reported the onset of gastrointestinal symptoms in patients upon admission and some reported detection of virus nucleic acid in fecal samples of patients. SARS-CoV-2 RNA was also detected in oesophagus, stomach, duodenum and rectum specimens for both two severe patients. In contrast, only duodenum was positive in one of the four non-severe patients. These findings suggest a possible oral-fecal transmission route of infection (Lin et al., 2020; Hindson, 2020).
Tracking changes in the virus has shed bright light on its global spread. Indeed, viral sequencing proved key in establishing transmission where only imported cases from travelers had been suspected. How can we be sure? We briefly overview the methods that sustain these claims and why we can be confident that they, and existing evidence, more than suffice to teach us important lessons about this virus' global spread.
Evolutionary trees (phylogenies) illustrate relationships among biological lineages. The tools that build such trees “work backwards” from existing sequence data, inferring the genealogical or phylogenetic relationships among them, and reconstructing which and when changes arose. There are a few basic methodologies with a plethora of different implementations which are beyond the scope of this review to detail (Boussau and Daubin, 2010; Holder and Lewis, 2003). Nevertheless, the currently most popular and reliable methods are based either on maximum likelihood or Bayesian inference. Some of these methods can incorporate temporal information, at the tips and/or at internal nodes which, along with some restrictions on constancy of the evolutionary rates, global or local, can be used to date particular nodes, including the root or Most Recent Common Ancestor (MRCA) of the sequences in the tree. Differently from most other phylogenetic analyses, the time of sampling of most sequences derived in the molecular epidemiology analyses of SARS-CoV-2 is known. This allows the use of time-stamped phylogenetic trees (Neher and Bedford, 2018). In these, the purely phylogenetic information is enriched with sampling time data which improves the use of genetic information from pathogens to track their spread in time and space. When the analyses include hundreds or even thousands of sequences we face a problem in analyzing the data and visualizing the results. Some convenient, easy-to-use solutions have been provided by the NextStrain Platform (https://nextstrain.org). A complete description of the tree building method used in NextStrain is available elsewhere (Hadfield et al., 2018), but most readers need only understand that millions of alternative trees are considered and only the most compatible with the data is represented. Nothing guarantees this tree to be correct in every detail because other, similar trees enjoy nearly equivalent support.
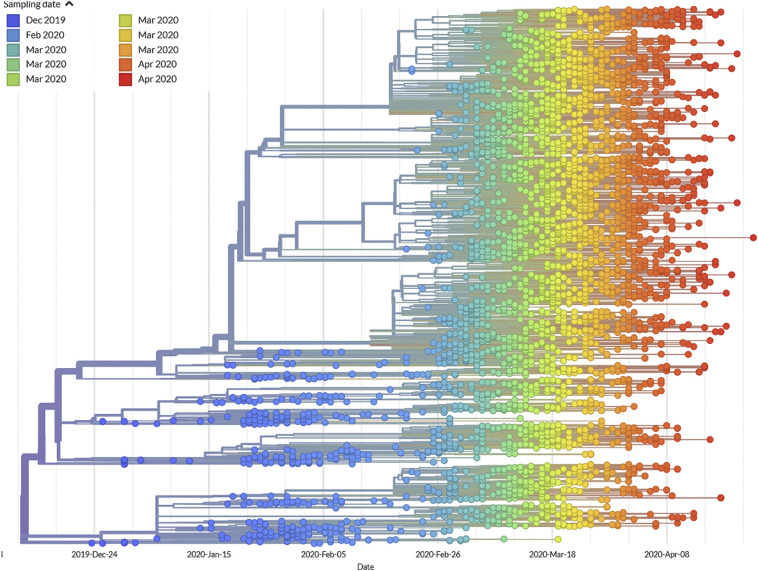
To fully appreciate how successfully the recent evolution of SARS-CoV-2 has been captured in the NextStrain platform, it is worth first reflecting on the strengths and weaknesses in the time-stamped tree. Fig. 3 describes the changes that SARS-CoV-2 viruses underwent since they became widely recognized as pathogenic in December 2019, through mid-April 2020. It depicts relationships among hundreds of viral sequences, sub-sampled from the thousands of sequences available at the time of writing, as rendered for real-time evaluation by the Nextstrain consortium (https://nextstrain.org/ncov) using data from GISAID.org.
Fig. 3.
Time-stamped maximum likelihood phylogenetic reconstruction of SARS-CoV-2 isolates deposited to GISAID.org and rendered by https://nextstrain.org/ncov. Isolates are represented by colored circles with the color code corresponding to time of sampling as detailed in the legend.
One feature establishes, without question, the usefulness of this tree: contemporaneously sampled viruses (each depicted by its own color) are perfectly chronologically organized. In this rendering, each sample (circle) was colored to indicate when it was sampled. The “rainbow” pattern means that each virus has undergone a nearly equal amount of change. Note, too, that the viruses do not “bunch up” at the tips of the tree, but are situated throughout. We need not guess when and where ancestors lived, because we can see them. Lastly, no branches protrude to any great extent, eliminating the fear of being misled by tree artifacts introduced by recombination. Although recombination contributed to the origins of this viral group (Ji et al., 2020; Boni et al., 2020), and future recombinations cannot be discounted, this does not appear to have been a major force over this five month period. By so faithfully reproducing these samples' known chronology, this tree should earn our trust.
The total number of points at any column (encompassing different time intervals) informs about the sampling success at those times. There are only a few sequences from the early weeks (before January 15) followed by a clear increase until the beginning of February (around February 5). Then, Chinese researchers (see below) almost completely ceased to deposit sequences in GISAID and only a few sequences were being obtained from other countries. This period corresponds to the silent spread of COVID-19 throughout most countries. Asymptomatic infections were not detected, hence no virus samples were available for sequencing and we lack detailed information on how the virus was spreading. This period also corresponds to a bout of diversification that can be observed in the upper part of the tree, with many new sublineages stemming from pre-existing lineages. The times of these diversification events are inferred from the application of a constant rate of evolution of 8 × 10−4 substitutions per site per year. The constancy of this evolutionary rate is difficult to be evaluated at this stage in the evolutionary history of the virus, given the very short time elapsed since its appearance. By the end of February, the situation changed drastically, as many more sequences became available for analysis when the pandemic exploded and cases accumulated rapidly in many countries.
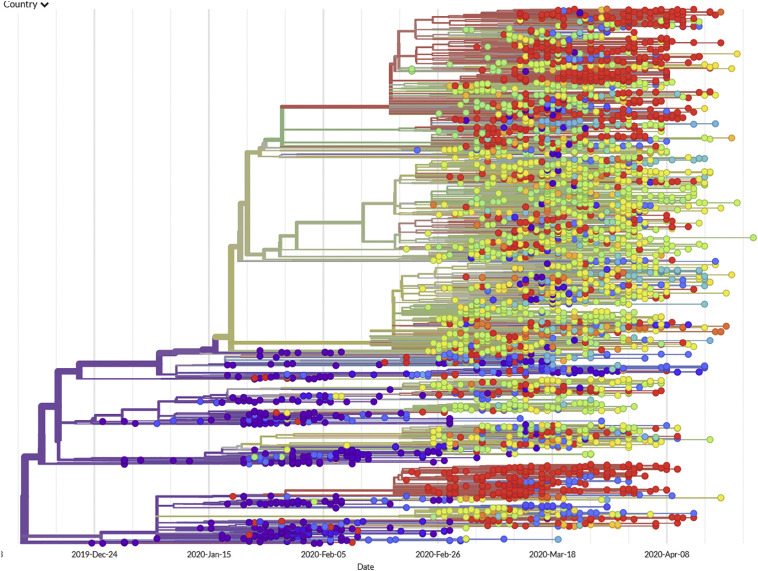
See below the same tree (Fig. 4 ), now colored to show each sample's geographical origins (as shown on the accompanying map). Note that isolates from China dominate the oldest branches of the tree. The first isolates sequenced came from Wuhan, China (where the epidemic was first recognized); therefore, it is logical that they should appear first.
Fig. 4.
The same reconstruction of SARS-CoV-2 phylogeny, now denoted by geography. Isolates originated and initially diversified in China (purple), followed by multiple and independent introductions to Oceania (blue), Europe (green and yellow), and North America (red). Less information is known about Africa, India, South America, and other populations of major concern in the Global South. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
What other epidemiological inferences can we draw from tracing the path of this virus' evolution?
Human infections may have commenced months before the recognized outbreak in Wuhan. Most human pathogens originate as pathogens of other animals; they occupy a spectrum, emerging from those exclusively transmitted among animals to those primarily transmitted among other animals, to those that have lost their dependence on non-human reservoirs and, ultimately, those no longer capable of being transmitted excepting from person to person (Wolfe et al., 2007). The human drama now unfolding began, in earnest, in the latter months of 2019; the tree above records the virus's subsequent proliferation and spread. But, what can be said about preceding events?
