Abstract
Acute lung injury (ALI) and its more severe form, acute respiratory distress syndrome (ARDS) are common lung disorders characterized by alveolar-capillary barrier disruption and dyspnea, which can cause substantial morbidity and mortality. Currently, a cluster of acute respiratory illnesses, known as novel coronavirus (2019-nCoV)-infected pneumonia (NCIP), which allegedly originally occurred in Wuhan, China, has increased rapidly worldwide. The critically ill patients with ARDS have high mortality in subjects with comorbidities. Previously, the excessive recruitment and activation of neutrophils (polymorphonuclear leukocytes [PMNs]), accompanied by neutrophil extracellular traps (NETs) formation were reported being implicated in the pathogenesis of ALI/ARDS. However, the direct visualization of lung epithelial injuries caused by NETs, and the qualitative and quantitative evaluations of this damage are still lacking. Additionally, those already reported methods are limited for their neglect of the pathological role exerted by NETs and focusing only on the morphological features of NETosis. Therefore, we established a cell-based assay for detecting NETs during lung epithelial cells-neutrophils co-culture using the xCELLigence system, a recognized real-time, dynamic, label-free, sensitive, and high-throughput apparatus. Our results demonstrated that lung epithelial injuries, reflected by declines in cell index (CI) values, could be induced by lipopolysaccharide (LPS)-activated PMNs, or NETs in a time and dose-dependent manner. NETs generation was verified to be the major contributor to the cytotoxicity of activated PMNs; protein components of NETs were the prevailing cytotoxic mediators. Moreover, this cell-based assay identified that PMNs from severe pneumonia patients had a high NETs formative potential. Additionally, acetylsalicylic acid (ASA) and acetaminophen (APAP) were discovered alleviating NETs formation. Thus, this study not only presents a new methodology for detecting the pathophysiologic role of NETs but also lays down a foundation for exploring therapeutic interventions in an effort to cure ALI/ARDS in the clinical setting of severe pneumonia, including the emerging of NCIP.
Keywords: Neutrophil extracellular traps, Lung epithelial cells, xCELLigence, Acute lung injury, Severe pneumonia
Abbreviations: NSAIDs, Nonsteroidal anti-inflammatory drugs; INM, Indometacin; PXM, Piroxicam; KTC, Ketorolc tromethamine; ASA, Acetylsalicylic acid; APAP, Acetaminophen; KP, Ketoprofen; DXM, Dexamethasone
Highlights
-
•
A real-time, dynamical and label-free assay for detecting NETs is established using the xCELLigence system.
-
•
This establishment relies on the co-culture of lung epithelia and neutrophils, focusing on evaluating NETs’ effects.
-
•
This cell-based assay has feasibility and practicality in clinical applications.
-
•
This methodology builds a solid foundation for exploring therapies for ALI/ARDS, including the emerging NCIP.
1. Introduction
An outbreak of pneumonia associated with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) started in December 2019 in Wuhan, China. This coronavirus disease (COVID-19) is quickly spreading across the world with continuously increasing morbidity. According to the latest reports of Chinese clinical data, the mortality of novel coronavirus (2019-nCoV)-infected pneumonia (NCIP) was relatively low compared to severe acute respiratory syndromes (SARS), with most of the deaths seen in the elderly admitted in intensive care units [1,2]. More remarkable, those Critical NCIP patients with multiple organ dysfunction, especially acute respiratory distress syndrome (ARDS), ultimately succumbed to respiratory failure [1,3,4]. Acute lung injury and its more severe form acute respiratory distress syndrome (ALI/ARDS) are common complications associated with pneumonia and sepsis [5]. Moreover, increased neutrophils (polymorphonuclear leukocytes [PMNs]) transepithelial migration and intense alveolar epithelial disruption are reported to be the important pathological features of ALI/ARDS [6,66]. All of these suggest that the recruitment and activation of PMNs may exacerbate the progression of NCIP in patients with ARDS.