Serological evidence suggests that people living the vicinity of bats harboring SARS-like viruses are frequently exposed to such infections (Wang et al., 2018). More surveillance will be needed to truly understand how often, and under what circumstances, such exposures take place. Nor can we yet appreciate what proportion of such exposures eventually lead to a virus becoming established as a human pathogen.
Acknowledging this broader context, what do the phylogenetic data say about the proximate origins of SARS-CoV-2? More surveillance among the viruses of suspected animal reservoirs (bats, most especially) will be needed to confidently estimate when and where this virus first took a firm foothold in people. Early reports (Lam et al., 2020; Zhou et al., 2020a; Zhou et al., 2020b) identified BatCoV RaTG13 and RmYN02 as the closest isolates to SARS-CoV-2 and it has been proposed that these lineages diverged between 40 and 70 years ago (Boni et al., 2020). Assuming constancy in the evolutionary rate, which is also problematic as discussed above, a few months may have elapsed between human acquisition of the virus and its recognition by the medical and public health communities in December 2019. Molecular clock analyses suggest a time for the most recent common ancestor of all current SARS-CoV-2 lineages in October–December 2019 (van Dorp et al., 2020) If further sampling identifies more genetically proximate enzootic viruses, then the estimated times for separation and diversification may likewise change. Lacking such additional sampling, it seems fair to suspect from the available evidence that an exclusively human chain of transmission may have gone undetected for some months before galvanizing public attention.
Some lineages took root outside of China. Note that clusters of viruses define foci of transmission. Some of these represent expansions outside of China. For example, a group almost exclusively composed of red dots (near the bottom of the figure) indicate a viral lineage that took root in the United States. These mostly came from the state of Washington (although Wyoming, Virginia, and other states are included in the group). Within this cluster, a few dots different to red indicate likely exports from the USA to other countries, such as Australia, Taiwan, Iceland and India. Nevertheless, inferences about epidemiological processes drawn exclusively from genome information must be taken very cautiously (Villabona-Arenas et al., 2020).
Long-distance travel spread closely related viral isolates. Although most of the isolates in the group discussed above were sampled from patients in the American Pacific Northwest, a few blue dots in that group denote Australian isolates, demonstrating air travel as a powerful disseminating force. Specific introductions [i.e., from China to Germany, from Germany to Italy, from Italy to Iceland (Gudbjartsson et al., 2020)] that accord with known travel histories are recapitulated in phylogenetic networks reconstructed from viral RNA sequences. For example, the phylogeny accords with other evidence that the virus was introduced to Europe by early January (Olsen et al., 2020; Spiteri et al., 2020; Stoecklin et al., 2020).
Community transmission occurred weeks before local outbreaks were recognized. Early efforts in the United States to manage the virus focused on limiting travel from suspected endemic regions, and early testing was limited to travelers returned from such places. Meanwhile, the virus had already established footholds, as discovered when nasal swabs collected for monitoring community transmission of influenza were discovered to harbor the RNA of SARS-CoV-2. Nearly identical viruses were found in residents of Seattle who had no direct contact with one another. This sounded the alarm that the virus was actively circulating in the community, a fact that would soon be tragically borne out by a critical surge in illness and death (Bedford et al., 2020; Fauver et al., 2020).
Multiple, independent introductions of the virus. A parallel process was soon to play out in New York. Based on the viral phylogeny, it appears the virus was established in New York at least seven times, primarily from Europe, while travel restrictions focused on perceived threats from China, South Korea, Italy and Iran (Gonzalez-Reiche et al., 2020). The occurrence of red dots in nearly every clade of the tree (even clades composed mostly of European isolates) underscores this point. This is equally true for Europe, because nearly every clade of the tree includes green dots (denoting samples of European origin). This tree includes relatively few isolates from Australia (blue dots), but the same phenomenon is evident: Australia received virtually every basic type of SARS-CoV-2.
The Global South has not been sampled nearly enough. A complete description of the virus evolution would necessitate vast and representative sampling. With diagnosed cases (as of mid-April) numbering in the millions, and in the face of untold numbers of undiagnosed cases, how much confidence should we place in trees reconstructed from only hundreds or thousands of isolates? Biologists have means to assess internal consistency, enabling many facets of the recorded history to be viewed by now as impervious to egregious sampling error. Nonetheless, it is clear that the available evidence neglects to inform us much about the epidemic. This is for lack of attention, not for lack of virus. Even in the most affluent of societies, the virus is exploiting preexisting social inequities (Yancy, 2020). As the virus takes root in Africa, South America, and other under-resourced regions, evolutionary epidemiologists should assume responsibility for understanding and communicating all that can be learned about how, where, when, and why the pandemic is unfolding there. The toll of this disease will be multiplied where patients cannot access therapies and where clinicians cannot protect themselves. There, the behavior of the virus may or may not parallel patterns so far described. We will not know if we do not look.
6. Diagnostic testing for SARS–CoV-2/COVID-19
SARS-CoV-2 was first detected in human bronchoalveolar lavage (BAL) specimens by unbiased Illumina and nanopore sequencing technologies, and a real-time RT-PCR assay using pan-betacoronavirus degenerate primers targeting a highly conserved RdRp region (Zhu et al., 2020). The SARS-CoV-2 isolation was performed in different cell lines, including human airway epithelial cells, Vero E6, and Huh-7. Cytopathic effects were observed in human airway epithelial cells 4 days after inoculation, but not in Vero E6 and Huh-7 (Zhu et al., 2020). For safety, CDC recommends that clinical virology laboratories should not attempt viral isolation from clinical specimens collected from COVID-19 patients under investigation (PUIs).
Because SARS-CoV-2 is a newly discovered virus and its genome is very divergent from those of HCoV-229E, -NL63, -OC43, and -HKU1, several commercially available multiplex NAAT tests (BioFire FilmArray Respiratory Panel, ePlex Respiratory Pathogen Panel, NxTAG Respiratory Pathogen Panel, RespiFinderSmart22kit, and others.) for the detection of respiratory organisms in clinical virology laboratories were predicted no cross-reactivity with SARS-CoV-2 (Phan, 2020). At that time, several different protocols for laboratory-developed tests (LDTs) were developed, and they were available on the WHO website (https://www.who.int/who-documents-detail/molecular-assays-to-diagnose-covid-19-summary-table-of-available-protocols). While the CDC assay was designed for specific detection of three different regions of the N gene, the assay developed by Corman et al. (2020) aimed to amplify three different regions of the RdRp, N and E genes.
Since SARS-CoV-2 continues to spread around the world, and the number of potential cases increases rapidly, faster and more-accessible testing is extremely needed. Thus, many companies are racing against the clock to develop commercial test kits to detect SARS-CoV-2 more quickly and accurately. More than fifty RT-PCR-based diagnostic tests are currently available in the US that have been granted the FDA's Emergency Use Authorization (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd). These tests can qualitatively identify SARS-CoV-2's RNA in the lower respiratory tract (bronchoalveolar lavage, sputum, and tracheal aspirate), and upper respiratory tract (nasopharyngeal and oropharyngeal swabs). Recently, the FDA approved emergency use for a portable, fast, swab test for SARS-CoV-2 which can provide results in less than 15 min. The IDNOW COVID-19 (Abbott, Illinois) can be used at a point-of-care in doctor's offices, urgent care and hospitals. Lateral flow immunoassays are another rapid, point-of-care diagnostic test, which has been widely used. The Sofia 2 SARS Antigen FIA is the first COVID-19 antigen test to be granted the FDA's Emergency Use Authorization. This lateral flow immunofluorescent sandwich assay is used with the Sofia 2 instrument intended for the qualitative detection of the nucleocapsid protein antigen from SARS-CoV-2 in nasopharyngeal and nasal swabs.
Another diagnostic approach would be to devise blood tests for antibodies against SARS-CoV-2. Many companies around the world have raced to develop antibody tests. The qSARS-CoV-2 IgG/IgM Rapid Test (Cellex Inc.) was the first antibody test being approved by the FDA. This test is intended to qualitatively detect IgG and IgM antibodies against the SARS-CoV-2 in human serum, plasma, and whole blood. Together with increased availability of commercial RT-PCR-based diagnostic tests, important questions about immunity to the novel coronavirus will be answered soon. The use of serological tests is complementary to those based on direct detection of viral RNA because they indicate that the individual has developed specific immunological response to the virus, which usually takes a few days after infection; hence, a negative result does not necessarily mean that a person is not infected - this might have occurred too recently to have developed the immune response, and a positive results does not necessarily mean that the person has an active infection. Consequently, these tests are essential to obtain a more precise estimate of the total number of infections in a population and in which proportion they are expected to be immune to a future infection by the virus. Its main use is in epidemiological surveillance rather than in clinical diagnostics. Early application of community surveys for seroprevalence have suggested far more widespread community exposure than has been supposed based merely on the reporting of diagnosed cases, suggesting for example that by mid-April as many as one in five residents of New York City may have been exposed, with diminishing exposure rates less proximate to the outbreak's epicenter.
Recently, clustered regularly interspaced short palindromic repeats (CRISPR) based nucleic acid detection technology has emerged as a powerful tool with the advantages of rapidity, simplicity and a low cost (Wang et al., 2020). A recent study reported that the CRISPR-based assay was used to detect SARS-CoV-2 in the patient's pharyngeal swab in China (Ai et al., 2020). The turn-around time (TAT) of CRISPR was 2 h, much faster than RT–PCR (3 h) and mNGS (24 h) (Ai et al., 2020). The FDA authorized a COVID-19 test that uses the gene-editing technology CRISPR. Sherlock CRISPR SARS-CoV-2 Kit has been developed by Sherlock Biosciences Inc.
7. Human susceptibility to SARS-CoV-2
Knowledge of genetic variation, at both individual and population levels, may further our understanding of disease transmission and pathogenesis, enabling identification of individuals at high risk of infection and sequelae. More broadly, this may inform drug design and vaccine development. However, previous coronavirus epidemics of recent years, namely SARS-CoV in 2003 and MERS-CoV in 2012, did not lend themselves to such evaluation owing to their rapid progression, acute onset, and relatively small reach. The current SARS-CoV-2 pandemic appears unfettered by any population-level protective immunity to cross-reactive epitopes. The much greater number of individuals affected, together with the availability of high throughput technologies, will stimulate rapid research on variation in host susceptibility. Indeed, the number of preprints appearing online, prior to peer review, is unprecedented. Other respiratory viruses where host susceptibility has received attention include Respiratory Syncytial Virus (RSV) (Tahamtan et al., 2019) and Influenza A (Nogales and DeDiego, 2019; Clohisey and Baillie, 2019). However, diseases caused by these viruses are sufficiently different that inferences concerning the role of genetics with respect to coronavirus infection cannot be directly applied.
7.1. Genetic control of susceptibility and approaches to study
Although host genetics is likely to be important for the outcome of infection, because of the nature of the disease, we only have preliminary information on the heritability of susceptibility to coronavirus infections, i.e. estimates of the proportion of phenotypic variability due to genetic factors. Heritability, a time and place-specific measurement for a population, provides an indication of likely success in the hunt for susceptibility loci, and low values for heritability of <20% would indicate that very large sample sizes might be needed to detect genetic effects. Williams et al. (2020) have reported high heritability, in the range 40–50%, of COVID-19 symptoms in a classical twin study (Williams et al., 2020). In addition, the apparent differences in susceptibility seen between ethnic groups could be due, in part, to human genetic variability. Niedzwiedz et al. (2020) report a higher risk of confirmed SARS-CoV-2 infection, not accounted for by variables such as socioeconomic differences, in some minority ethnic groups as included in the UK Biobank (Niedzwiedz et al., 2020). The genetic contribution to the outcome of infection will be complex, since some of the underlying health conditions associated with death from coronavirus infection are known to have a significant genetic contribution.
The difficulties of family-based studies, whilst controlling for exposure, leaves us with case control association methodology. Ultimately, the high numbers of affected people will facilitate genome wide association studies (GWAS), or even studies of whole genome or exome sequence, particularly where genotypic information is already available, as is the case for UK Biobank participants. In the short term, we are likely to see a number of candidate gene studies. For susceptibility to coronavirus infection per se, loci coding for viral receptors provide obvious candidate genes. Nevertheless, the major question is why some individuals develop life-threatening immune-mediated pathologies, where knowledge of genetic variation may contribute to our understanding of key mediators of the immune response.
Most relevant, albeit limited, information on the allelic diversity contributing to susceptibility/resistance to date has used sampling from the SARS CoV 2003 epidemic, and a little information has been gleaned from the MERS-CoV 2012 epidemic. Sample sizes are small, and results sometimes conflicting, hence no firm conclusions can be drawn. There are several studies identifying genes of interest from mouse models (Kane and Golovkina, 2019) and some information from interspecies comparisons (Hou et al., 2010; Sironi et al., 2015). To date, candidate genes for published human studies have included pathogen receptor genes and loci controlling innate and adaptive immunity.
7.2. Receptors
S1 spike proteins of SARS-CoV and SARS-CoV-2 bind to angiotensin converting enzyme 2 (ACE2) on cell membranes, catalysing the cleavage of the vasoconstrictor angiotensin II and countering the activity of ACE. MERS-CoV binds to the dipeptidyl peptidase 4 (DPP4, CD26) an immunoregulatory serine exopeptidase. Particularly considering indications of relevance from cross-species comparisons (Sironi et al., 2015), there has been a surprising lack of interest in these 2 candidate loci for susceptibility to infection per se. A small, low-powered, case control study, with information on anti-SARS-CoV antibody status, did not show any associations between SARS phenotypes and ACE2 polymorphisms in a Vietnamese population (Itoyama et al., 2005). Genes coding for functionally associated molecules such as transmembrane serine protease 2 (TMPRSS2), which cleaves and activates viral spike glycoproteins, are also worthy of study (Iwata-Yoshikawa et al., 2019). Lopera et al. (2020) have just reported a PheWAS of 178 quantitative phenotypes, including cytokine and cardio-metabolic markers but not specific SARS-CoV-2 infection markers, in relation to ACE2 and TMPRSS2 variation (Lopera et al., 2020).
7.3. MHC
Amongst immune response related loci, MHC class I and class II allelic associations are to be expected, particularly through MHC class I restriction of CD8+ T cells (Lin et al., 2003; Ng et al., 2004; Wang et al., 2011; Keicho et al., 2009). MHC associations are relevant for susceptibility to disease per se, disease pathogenesis and response to vaccination. There are a few follow up studies e.g. of HLA A*0201 restricted SARS-CoV epitopes for CD8+ T cells (Zhao et al., 2011), but investigation of such highly polymorphic loci needs to be more comprehensive.
7.4. Other loci
Other loci, with some information on allelic associations, include those coding for ICAM3 (DC-SIGN, CD209) (Chan et al., 2007; Chan et al., 2010) and DC-SIGNR (CD209L) (Li et al., 2008) and widely studied loci such as MBL (Zhang et al., 2005; Ip et al., 2005) and CD14. In particular, the inflammatory response associated with SARS suggests a number of candidate loci e.g. AHSG (Zhu et al., 2011), IFNG (Chong et al., 2006), CD14 (Yuan et al., 2007) and CCL5 (Rantes) (Ng et al., 2007). Nevertheless, some relatively small studies have resulted in some conflicting findings being noted e.g. for MBL (Yuan et al., 2005) and DC-SIGNR (Li et al., 2008).
7.5. And from mice
More recently, loci of interest have been identified using mouse models, after infection with SARS-CoV, where pathology can be well studied. These include Trim55 and Ticam2 (Kane and Golovkina, 2019). Trim55 codes for an E3 ubiquitin ligase present in smooth muscle around blood vessels, affecting lung pathology by controlling airways and immune cell infiltration. Deficiency was relevant to lung injury although susceptibility alleles were not reported (Gralinski et al., 2015). Ticam2 knockout mice were highly susceptible to disease with some evidence of allelic heterogeneity. Ticam2 is an adaptor for MyD88-independent TLR4 signaling contributing to innate immunity (Gralinski et al., 2017). These genes require complementary studies in human populations.
7.6. Choice of phenotypes and genotypes
To date, phenotypes employed for human genetics have been limited: susceptibility to infection per se, some measures of morbidity and mortality. For both candidate gene studies and GWAS, finding totally ‘resistant’ individuals, for an unaffected control group, is hard since for COVID19, despite the occurrence of asymptomatic individuals, many show very mild symptoms of disease. There is scope for development of phenotypes, particularly relating to the most pertinent issues of pathogenesis and control of immune responsiveness. Perhaps immediate research could focus on critical and potentially useful phenotypes e.g. severity of disease sufficient to require a ventilator, or antibody responses.
Identification of a susceptibility gene with no a priori hypothesis is difficult for an active acute viral infection, but candidate genes, potentially controlling these phenotypes, would include those from both innate and adaptive immunity. Published GWAS, which might be of interest for suggesting susceptibility loci, tend to have used phenotypes from chronic rather than acute conditions e.g. COPD. Whether conducting genotype/phenotype studies or simply measuring immune responses, age and sex are clearly major factors in the outcome of infection with SARS-CoV-2, and will require careful consideration in all analyses.