Inflammation is crucial for immune defense against pathogens including viruses and bacteria. However, when dysregulated, the cytokines that normally mediate protective immunity and promote recovery can cause a systemic hyperactivated immune state known as cytokine release syndrome (CRS), releasing a huge excess of cytokines, which can cause more harm than good, particularly in the lungs [67]. Before the COVID-19 outbreak, there was evidence to support the frequent involvement of exaggerated pro-inflammatory activation of PMNs, accompanied by the release of cytotoxic neutrophil extracellular traps (NETs), in the pathogenesis of ALI/ARDS [68,10]. Recently, as the clinical characteristics of NCIP patients have shown, the severe cases were in a high inflammatory response state, accompanied by the occurrence of CRS, causing the activation and recruitment of PMNs [1,2]. Although the involvement of NETs in lung injury in NCIP patients is yet to be corroborated by further pathological analysis of autopsy specimens, the qualitative and quantitative evaluation of NETs influencing lung epithelial function during this critical period is promising and significant for the development of treatment for this disease.
NETs are composed of extracellular chromatin fibers decorated with antimicrobial proteins, including neutrophil elastase (NE) and myeloperoxidase (MPO) [69]. Diverse stimuli have been reported to induce the NETs formation, including common pathogens (S. aureus, S. pneumoniae, influenza A virus, respiratory syncytial virus) of pulmonary infection [[13], [14], [15], [69]], inflammatory cytokines (IL8, IL-6, IL-1α/β) [69,70,17], and biochemicals (PMA, LPS[remain controversial]) [69,17,71]. Many methods such as fluorescent/immunofluorescent staining, scanning electron microscopy (SEM) [69,19], spectrophotometric analysis [72], colorimetric assays [10,21], and flow cytometry [22,23], have been documented for visualizing NETs or counting NETting PMNs in ALI/ARDS or other disease conditions. Among these, immunofluorescence microscopy characterizes NETs in lung tissue sections [24,25] or peripheral blood PMNs [68], based on the co-localization of extracellular DNA and neutrophil-derived proteins. Soluble NETs remnants in fluid samples, such as serum [26], bronchoalveolar lavage buffer [10,73], and cell culture medium [72] are tested by measuring the fluorescent values of extracellular DNA after staining with highly sensitive PicoGreen or membrane-impermeable SYTOX Green. The protein components in NETs, such as citrullinated histone 3 (HiCt3) [72], NE [21], and MPO [10,13] can also be examined by western blotting or enzyme-linked immunochemistry assays.
Although the aforementioned methods identified the biological relevance of NETs in the pathogenesis of ALI/ARDS, particularly that of NETs-bound components, direct evidence of lung epithelial injuries induced by NETs is still lacking. Those methods use only one or a few components of NETs as an indicator of the whole NETs, which is incorrect and misleading. Moreover, they only focus on the morphological changes in NETs formation and do not evaluate the pathological role exerted by NETs in disease microenvironments. These make them insufficient for deepening and expanding our understanding of the effects of NETs on the initiation and progression of ALI/ARDS. Hence, there is a need for a simpler, objective, and quantitative approach for detecting NETs, as well as evaluating lung epithelial disruption caused by NETs.
In this study, we described the optimization and validation of a novel lung epithelial cell-based assay to analyze the damage caused by NETs on lung epithelial barriers using the xCELLigence system. Till now, this label-free, real-time, and dynamically analytic apparatus has been widely used in evaluating Chinese compound medicines [29] or modern pharmaceuticals [74], monitoring epithelial barrier function [31], and detecting T cell activation [32] or NK cell killing [33]. The advantages of this system are its high sensitivity, accuracy, consistency, and repeatability. Comprehensive information about target cells, including proliferation, apoptosis, adhesion, and migration, is continually collected as cell index (CI) values. Suspension cells, such as PMNs, cannot directly generate CI due to their nonattachment to the metal chips embedded in the bottom, which makes the co-culture of adherent lung epithelial cells and suspended PMNs feasible. Our results demonstrated that lung epithelial injuries caused by lipopolysaccharide (LPS)-activated PMNs or isolated NETs showed time and number/concentration dependence. The cytotoxicity of activated PMNs was mainly ascribed to the generation of NETs, and the inhibition of NETs generation could relieve the destructive effects. Moreover, the protein components of NETs were the prevailing cytotoxic mediators.