7.7. Immunity and ageing
Immunity and pathology will be influenced by many factors including genetics, age, sex, viral dose and underlying health problems. The interactions between these factors will also be important (Gubbels Bupp et al., 2018). COVID19 particularly affects the elderly, with age a key consideration in any study design, as are sex differences, which are known to be important for respiratory infections in general (Kadel and Kovats, 2018). Although both innate and adaptive immunity are required to mount an effective immune response, and despite the tools for investigation of the relevant loci being available, the lack of candidate gene studies for coronavirus infection even prior to COVID19, including information on MHC associations, has been noted. Not only will variability in amino acid sequence be important, such as that influencing MHC restriction, but also the levels of immune mediators, possibly driven by promoter variation. Timing of responses is another area for exploration. Rockx et al. (2009) discuss molecular mechanisms of age related susceptibility in coronavirus infected mice, and development of pathology via the host response. Aged mice showed a greater number of differentially expressed genes when infected than young mice, including elevated interferon and cytokine genes, indicating a different host response kinetics in old versus young mice.
A number of immune response molecules/genes are of interest, including Toll-like receptors (TLRs), proinflammatory cytokines and the appropriate signalling pathways. Both adaptive and innate immunity are recognized to decline with age (Goronzy and Weyand, 2013; Nikolich-Žugich, 2018), and although, in the past, adaptive immunity has been the focus of attention, there is now cumulative evidence for a decline in innate immunity with age. Changes occur in neutrophil, macrophage, dendritic cell and NK populations, and there are changes in TLR expression, with an overall distinct but not uniformly lower expression in later life, particularly in macrophages, and consequent major changes in TLR mediated responses (Solana et al., 2012). The innate immune system of the frail elderly is often in a state of heightened inflammation, but this higher basal production of proinflammatory cytokines does not necessarily translate to a generally elevated and/or effective response (Fulop et al., 2018). In particular, the incidence of and mortality from lung infections increase sharply with age, with such infections often leading to worse outcomes, prolonged hospital stays and life-threatening complications, such as sepsis or acute respiratory distress syndrome. The review by Boe et al. (2017) covers research on bacterial pneumonias and pulmonary viral infections and discusses age-related changes in innate immunity that contribute to the higher rate of these infections in older populations.
Regulation of adaptive immune function appears to be diminished in old versus young adults, and decreased TLR responses are associated with the inability to mount protective antibody responses to flu vaccine (Kollmann et al., 2012). In addition, the pre-existing B cell repertoire restricts the quantitative response in the elderly (Goronzy and Weyand, 2013). B cell maintenance and function in aging has been well reviewed and is of particular relevance when considering responsiveness to vaccines evoking primary versus memory humoral responses (Kogut et al., 2012). The age-related rise in proinflammatory cytokines is also associated with reduced response to vaccination (Oh et al., 2019). It is known that vaccination with TLR5 targeting adjuvants in elderly patients enhances flu vaccine responsiveness without increasing inflammation (Goronzy and Weyand, 2013).
We know multiple host factors, including age and sex, are important for disease outcome of coronavirus infection and a systems approach has been suggested for the study of disease pathogenesis (Schäfer et al., 2014). However, to date, we do not know the relative importance of factors such as host and pathogen genetic variability. Whilst we can draw some comparisons with other similar acute respiratory infections such as influenza, SARS-CoV-2 presents unique and urgent challenges. There is an unprecedented opportunity to obtain large sample sizes and the need to understand the role of human genetics in the outcome of infection with SARS-CoV-2 is well recognized (Kaiser, 2020). Information on infection with SARS-CoV-2 is being added to health data e.g. inclusion in the UK Biobank with its 500,000 volunteers (www.ukbiobank.ac.uk ). The scale of the current pandemic is stimulating free access to human genetic studies as they progress and data sharing for combined analyses, with multiple, diverse partners from academia and industry, e.g. The COVID19 Host Genetics Initiative (www.covid19hg.org ). Hopefully, useful information will be produced at a rate not previously seen for a transmissible disease.
8. Host response and considerations for vaccine development
There is limited knowledge on the mechanisms driving immune response against COVID-19. A recent work (Thevarajan et al., 2020) showed that a strong adaptive response across B and T cells was mounted during symptomatic phase in a 47-year-old woman from Wuhan, Hubei province in China, that successfully cleared the virus within two weeks. Increased antibody-secreting cells (ASCs), follicular helper T cells (TFH cells), activated CD4+ T cells and CD8+ T cells and immunoglobulin M (IgM) and IgG antibodies that bound the COVID-19-causing coronavirus SARS-CoV-2 were detected in blood before symptomatic recovery and with a peak around day 7–9 since onset of symptoms. These immunological changes persisted after full resolution of symptoms at day 20. These findings, based on a single subject, support the hypothesis that a rapid multi-factorial immune response against COVID-19 can be mounted within a week and with evidence of recruitment of T and B cells as well as macrophages before resolution of symptoms, and that this rapid response may correlate with positive clinical outcome.
Severe cases, on the other hand, seem to have disrupted immune responses. Several early case reports have demonstrated the presence of cytokines release storms, which demonstrate the presence of severe inflammatory responses in these patients characterised by increased interleukin (IL)-2, IL-7, granulocyte-colony stimulating factor, interferon-γ inducible protein 10, monocyte chemoattractant protein 1, macrophage inflammatory protein 1-α, and tumor necrosis factor-α.6 (Mehta et al., 2020).
Immunogenomics analysis of Bronchoalveolar Lavage Fluid (BALF) and also from blood samples across two different studies from China (Wen et al., 2020; Liao et al., 2020) also revealed clear immunological features characterizing severe cases. The overall message based on these early studies point towards the polarization of a macrophage response in the lungs associated with severe disease. Conversely, patients that recovered had increased cytotoxic T cell response compared to severe cases. Specifically, analysis of bronchoalveolar lavage fluid (BALF) from 3 severe and 3 mild COVID-19 patients using single-cell RNA sequence (scRNA-seq) combined with T cell receptor sequences revealed a distinct population of monocyte-derived FCN1+ macrophages in BALF samples from patients with severe disease, while patients with mild disease presented increased levels of FABP4+ alveolar macrophages (Liao et al., 2020). These cells were found highly inflammatory and strongly associated with cytokine storms. Notably, immunogenomics analysis on blood samples from 5 patients that had early recovery versus 5 patients that recovered late (Wen et al., 2020) confirmed the findings in BALF, namely that mild disease was associated to clonally expanded cytotoxic CD8+ T cells, which suggest that a specific T cell response may be at play in controlling COVID-19 infection in the lungs.
Several vaccine trials are underway, carrying great promise for sustainable solutions to future epidemics (https://www.who.int/blueprint/priority-diseases/key-action/list-of-candidate-vaccines-developed-against-sars.pdf) with so far 33 vaccines listed (accessed on April 25th, 2020).
From a vaccine point of view, it remains to be seen whether a strong humoral response mediated by neutralizing antibodies can be successfully mounted without side effects, and whether T cell responses could be also generated by a vaccine. Based on current knowledge it appears that severe cases are associated with a sustained, virus-specific immune response, with evidence of a hyper-inflammatory response causing severe lung damage. Notably, recent work revealed that SARS-Covid-19 specific CD4+ and CD8+ T cell responses are identified in 70% and 100% of convalescent patients, respectively, thus supporting the notion that viral specific adaptive response can be mounted and maintained (Griffoni et al., 2020). The fact that all arms of the immune responses are activated in patients that clear the virus, while in severe disease cytokine storms occur along with lymphopenia, suggests that a broad T and B cell-based vaccine should be considered (Shi et al., 2020b).
Data on SARS-CoV-2 so far revealed that several epitopes can be targeted by neutralizing antibodies response. Previous work on a SARS-CoV/macaque models investigating host response in the productively infected lungs revealed unexpected interactions between the presence of anti–spike IgG (S-IgG) prior to viral clearance and the Alveolar macrophages (Liu et al., 2019). The latter population was found to undergo functional polarization in acutely infected macaques, demonstrating simultaneously both proinflammatory and wound-healing characteristics. However, the presence of S-IgG prior to viral clearance, abrogated wound-healing responses and promoted MCP1 and IL-8 production and proinflammatory monocyte/macrophage recruitment and accumulation. The authors commented that patients who died of SARS displayed similarly accumulated pulmonary proinflammatory, absence of wound-healing macrophages, and faster neutralizing antibody responses, whereas blockade of FcγR reduced such effects. Therefore, vaccine development should consider possible host responses that exacerbate disease, and alternative therapies should be also considered in combination with vaccines to avoid virus-mediated lung injury.
9. Perspectives and conclusions
The current COVID-19 pandemic has impacted on many aspects of social, economic, personal, and behavioral matters of our daily life. We still do not know when we will completely recover from these effects and when life will return to be “normal”. It may never be the same as it used to be or, with luck, an effective vaccine becomes widely available and the return takes only a few months. But the pandemic has also exposed scientists and the processes of scientific advancement, by focusing on what the experts had to say about the new virus, how to control its spread, how to treat those infected, or what to expect if different social measures should be taken. It has shown that science advances through uncertainties and that different opinions and interpretations are frequent, especially when moving through unexplored fields. And a new pathogen is almost inevitably one such field and when it has such a dramatic impact on the health of so many people, the need for urgent answers puts an extraordinary, additional pressure on the daily work of scientists.
In little more than four months, we have learned a lot about SARS-CoV-2 and COVID-19. We know that it is a new coronavirus, closely related to those usually found in bats and, quite oddly, in an endangered species, Malayan pangolins. But we do not know how the jump from a yet unknown intermediate species to humans occurred and how the virus was capable of being so easily and efficiently transmitted among individuals of our species. From what we have learned from other zoonotic spillovers, it seems clear that several factors have concurred in this jump, including ecological, cultural, and possible behavioral. We know that recombination is frequent in coronaviruses, including sarbecoviruses, but we do not know whether this process played a significant role in the emergence of SARS-CoV-2 as a new pathogen. All the evidence gathered so far undermines the possibility that the virus was created in, or escaped from, a laboratory proximate to the city where the initial infections were detected.
SARS-CoV-2 is highly transmissible, even by asymptomatic and presymptomatic infected persons, which makes its spread very difficult to control. This can be observed in its fast expansion in just a few weeks to almost every country in the world and the poor efficiency of border closures, as implemented by most governments, to stop it. When these have been put in action the virus was already circulating within borders. Genomic epidemiology of the virus is allowing a “live” monitoring of the viral spread and has revealed differentiation in several lineages, some clearly resulting from ongoing local circulation, which also show the frequent exchange across borders even from distant countries and continents. Globalization shows its dark side in this fast spreading pandemic.
There are substantial differences in the natural history of infection. These were observed from the very start of the epidemic, with a much more severe presentation in aged persons and those with additional pathologies. Those differences have persisted but, as the number of infected persons has grown continuously, the cases of serious, even fatal, infections in younger persons have become more frequent. Nevertheless, there are still many unknowns around the differences in susceptibility, progression, and outcome of the infection by SARS-CoV-2 that, as in other infectious diseases, probably depend on a complex interaction of host, pathogen, and environmental factors. Several large-scale analyses encompassing all three kinds of factors are underway and hopefully will shed light on this critical point.
Currently, there are many additional unknowns highly relevant for the possibility of recovering a more or less “normal” lifestyle. There is no specific and highly efficient treatment yet, there are several vaccine candidates in the making but we are still at least months away from their availability for the general population. We do not even know whether those infected by SARS-CoV-2 will develop a strong immune response or a weak one and how long it will last nor whether the virus will have a seasonal behavior or will circulate all year round. In the meantime, the most important measures to prevent a generalized spread of the virus and a collapse of health systems, the main reason behind the social and economic disruption caused by COVD-19, are still social distancing, contact tracing and testing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Michel Tibayrenc (editor-in-chief Infection, Genetics and Evolution) for launching the project of this article, and for inspiring its conception. . Tung Phan acknowledges support from the Division of Clinical Microbiology, University of Pittsburgh Medical Center, USA. Manuela Sironi was supported by the Italian Ministry of Health (“Ricerca Corrente 2019-2020”). Fernando González-Candelas was supported by project BFU2017-89594R from MICIN (Spanish Government).
References
- Ai Jing-Wen, Zhang Yi, Zhang Hao-Cheng, Xu Teng, Zhang Wen-Hong. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microb. Infect. 2020;9(1):597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen Toph, Murray Kris A., Zambrana-Torrelio Carlos, Morse Stephen S., Rondinini Carlo, Di Marco Moreno, Breit Nathan, Olival Kevin J., Daszak Peter. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017;8(1):1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen Kristian G., Andrew Rambaut, Ian Lipkin W., Holmes Edward C., Garry Robert F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyamba Assaf, Chretien Jean-Paul, Britch Seth C., Soebiyanto Radina P., Small Jennifer L., Jepsen Rikke, Brett M. Forshey, et al. Global disease outbreaks associated with the 2015–2016 El Niño event. Sci. Rep. 2019;9:1930. doi: 10.1038/s41598-018-38034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford Trevor, Greninger Alexander L., Roychoudhury Pavitra, Starita Lea M., Famulare Michael, Huang Meei-Li, Nalla Arun. Cryptic transmission of SARS-CoV-2 in Washington State. medRxiv. 2020 doi: 10.1101/2020.04.02.20051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe D.M., Boule L.A., Kovacs E.J. Innate immune responses in the ageing lung. Clin. Exp. Immunol. 2017;187:16–25. doi: 10.1111/cei.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni Maciej F., Lemey Philippe, Jiang Xiaowei, Lam Tommy Tsan-Yuk, Perry Blair, Castoe Todd, Rambaut Andrew, Robertson David L. Evolutionary origins of the SARS-CoV-2 Sarbecovirus lineage responsible for the COVID-19 pandemic. bioRxiv. 2020 doi: 10.1101/2020.03.30.015008. [DOI] [PubMed] [Google Scholar]
- Borremans Benny, Faust Christina, Manlove Kezia R., Sokolow Susanne H., Lloyd-Smith James O. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019;374(1782) doi: 10.1098/rstb.2018.0344. 20180344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau Bastien, Daubin Vincent. Genomes as documents of evolutionary history. Trends Ecol. Evol. 2010;25(4):224–232. doi: 10.1016/j.tree.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Cagliani Rachele, Forni Diego, Clerici Mario, Sironi Manuela. Computational inference of selection underlying the evolution of the novel coronavirus, SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00411-20. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha Mandeep S., Comer James A., Lowe Luis, Rota Paul A., Rollin Pierre E., Bellini William J., Ksiazek Thomas G., Mishra Akhilesh. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 2006;12(2):235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Kelvin Y.K., Johannes C.Y., Ching M.S. Xu, Cheung Annie N.Y., Yip Shea-ping, Yam Loretta Y.C., Lai Sik-to. Association ofICAM3Genetic variant with severe acute respiratory syndrome. J. Infect. Dis. 2007;196:271–280. doi: 10.1086/518892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.Y.K., Xu M.S., Ching J.C.Y., Chan V.S., Ip Y.C., Yam L., Chu C.M. Association of a single nucleotide polymorphism in the CD209 (DC-SIGN) promoter with SARS severity. Hong Kong Medical Journal = Xianggang Yi Xue Za Zhi / Hong Kong Academy of Medicine. 2010;16(5 Suppl 4):37–42. [PubMed] [Google Scholar]