We compared the consistency of this novel cell-based NETs detection methodology with other common analytical approaches. More encouragingly, the feasibility and practicality of this assay helped to distinguish NETs formation differences between PMNs from severe pneumonia patients and healthy controls, and seemed to discover the mitigative effects of acetylsalicylic acid (ASA) and acetaminophen (APAP) on NETosis.
Overall, this novel lung epithelial cell-based NETs detection assay using the xCELLigence system may not only advance our understanding of the role of NETs in the pathogenesis of ALI/ARDS, but might also provide a powerful methodology to develop new therapeutic strategies against this devastating lung disorder.
2. Materials and methods
2.1. Cell culture
A549 (human alveolar epithelial cells), HBEC (human bronchial epithelial cells), and MLE-12 (mouse alveolar epithelial cells) were purchased from ATCC. RPMI 1640 and DMEM media containing 10% FBS with 50 U/mL penicillin and 50 U/mL streptomycin were used for A549 and HBEC culture, respectively. DME/F-12 medium containing 2% FBS with 50 U/mL penicillin and 50 U/mL streptomycin was used for MLE-12 culture. RPMI 1640 medium without phenol red (GNM-11835, GENOM) containing 2% FBS was used for PMNs culture.
2.2. Isolation of PMNs
Blood samples were collected from severe pneumonia patients (admitted in Sir Run Run Shaw Hospital respiratory medicine and intensive care units, Zhejiang University School of Medicine) or healthy volunteers. A written informed consent was obtained from all the participants and this study was approved by the Human Ethics Committee at Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. Human PMNs were isolated from the peripheral blood by density-gradient centrifugation using a Neutrophil Isolation Kit (LZS11131, TBD Science), according to the manufacturer's instructions. The purity of PMNs labeled with anti-Ly6G antibody (ab25024, Abcam) was analyzed by flow cytometry, and cell morphology was determined by Giemsa staining.
All animal care and surgical procedures were approved by the Animal Ethics Committee at Zhejiang University. The mouse peritoneal PMNs separation procedure was slightly modified from that described in previous studies [34]. The mouse was intraperitoneally injected with 1 mL 3% thioglycolate (MERCK). After 4 h, PMNs were harvested from the peritoneal cavity by repeated lavage in 5 mL sterile PBS. After filtering from a 40-μm cell-filter and washing in red blood cell lysis buffer (NH4CL2009, TBD Science), the PMNs were suspended in RPMI 1640 medium without phenol red.
2.3. Preparation of NETting PMNs
Freshly isolated human or mouse PMNs diluted to designated densities were incubated in microfuge tubes with LPS (1 μg/mL) stimulation at 37 °C, 5% CO2 for 2 h. Then, the PMN suspensions were centrifuged at 300 g for 5 min to pellet the stimulated PMNs, and the culture supernatant was discarded. These LPS-activated PMNs were re-suspended with fresh RPMI 1640, DMEM, or DME/F-12 media, depending on the subsequent co-cultured epithelial cells.
2.4. xCELLigence system
xCElligence (Roche Applied Science) is composed of a real-time impedance analyzer, a computer with cell analysis software for controlling operation, a 96-well electronic microtiter plate (E-Plate), and a station placed inside a culture incubator (37 °C, 5% CO2). Cell presence sensed by the chips at the bottom can create electronic impedance, which is converted into CI values.
The application of the xCELLigence system was modified from that described previously [[29], [31], [32], [33], [74]]. To obtain background reading, 50 μL of culture medium was added into the 96-well E-plate, followed by the loading of 100 μL cell suspensions. The E-plate was incubated at room temperature for 30 min before being placed in the station. CI was recorded every 5 min. Special care was taken to limit cell perturbation during the manipulation, as even mild shaking could result in signal fluctuations. When the CI values almost reached the stable phase, follow-up testing could start. Volumes of 100 μL supernatant per well were carefully discarded from the E-plate and then effectors, such as PMNs, NETs, or drugs equilibrated in 100 μL medium, were added. Meanwhile, the time point of loading was normalized to 0, and the CI at loading point was normalized to 1. The normalized CI continued to be automatically monitored at 30 s intervals.
2.5. Scanning electron microscopy
A549 or PMN samples were subjected to washing three times with 0.05 M sodium cacodylate buffer and fixing in 2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer for 4 h. Secondary fixation with 0.1% osmium tetroxide was performed for 15 min prior to sequential dehydration with increasing concentrations of ethanol. Samples were dried at the critical point using a CO2 drier, mounted onto an aluminum stub, and sputter-coated with 80/20 gold-palladium. A thin strip of colloidal silver was painted at the sample edge to dissipate sample charging. Samples were imaged with a field emission scanning electron microscope Nova Nano 450 (Thermo FEI).
2.6. Immunofluorescence
PMNs after treatment with Escherichia coli O128: B12 endotoxin (LPS, Sigma-Aldrich), or lung epithelial cells after being co-cultured with PMNs for the designated time, were washed twice with PBS. Especially for the lung epithelial cells-PMNs co-culture samples, PMNs or NETs were cleared away as much as possible to avoid staining. After fixation with 4% PFA and permeabilization with 0.2% Triton x-100, the samples were blocked with 5% BSA. PMNs were then incubated with antibodies against NE (dilution 1:250; ab21595, Abcam) and MPO (dilution 1:1000; ab25989, Abcam), followed by secondary antibodies conjugated with a green fluorescence Alexa Fluor 488 dye and a red fluorescence Alexa Fluor 555 dye (Invitrogen), respectively. Lung epithelial cells were subsequently incubated with the red fluorescence Alexa Fluor™ 555 Phalloidin (dilution 1:1000; A34055, Thermo Fisher) to stain F-actin, and DAPI to stain nuclear DNA. In another co-culture experiment, A549 cells were additionally stained with cleaved caspase-3 (dilution 1:400; 9661S, Cell Signaling Technology), followed by secondary antibody conjugated with a green fluorescence Alexa Fluor 488 dye. The co-cultured PMNs were reserved and stained with an antibody against MPO (dilution 1:1000; ab25989, Abcam), followed by secondary antibody conjugation with an orange fluorescence Alexa Fluor 645 dye (Invitrogen). After washing with PBS, all the samples were mounted with antifade mounting medium (Beyotime) before taking images under an inverted Nikon A1R confocal microscope.
2.7. Live-cell imaging
This experiment was conducted using a Nikon A1R confocal microscope equipped with phase-contrast microscopy and a temperature-control to maintain incubation at 37 °C. PMNs with different treatments (medium, 1 μg/mL LPS, 1 μM DPI, and 1 μg/mL LPS + 1 μM DPI) were re-suspended in RPMI 1640 without phenol red containing 5 μM SYTOX Green (S7020, Thermo Fisher). Then, these suspensions were cultured in a 35 mm 4-chamber glass-bottom dish (D35C4-20-1.5-N, Cellvis) (1 × 105/mL, 500 μL/chamber). After 30 min, images were randomly taken under low-light illumination at 15 min intervals for a total of 4 h. Video images were controlled by NIS elements 4.3.0 software to generate Quick-Time 3.0 movies in real-time at a frame speed of five frames per second.
2.8. Calcein-AM/PI double staining
This assay was processed using the Calcein-AM/PI double stain Kit (40747ES76, YEASEN) as described previously [75]. A549 (1 × 105/mL, 500 μL/chamber) were seeded into a 35 mm 4-chamber glass-bottom dish overnight. Then, culture media were discarded and 500 μL of untreated PMNs or NETting PMN suspensions (dyed with Hoechst 33258, 4 × 105/mL) were added into the chambers. In another experiment, different concentrations of NETs solution were added. After co-incubation for 6 h, the media were carefully removed and the chambers were washed to clear PMNs or NETs remnants as much as possible. Subsequently, A549 were double-stained with calcein acetoxymethyl ester (AM-Calcein)/propidium iodide (PI) fluorescein following the manufacturer’s instructions. After washing twice with icy PBS, living (green) and dead (red) A549 cells were observed by confocal microscopy.
2.9. SYTOX green-based kinetic plate assay
The procedure used in this study was modified from that described previously [[36], [37], [38]]. PMNs (3 × 104/mL) stimulated by different concentrations of LPS were re-suspended in RPMI 1640 without phenol red containing 5 μM membrane-impermeable DNA dye SYTOX Green (S7020, Thermo Fisher). Then, these suspensions were seeded in a 96-well black plate (WHB-96, WHB) (100 μL/well), usually in quadruplicates, and cultured at 37 °C in the dark. Extracellular DNA released from PMNs were quantified by measuring fluorescence values at sequential time points in a microplate fluorescence reader at 488/523 nm.