- Chan Jasper Fuk-Woo, Yuan Shuofeng, Kok Kin-Hang, Kelvin Kai-Wang To, Chu Hin, Yang Jin, Xing Fanfan. A Familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Chong Wai Po, Eddie Ip W.K., Tso Gloria Hoi Wan, Ng Man Wai, Wong Wilfred Hing Sang, Law Helen Ka Wai, Raymond W. H. Yung, et al. The Interferon gamma gene polymorphism 874 A/T is associated with severe acute respiratory syndrome. BMC Infect. Dis. 2006;6:82. doi: 10.1186/1471-2334-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Crameri G., Hyatt A., Yu M., Tompang M.R., Rosli J., McEachern J. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. 2007;104:11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Kaw Bing, Voon Kenny, Crameri Gary, Tan Hui Siu, Rosli Juliana, McEachern Jennifer A., Suluraju Sivagami, Yu Meng, Wang Lin-Fa. Identification and characterization of a new orthoreovirus from patients with acute respiratory infections. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clohisey Sara, Baillie John Kenneth. Host susceptibility to severe influenza a virus infection. Critical Care / the Society of Critical Care Medicine. 2019;23(1):303. doi: 10.1186/s13054-019-2566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the ICTV The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:236–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten Matthew, Watson Simon J., Zumla Alimuddin I., Makhdoom Hatem Q., Palser Anne L., Ong Swee Hoe, Al Rabeeah Abdullah A. Spread, circulation, and evolution of the middle east respiratory syndrome coronavirus. mBio. 2014;5(1):1062. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Jie, Li Fang, Shi Zheng-Li. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., Zambrana-Torrelio C., Bogich T.L., Fernandez M., Epstein J.H., Murray K.A., Hamilton H. Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. Proc. Nat. Acad. Sci. USA. 2013;110(Supp. 1):3681–3688. doi: 10.1073/pnas.1201243109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquerroy S., Vigouroux A., Rottier P.J.M., Rey F.A., Bosch B.J. Post-fusion hairpin conformation of the Sars coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver Joseph R., Petrone Mary E., Hodcroft Emma B., Shioda Kayoko, Ehrlich Hanna Y., Watts Alexander G., Chantal B. F. Vogels, et al. Coast-to-coast spread of SARS-CoV-2 in the early epidemic in the United States. Public and Global Health. Cell. 2020 doi: 10.1016/j.cell.2020.04.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni Diego, Filippi Giulia, Cagliani Rachele, De Gioia Luca, Pozzoli Uberto, Al-Daghri Nasser, Clerici Mario, Sironi Manuela. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci. Rep. 2015;5(September):14480. doi: 10.1038/srep14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni Diego, Cagliani Rachele, Clerici Mario, Sironi Manuela. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M.J. WHO’s blueprint list of priority diseases. JAMA. 2018;319:1973. doi: 10.1001/jama.2018.5712. [DOI] [PubMed] [Google Scholar]
- Fulop Tamas, Witkowski Jacek M., Olivieri Fabiola, Larbi Anis. The integration of inflammaging in age-related diseases. Semin. Immunol. 2018;40:17–35. doi: 10.1016/j.smim.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Geoghegan Jemma L., Holmes Edward C. Predicting virus emergence amid evolutionary noise. Open Biol. 2017;7:170189. doi: 10.1098/rsob.170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles John R., Eby Peggy, Parry Hazel, Peel Alison J., Plowright Raina K., Westcott David A., McCallum Hamish. Environmental drivers of spatiotemporal foraging intensity in fruit bats and implications for Hendra virus ecology. Sci. Rep. 2018;8(1):9555. doi: 10.1038/s41598-018-27859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Lang, Li Jie, Zhou Qingfeng, Xu Zhichao, Chen Li, Zhang Yun, Xue Chunyi, Wen Zhifen, Cao Yongchang. A new bat-HKU2–like coronavirus in Swine, China, 2017. Emerg. Infect. Dis. 2017;23:16071609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reiche Ana S., Hernandez Matthew M., Sullivan Mitchell, Ciferri Brianne, Alshammary Hala, Obla Ajay, Fabre Shelcie. Introductions and early spread of SARS-CoV-2 in the New York City Area. medRxiv. 2020 doi: 10.1101/2020.04.08.20056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy Jörg J., Weyand Cornelia M. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 2013;14:428–436. doi: 10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski Lisa E., Ferris Martin T., Aylor David L., Whitmore Alan C., Green Richard, Frieman Matthew B., Deming Damon. Genome wide identification of SARS-CoV susceptibility Loci using the collaborative cross. PLoS Genet. 2015;11(10) doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski Lisa E., Menachery Vineet D., Morgan Andrew P., Totura Allison L., Beall Anne, Kocher Jacob, Plante Jessica. Allelic variation in the toll-like receptor adaptor protein Ticam2 contributes to SARS-coronavirus pathogenesis in mice. G3: Genes|Genomes|Genetics. 2017;7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffoni Alba, Weiskopf Daniela, Ramirez Sydney I., Mateus Jose, Dan Jennifer M., Moderbacher Carolyn Rydyznski. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels Bupp Melanie R., Potluri Tanvi, Fink Ashley L., Klein Sabra L. The confluence of sex hormones and aging on immunity. Front. Immunol. 2018;9:1269. doi: 10.3389/fimmu.2018.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson Daniel F., Helgason Agnar, Jonsson Hakon, Magnusson Olafur T., Melsted Pall, Norddahl Gudmundur L., Saemundsdottir Jona. Early spread of SARS-Cov-2 in the Icelandic population. Epidemiology. medRxiv. 2020 doi: 10.1101/2020.03.26.20044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon Stéphane, Gascuel Olivier. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hadfield James, Megill Colin, Bell Sidney M., Huddleston John, Potter Barney, Callender Charlton, Sagulenko Pavel, Bedford Trevor, Neher Richard A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;43:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema Bert Jan, Volders Haukeliene, Rottier Peter J.M. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77(8):4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Qingmei, Lin Qingqing, Ni Zuowei, You Liangshun. Uncertainties about the transmission routes of 2019 Novel coronavirus. Influenza Other Respir. Viruses. 2020 doi: 10.1111/irv.12735. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson Jordan. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder Mark, Lewis Paul O. Phylogeny estimation: traditional and bayesian approaches. Nat. Rev. Genet. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Hou Yuxuan, Cheng Peng, Yu Meng, Li Yan, Han Zhenggang, Li Fang, Wang Lin-Fa, Shi Zhengli. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV Entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Chaolin, Wang Yeming, Li Xingwang, Ren Lili, Zhao Jianping, Hu Yi, Zhang Li. Clinical features of patients infected with 2019 Novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip W., Eddie K., Eddie Ip W.K., Chan Kwok Hung, Helen K. W. Law, Gloria H. W. Tso, Eric K. P. Kong, Wilfred H. S. Wong, et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191:1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoyama Satoru, Keicho Naoto, Hijikata Minako, Quy Tran, Phi Nguyen Chi, Long Hoang Thuy, Le Dang Ha. Identification of an alternative 5′-untranslated exon and new polymorphisms of angiotensin-converting enzyme 2 gene: lack of association with SARS in the Vietnamese population. Am. J. Med. Genet. 2005;136A:52–57. doi: 10.1002/ajmg.a.30779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa Naoko, Okamura Tadashi, Shimizu Yukiko, Hasegawa Hideki, Takeda Makoto, Nagata Noriyo. TMPRSS2 contributes to virus spread and immunopathology in the airways of Murine models after coronavirus infection. J. Virol. 2019;93:e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Wei, Wang Wei, Zhao Xiaofang, Zai Junjie, Li Xingguang. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Christine K., Hitchens Peta L., Pandit Pranav S., Rushmore Julie, Evans Tierra Smiley, Young Cristin C.W., Doyle Megan M. Global shifts in Mammalian population trends reveal key predictors of virus spillover risk. Proc. Biol. Sci. /R. Soc. 2020;287(1924) doi: 10.1098/rspb.2019.2736. 20192736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Kate E., Patel Nikkita G., Levy Marc A., Storeygard Adam, Balk Deborah, Gittleman John L., Daszak Peter. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadel Sapana, Kovats Susan. Sex hormones regulate innate immune cells and promote sex differences in respiratory virus infection. Front. Immunol. 2018;9:1653. doi: 10.3389/fimmu.2018.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser Jocelyn. How sick will the coronavirus make you? The answer may be in your genes. Science. 2020 doi: 10.1126/science.abb9192. [DOI] [Google Scholar]
- Kane Melissa, Golovkina Tatyana V. Mapping viral susceptibility loci in mice. Ann. Rev. Virol. 2019;6(1):525–546. doi: 10.1146/annurev-virology-092818-015544. [DOI] [PubMed] [Google Scholar]