2.10. Protein extraction and western blot
Following PMNs incubation in a 6-well plate (1 × 108/mL, 1 mL/well), 900 μL culture medium per well was carefully aspirated and 10 μL 10 × RIPA buffer (9806S, Cell Signaling Technology) was added into the wells supplemented with EDTA-free protease inhibitor tablets and phosphatase inhibitors (Roche Diagnostics). After lysis on ice for 5 min, the total PMN lysates were collected and sonicated on ice for 15 s. The protein concentrations were quantified using the BCA assay (Beyotime) before solubilizing in 6 × loading buffer, and were boiled for 8 min. Equal amounts of protein lysates per lane were resolved using 13% SDS-PAGE, transferred by electrophoresis onto nitrocellulose membranes (Pall, Port Washington, NY), blocked with 5% BSA for 1 h at room temperature and probed with an anti-HiCt3 primary antibody (dilution 1:1000; ab5103, Abcam), and an anti-β-actin antibody (dilution 1:5000; M1210-2, Huaan Biosciences) at 4 °C overnight. This was followed by incubation with IRDye 680LT/IRDye 800CW secondary Abs (LI-COR Biosciences) for 1 h at room temperature. Finally, the signals were detected by an Odyssey two-color infrared imaging system.
2.11. Flow cytometry
PMNs after LPS (1 μg/mL) or DPI (1 μM; HY-100965, MCE) treatments, or A549 cells after NETs incubation were washed twice with PBS and re-suspended in 100 μL PBS. PMNs were stained with DCFH-DA probes (final concentration 10 μM; S0033, Beyotime). In another experiment, A549 cells were stained with PI fluorescein (final concentration 5 μM; 40747B, YEASEN). Subsequently, these samples were incubated at 37 °C in the dark for 1 h. After washing twice with icy PBS, PMNs or A549 cells were suspended in 100 μl of PBS containing 0.05% EDTA for FACS analysis performed on ACEA NovoCyte Flow Cytometer. Data were analyzed using FlowJo software.
2.12. Isolation and dilution of NETs
The procedure of NETs isolation was modified from that described previously [72,76]. Freshly isolated PMNs (1 × 107/mL) suspended in RPMI 1640 medium were seeded into a 6-well plate with 1 mL per well and stimulated with LPS (1 μg/mL) for 4 h in an incubator at 37 °C. The culture medium was aspirated gently, and PBS was slowly added to the sides of the wells for two washes. Thereafter, 100 μL of 1640 medium per well was added into the culture plate, followed by repeated pipetting. The entire content of the wells was then transferred into microfuge tubes and centrifuged at 300 g for 5 min to pellet cellular debris. The supernatant containing NETs was collected and stored at −80 °C.
The concentration of original NETs extracted from 1 × 107 PMNs dissolved in 100 μL RPMI 1640 medium was assumed to be 100%. Diluted NET samples were prepared by adding the medium.
2.13. Measurement of NE concentration in NETs
The concentrations of NE in prepared NET samples were examined by the NETosis Assay Kit (601010, Cayman) following the manufacturer’s instructions [21].
2.14. Cell Counting Kit-8 assay
A549 (3 × 104/mL, 100 μL/well) were seeded into a 96-well plate overnight. 100 μL of the supernatant per well was carefully discarded, followed by the addition of 100 μL original NETs or the diluted NET solutions. After incubation for 6 h, the vitality of A549 was characterized using the CCK-8 (CK04, Dojindo) assay following the manufacturer’s instructions.
2.15. Purification of NETs-DNA and NETs-proteins
NETs-DNA was acquired after 500 μL of original NETs being incubated with 5 μL proteinase K (25 mg/mL; 539480, MERCK) at 37 °C for 15 min to digest protein components, and then purified using a DNA extraction kit (AP-MN-MS-GDNA-50, Axygen), according to the manufacturer's instructions.
NETs-proteins were acquired by digesting the DNA according to the description in previous studies [40]. Equal amount of NETs were incubated with different volumes (0.5, 1, 1.5, and 2 μL) of micrococcal nuclease (3.2 × 105 gel units; M0247S, BioLabs) at 37 °C for 15 min. The DNA degradation efficiency was identified by agarose electrophoresis.
2.16. Statistical analysis
CI values recorded for the real-time cell analysis were calculated automatically by the RTCA Software Package 1.2 of the xCELLigence system. Numerical data were expressed as mean ± SEM and were analyzed using GraphPad Prism 5.0 software. Intergroup differences were evaluated using One-way analysis of variance (ANOVA), and p < 0.05 was considered to indicate statistical significance.
3. Results
3.1. Optimization of the xCELLigence system for detecting lung epithelial injuries induced by LPS-activated PMNs
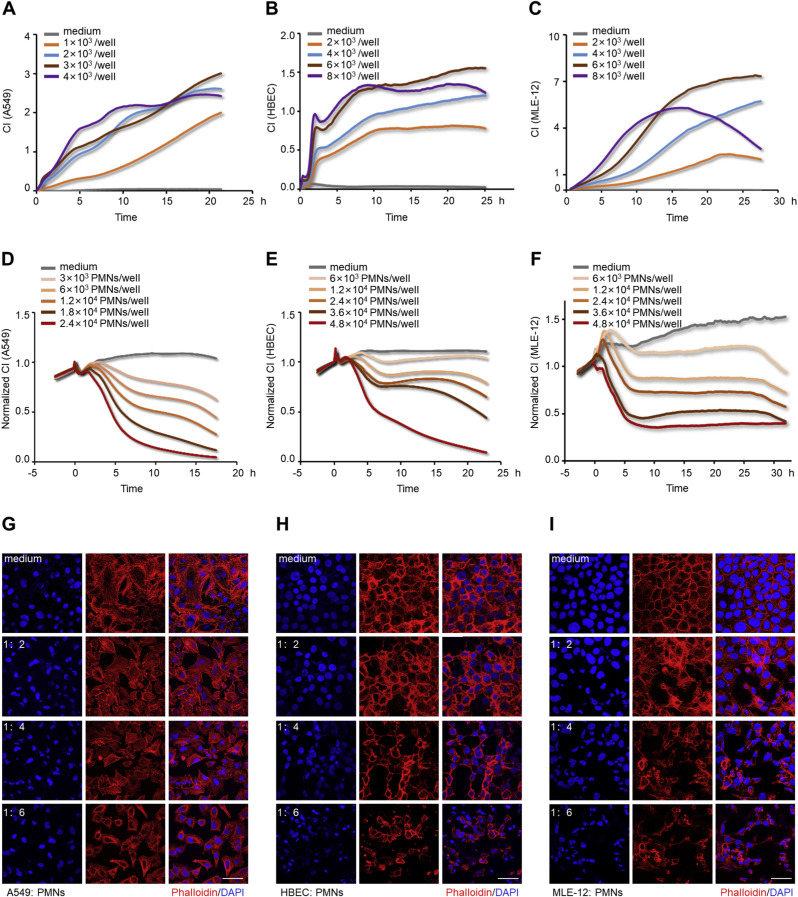
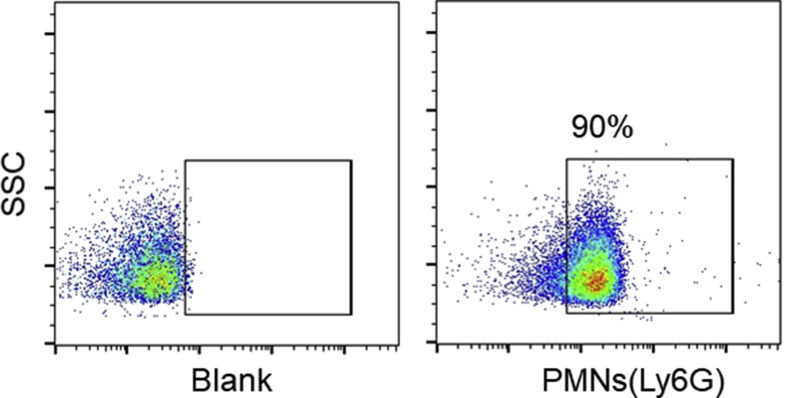
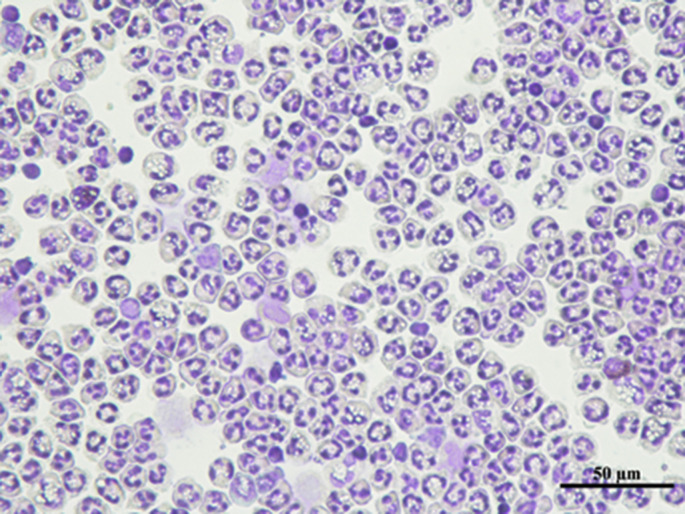
Although the role of activated PMNs in the pathogenesis of lung epithelial dysfunction has been documented in ALI [5,66], there is a paucity of evidence quantifying it. In this study, we evaluated the cytotoxic effects of LPS-activated PMNs in the xCELLigence system. The purity of isolated human PMNs detected by flow cytometry was 90% (Fig. Supplemental Fig. S1), and their morphology was polymorphonuclear (Fig. Supplemental Fig. S2). A549, HBEC, and MLE-12 cells were selected