- Keesing F., Holt R.D., Ostfeld R.S. Effects of species diversity on disease risk. Ecol. Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- Keesing Felicia, Belden Lisa K., Daszak Peter, Andrew Dobson, Drew Harvell C., Holt Robert D., Hudson Peter. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468(7324):647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keicho Naoto, Itoyama Satoru, Kashiwase Koichi, Phi Nguyen Chi, Long Hoang Thuy, Le Dang Ha, Van Ban Vo. Association of human leukocyte antigen class ii alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009;70:527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby Marie E., Biggs Holly M., Midgley Claire M., Gerber Susan I., Watson John T. Middle east respiratory syndrome coronavirus transmission. Emerg. Infect. Dis. 2020;26(2):191–198. doi: 10.3201/eid2602.190697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Yuri, Cheon Shinhye, Min Chan-Ki, Sohn Kyung Mok, Kang Ying Jin, Cha Young-Je, Kang Ju-Il. Spread of mutant middle east respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio. 2016;7(2) doi: 10.1128/mBio.00019-16. e00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Yeon-Sook, Aigerim Abdimadiyeva, Park Uni, Kim Yuri, Rhee Ji-Young, Choi Jae-Phil, Park Wan Beom. sequential emergence and wide spread of neutralization escape middle east respiratory syndrome coronavirus Mutants, South Korea, 2015. Emerg. Infect. Dis. 2019;25(6):1161–1168. doi: 10.3201/eid2506.181722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Young-Li, Kim Seong-Gyu, Kim Se-Mi, Kim Eun-Ha, Park Su-Jin, Yu Kwang-Min, Chang Jae-Hyung. Infection and rapid transmission of SARS-CoV-2 in Ferrets. 2020. Cell Host Microbe. 2020;27 doi: 10.1016/j.chom.2020.03.023. 704–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber Hannah, Elzayat Mahmoud Tarek, Wang Lingshu, Graham Barney S., Müller Marcel A., Drosten Christian, Pöhlmann Stefan, Hoffmann Markus. Mutations in the spike protein of middle east respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 2019;93(2) doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut Igor, Scholz Jean L., Cancro Michael P., Cambier John C. B cell maintenance and function in aginG. Semin. Immunol. 2012;24:342–349. doi: 10.1016/j.smim.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Kollmann Tobias R., Levy Ofer, Montgomery Ruth R., Goriely Stanislas. Innate immune function by toll-like receptors: distinct responses in newborns and the elderly. Immunity. 2012;37:771–783. doi: 10.1016/j.immuni.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species Barrier. J. Virol. 2000;74(3):1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Sai Kit, Chua Kaw Bing. Nipah virus encephalitis outbreak in Malaysia. Clin. Infect. Dis. 2002;34:S48–S51. doi: 10.1086/338818. [DOI] [PubMed] [Google Scholar]
- Lam Tommy Tsan-Yuk, Shum Marcus Ho-Hin, Zhu Hua-Chen, Tong Yi-Gang, Ni Xue-Bing, Liao Yun-Shi, Wei Wei. Identifying SARS-CoV-2 related coronaviruses in Malayan Pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. in press. [DOI] [PubMed] [Google Scholar]
- Lan Jun, Ge Jiwan, Yu Jinfang, Shan Sisi, Zhou Huan, Fan Shilong, Zhang Qi. Structure of the SARS-CoV-2 Spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. in press. [DOI] [PubMed] [Google Scholar]
- Lau Susanna K.P., Feng Yun, Chen Honglin, Hayes K. H. Luk, Wei-Hong Yang, Kenneth S. M. Li, Yu-Zhen Zhang, et al. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89(20):10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tang N.L.-S., Chan P.K.-S., Wang C.-Y., Hui D.S.-C., Luk C., Kwok R. Polymorphisms in the C-type lectin genes cluster in chromosome 19 and predisposition to severe acute respiratory syndrome coronavirus (SARS-CoV) infection. J. Med. Genet. 2008;45:752–758. doi: 10.1136/jmg.2008.058966. [DOI] [PubMed] [Google Scholar]
- Liang Wan-Nian, Huang Yong, Zhou Wan-Xin, Qiao Lei, Huang Jian-Hui, Zheng-Lai Wu. Epidemiological characteristics of an outbreak of severe acute respiratory syndrome in Dongcheng District of Beijing from March to May 2003. Biomed. Environ. Sci.: BES. 2003;16(4):305–313. [PubMed] [Google Scholar]
- Liao Minfeng, Yang Liu, Yuan Jin, Wen Yanling, Xu Gang, Zhao Juanjuan, Lin Chen. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 doi: 10.1101/2020.02.23.20026690. [DOI] [Google Scholar]
- Lin Marie, Tseng Hsiang-Kuang, Trejaut Jean A., Lee Hui-Lin, Loo Jun-Hun, Chu Chen-Chung, Chen Pei-Jan. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Lu, Jiang Xiayang, Zhang Zhenling, Huang Siwen, Zhang Zhenyi, Fang Zhaoxiong, Zhiqiang Gu. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;4:9. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Liu Li, Wei Qiang, Lin Qingqing, Fang Jun, Wang Haibo, Kwok Hauyee, Tang Hangying. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopera Esteban, van der Graaf Adriaan, Lanting Pauline, van der Geest Marije, Study Lifelines Cohort, Fu Jingyuan, Swertz Morris. Lack of association between genetic variants at ACE2 and TMPRSS2 genes involved in SARS-CoV-2 infection and human quantitative phenotypes. medRxiv. 2020 doi: 10.1101/2020.04.22.20074963. 2020.04.22.20074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Guangwen, Wang Qihui, Gao George F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23(8):468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray Paul B., Jr., Pewe Lecia, Wohlford-Lenane Christine, Hickey Melissa, Manzel Lori, Shi Lei, Netland Jason. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane Rosemary, Becker Niels, Field Hume. Investigation of the climatic and environmental context of hendra virus spillover events 1994-2010. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta Puja, McAuley Daniel F., Brown Michael, Sanchez Emilie, Tattersall Rachel S., Manson Jessica J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery Vineet D., Yount Boyd L., Jr Kari Debbink, Agnihothram Sudhakar, Gralinski Lisa E., Plante Jessica A., Rachel L. Graham, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore Michael J., Dorfman Tatyana, Li Wenhui, Wong Swee Kee, Li Yanhan, Kuhn Jens H., Coderre James. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78(19):10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand Serge, Owers Katharine A., Agnes Waret-Szkuta, Marie McIntyre K., Baylis Matthew. Climate variability and outbreaks of infectious diseases in Europe. Sci. Rep. 2013;3:1774. doi: 10.1038/srep01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller Marcel A., Corman Victor Max, Jores Joerg, Meyer Benjamin, Younan Mario, Liljander Anne, Bosch Berend-Jan. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg. Infect. Dis. 2014;20(12):2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth Doreen, Corman Victor Max, Roth Hanna, Binger Tabea, Dijkman Ronald, Gottula Lina Theresa, Gloza-Rausch Florian. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8(1):15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher Richard A., Bedford Trevor. Real-time analysis and visualization of pathogen sequence data. J. Clin. Microbiol. 2018;56(11) doi: 10.1128/JCM.00480-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Margaret H.L., Lau Kin-mang, Li Libby, Cheng Suk-hang, Chan Wing Y., Hui Pak K., Zee Benny, Leung Chi-bon, Sung Joseph J.Y. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Man Wai, Zhou Gangqiao, Chong Wai Po, Lee Loretta Wing Yan, Law Helen Ka Wai, Zhang Hongxing, Wong Wilfred Hing Sang. The association of RANTES polymorphism with severe acute respiratory syndrome in Hong Kong and Beijing Chinese. BMC Infect. Dis. 2007;7:50. doi: 10.1186/1471-2334-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls N. El Niño-Southern oscillation and vector-borne disease. Lancet. 1993;342:1284–1285. doi: 10.1016/0140-6736(93)92368-4. [DOI] [PubMed] [Google Scholar]
- Niedzwiedz Claire L., O’Donnell Catherine A., Jani Bhautesh D., Demou Evangelia, Ho Frederick K., Celis-Morales Carlos, Nicholl Barbara I. Ethnic and socioeconomic differences in SARS-CoV2 infection in the UK Biobank cohort study. medRxiv. 2020;(April) doi: 10.1186/s12916-020-01640-8. 2020.04.22.20075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich-Žugich Janko. The twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. 2018;19:10–19. doi: 10.1038/s41590-017-0006-x. [DOI] [PubMed] [Google Scholar]
- Nogales Aitor, DeDiego Marta L. Host single nucleotide polymorphisms modulating influenza A virus disease in humans. Pathogens. 2019;8:168. doi: 10.3390/pathogens8040168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile Dennis, Enserink Martin. SARS in China. Tracking the roots of a killer. Science. 2003;301(5631):297–299. doi: 10.1126/science.301.5631.297. [DOI] [PubMed] [Google Scholar]
- Oh Soo-Jin, Lee Jae Kyung, Shin Ok Sarah. Aging and the immune system: the impact of immunosenescence on viral infection, immunity and vaccine immunogenicity. Immune Network. 2019;19 doi: 10.4110/in.2019.19.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen Sonja J., Chen Meng-Yu, Liu Yu-Lun, Witschi Mark, Ardoin Alexis, Calba Clémentine, Mathieu Pauline. Early introduction of severe acute respiratory syndrome coronavirus 2 into Europe. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Tung. Novel coronavirus: from discovery to clinical diagnostics. Infect. Genet. Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright Raina K., Parrish Colin R., McCallum Hamish, Hudson Peter J., Ko Albert I., Graham Andrea L., Lloyd-Smith James O. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15(8):502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Population genetics and population biology: what did they bring to the epidemiology of transmissible diseases? An E-debate Infection. Genet. Evol. 2001;1:161–166. doi: 10.1016/s1567-1348(01)00023-5. [DOI] [PubMed] [Google Scholar]