for constructing alveolar or bronchial epithelial barriers. The seeding densities for the xCELLigence system were optimized (Fig. 1 A–C) for A549 (3 × 104/mL), HBEC (6 × 104/mL), and MLE-12 (6 × 104/mL), and these optimized densities were used in further experiments. In the desired state, the ideal A549 lung epithelial barrier was constructed as shown in the SEM image in (Fig. Supplemental Fig. S3). The appropriate time point for the addition of PMNs was selected as approximately 25 h after lung epithelial cells seeding. Subsequently, lung epithelial damage initiated by LPS-activated PMNs (subsequently verified as NETting PMNs), and the cross-talk between cells were monitored by the xCELLigence system. The CI values of lung epithelial cells incubated with these sensitized PMNs decreased in a time and number-dependent manner, while the CI values of the controls remained unchanged (Fig. 1D–F). Generally, the integrity of epithelial cells is closely associated with an intact cytoskeleton composed of actin filaments and microtubules [41]. As confirmed by immunofluorescence with phalloidin staining F-actin in the epithelial cells, diverse extents of epithelial cytoskeletal destruction were significantly induced when the cells were co-cultured with different numbers of activated PMNs (Fig. 1G–I). To sum up, PMNs activated by LPS can damage lung epithelial cells, which can be consistently detected by the xCELLigence system in a time- and number-dependent manner.
Fig. 1.