- Qu Xiu-Xia, Hao Pei, Song Xi-Jun, Jiang Si-Ming, Liu Yan-Xia, Wang Pei-Gang, Rao Xi. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx Barry, Baas Tracey, Zornetzer Gregory A., Haagmans Bart, Sheahan Timothy, Frieman Matthew, Matthew D. Dyer, et al. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/jvi.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx Barry, Donaldson Eric, Frieman Matthew, Sheahan Timothy, Corti Davide, Lanzavecchia Antonio, Baric Ralph S. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2010;201(6):946–955. doi: 10.1086/651022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr Jason R., Barrett Christopher B., Civitello David J., Craft Meggan E., Delius Bryan, DeLeo Giulio A., Peter J. Hudson, et al. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019;2(6):445–456. doi: 10.1038/s41893-019-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer Alexandra, Baric Ralph S., Ferris Martin T. Systems approaches to coronavirus pathogenesis. Curr. Opin. Virol. 2014;6:61–69. doi: 10.1016/j.coviro.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickli Jeanne H., Thackray Larissa B., Sawicki Stanley G., Holmes Kathryn V. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 2004;78(17):9073–9083. doi: 10.1128/JVI.78.17.9073-9083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvey Linda A., Wells Rachel M., McCormack Joseph G., Ansford Anthony J., Murray Keith, Rogers Russell J., Lavercombe Peter S., Selleck Paul, Sheridan John W. Infection of humans and horses by a newly described Morbillivirus. Med. J. Aust. 1995;162:642–645. doi: 10.5694/j.1326-5377.1995.tb126050.x. [DOI] [PubMed] [Google Scholar]
- Shi Jianzhong, Wen Zhiyuan, Zhong Gongxun, Yang Huanliang, Wang Chong, Huang Baoying, Liu Renqiang. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Yufang, Wang Ying, Shao Changshun, Huang Jianan, Gan Jianhe, Huang Xiaoping, Bucci Enrico, Piacentini Mauro, Ippolito Giuseppe, Melino Gerry. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi Manuela, Cagliani Rachele, Forni Diego, Clerici Mario. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015;16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Katherine F., Behrens Michael, Schloegel Lisa M., Marano Nina, Burgiel Stas, Daszak Peter. Ecology. Reducing the risks of the wildlife trade. Science. 2009;324(5927):594–595. doi: 10.1126/science.1174460. [DOI] [PubMed] [Google Scholar]
- Solana Rafael, Tarazona Raquel, Gayoso Inmaculada, Lesur Olivier, Dupuis Gilles, Fulop Tamas. Innate immunosenescence: effect of aging on cells and receptors of the innate immune system in humans. Semin. Immunol. 2012;24:331–341. doi: 10.1016/j.smim.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Spiteri Gianfranco, Fielding James, Diercke Michaela, Campese Christine, Enouf Vincent, Gaymard Alexandre, Bella Antonino. First cases of coronavirus disease 2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Eurosurveillance. 2020;25:178. doi: 10.2807/1560-7917.ES.2020.25.9.2000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin Sibylle Bernard, Rolland Patrick, Silue Yassoungo, Mailles Alexandra, Campese Christine, Simondon Anne, Mechain Matthieu. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Eurosurveillance. 2020;25:94. doi: 10.2807/1560-7917.es.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi Sonu, Rapin Noreen, Misra Vikram. Immune system modulation and viral persistence in bats: understanding viral spillover. Viruses. 2019;11:192. doi: 10.3390/v11020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan Alireza, Askari Fatemeh Sana, Bont Louis, Salimi Vahid. Disease severity in respiratory syncytial virus infection: role of host genetic variation. Rev. Med. Virol. 2019;29(2) doi: 10.1002/rmv.2026. [DOI] [PubMed] [Google Scholar]
- Thevarajan Irani, Thi H. O. Nguyen, Marios Koutsakos, Julian Druce, Leon Caly, Carolien E. van de Sandt, Xiaoxiao Jia, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valitutto Marc T., Aung Ohnmar, Tun Kyaw Yan Naing, Vodzak Megan E., Zimmerman Dawn, Yu Jennifer H., Win Ye Tun. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp Lucy, Acman Mislav, Richard Damien, Shaw Liam P., Ford Charlotte E., Ormond Louise, Christopher J. Owen, et al. Emergence of Genomic Diversity and Recurrent Mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;104351 doi: 10.1016/j.meegid.2020.104351. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwambeke Sophie O., Linard Catherine, Gilbert Marius. Emerging challenges of infectious diseases as a feature of land systems. Curr. Opin. Environ. Sustain. 2019;38:31–36. [Google Scholar]
- Villabona-Arenas Ch. Julián, Hanage William P., Tully Damien C. Phylogenetic interpretation during outbreaks requires caution. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0738-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls Alexandra C., Young-Jun Park, Alejandra Tortorici M., Wall Abigail, McGuire Andrew T., Veesler David. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, in press. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Sheng-Fan, Chen Kuan-Hsuan, Chen Marcelo, Li Wen-Yi, Chen Yen-Ju, Tsao Ching-Han, Yen Muh-Yong, Huang Jason C., Chen Yi-Ming Arthur. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- Wang Liang, Shuo Su, Bi Yuhai, Wong Gary, Gao George F. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26:466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xinjie, Ji Pinpin, Fan Huiying, Lu Dang, Wan Wenwei, Liu Siyuan, Li Yanhua. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020;3(1):62. doi: 10.1038/s42003-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Wen Wen, Su Wenru, Tang Hao, Le Wenqing, Zhang Xiaopeng, Zheng Yingfeng. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. medRxiv. 2020 doi: 10.1101/2020.03.23.20039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson David A., Marshall Jonathan C., French Nigel P., Hayman David T.S. Habitat fragmentation, biodiversity loss and the risk of novel infectious disease emergence. J. R Soc. Interf. / the R Soc. 2018;15:403. doi: 10.1098/rsif.2018.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Frances M.K., Freydin Maxim, Mangino Massimo, Couvreur Simon, Visconti Alessia, Bowyer Ruth C.E., Le Roy Caroline I. Self-reported symptoms of Covid-19 including symptoms most predictive of SARS-CoV-2 infection, are heritable. medRxiv. 2020;(April) doi: 10.1017/thg.2020.85. 2020.04.22.20072124. [DOI] [PubMed] [Google Scholar]
- Wolfe Nathan D., Dunavan Claire Panosian, Diamond Jared. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Kailang, Peng Guiqing, Wilken Matthew, Geraghty Robert J., Li Fang. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287(12):8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Yifei. Unveiling the origin and transmission of 2019-nCoV. Trends Microbiol. 2020;28(4):239–240. doi: 10.1016/j.tim.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancy Clyde W. COVID-19 and African Americans. JAMA, in press. 2020 doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- Yuan F.F., Tanner J., Chan P.K.S., Biffin S., Dyer W.B., Geczy A.F., Tang J.W., Hui D.S.C., Sung J.J.Y., Sullivan J.S. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66:291–296. doi: 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Fang F., Boehm Ingrid, Chan Paul K.S., Marks Katherine, Tang Julian W., Hui David S.C., Sung Joseph J.Y., Dyer Wayne B., Geczy Andrew F., Sullivan John S. High prevalence of the CD14-159CC genotype in patients infected with severe acute respiratory syndrome-associated coronavirus. Clin. Vaccine Immunol. 2007;14(12):1644–1645. doi: 10.1128/CVI.00100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki Ali M., van Boheemen Sander, Bestebroer Theo M., Albert D.M., Fouchier Ron A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang Hongxing, Zhou Gangqiao, Zhi Lianteng, Yang Hao, Zhai Yun, Dong Xiaojia, Zhang Xiumei, Xue Gao, Zhu Yunping, He Fuchu. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;192:1355–1361. doi: 10.1086/491479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Kai, Wang Hui, Changyou Wu. The immune responses of HLA-A*0201 restricted SARS-CoV S peptide-specific CD8 T cells are augmented in varying degrees by CpG ODN, PolyI:C and R848. Vaccine. 2011;29(38):6670–6678. doi: 10.1016/j.vaccine.2011.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Hong, Chen Xing, Hu Tao, Li Juan, Song Hao, Liu Yanran, Wang Peihan. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertins at th S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:1–8. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Peng, Yang Xing-Lou, Wang Xian-Guang, Hu Ben, Zhang Lei, Zhang Wei, Si Hao-Rui. A pneumonia outbreak associated with a new coronavirus of probable bat Origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Xiaohui, Wang Yan, Zhang Hongxing, Liu Xuan, Chen Ting, Yang Ruifu, Shi Yuling. Genetic variation of the human α-2-heremans-schmid glycoprotein (AHSG) gene associated with the risk of SARS-CoV infection. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Na, Zhang Dingyu, Wang Wenling, Li Xingwang, Yang Bo, Song Jingdong, Zhao Xiang. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsstag Jakob, Crump Lisa, Schelling Esther, Hattendorf Jan, Maidane Yahya Osman, Ali Kadra Osman, Muhummed Abdifatah. Climate change and one health. FEMS Microbiol. Lett. 2018;365 doi: 10.1093/femsle/fny085. fny085. [DOI] [PMC free article] [PubMed] [Google Scholar]