Detection of lung epithelial injuries induced by LPS-activated PMNs using the xCELLigence system. (A, B, and C) Optimization of lung epithelial cells seeding densities in the xCELLigence system. CI growth curves of A549 (A), HBECs (B), and MLE-12 (C). (D, E, and F) CI response curves of lung epithelial cells co-cultured with LPS-activated PMNs in the xCELLigence system. A549 (3 × 103/well) (D), HBEC (6 × 103/well) (E), and MLE-12 (6 × 103/well) (F) were co-cultured with different numbers of activated PMNs, and only medium (RPMI 1640, DMEM, or DME/F12) incubations were used as controls. (G, H, and I) Fluorescence staining of lung epithelial cells after co-culturing with activated PMNs at sequential number ratios for 10 h, and only medium incubations as controls. A549 (G), HBEC (H), and MLE-12 (I) co-stained with F-actin (phalloidin, red) and DNA (DAPI, blue); Scale bar: 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. LPS stimulation induces NETs formation
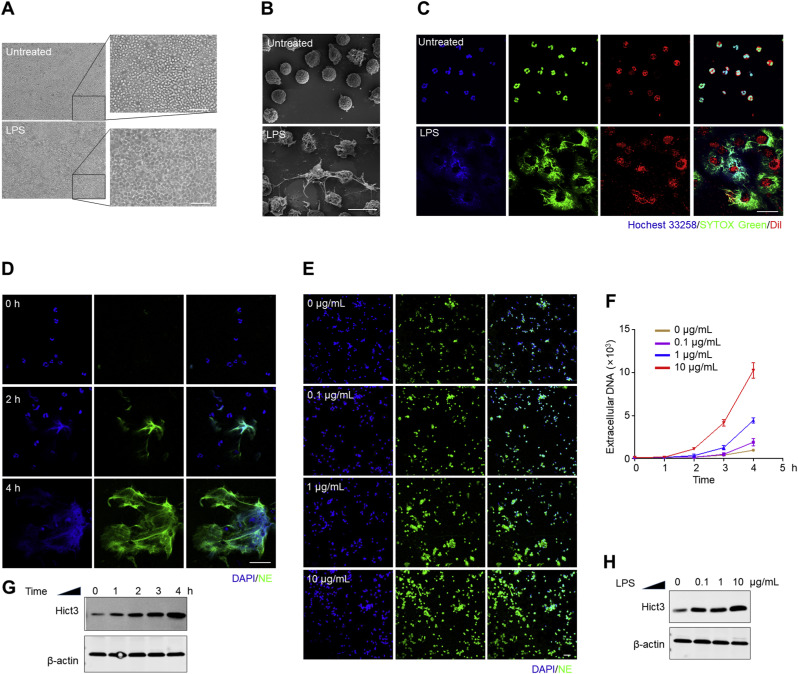
To explore the stimulation of PMNs by LPS, we compared the differences in PMNs with or without LPS treatment by multiple methods. Using an optical microscope, we observed the distinct morphological changes of LPS-stimulated PMNs compared to untreated PMNs: cell deformation with mucus tightly adhering to the plate bottom (Fig. 2 A). The morphological features of NETting PMNs stimulated by LPS were characterized using the high-resolution SEM: mesh-like filamentary structures and metamorphosis (Fig. 2B). Based on the fact that DNA is the major backbone of NETs, SYTOX Green, a plasma membrane-impermeable DNA-binding dye, has been popularly applied as an indicator for NETosis [26,[36], [37], [38]]. Here, as shown in Fig. 2C, we observed the disintegration of cytomembranes (stained by a recognized membrane lipid dye DiI), as well as the release of DNA (dyed with SYTOX Green) from cell bodies in the presence of LPS; and these extracellular DNA were surrounding PMN bodies. This was further determined in Fig. 2D and E, in which LPS-induced NETs formation was identified by the co-localization of extracellular fibrillar and web-like structures with DAPI as well as by the appearance of neutrophil-derived NE. The degree of membrane rupture and the number of NETting PMNs increased, depending on the exposure duration and concentration. This time and dose-dependent stimulation was also verified by measuring the fluorescent values of extracellular DNA using a recognized SYTOX Green-based kinetic plate assay (Fig. 2F). Additionally, HiCt3 (a specific marker for NETs formation) was used for exploring the mechanism of LPS-induced NETosis by western blotting. There were significantly higher levels of HiCt3 in LPS-treated PMNs compared to untreated PMNs, and the levels of HiCt3 in PMNs varied with the time and concentration of LPS stimulation (Fig. 2G and H). Thus, a combination of methods indicated that LPS stimulates NETs formation through histone H3 citrullination, and this induction is dose and time-dependent.
Fig. 2.
LPS stimulates NETs formation in a time and concentration-dependent manner. (A, B, and C) PMNs were treated with or without LPS (1 μg/mL) for 2 h. Optical micrographs and their higher-magnification views; Scale bar: 50 μm (A). High-resolution SEM micrographs; Scale bar: 10 μm (B). Immunofluorescence staining of the cytomembrane rupture and DNA release in LPS-stimulated PMNs co-stained by membrane lipid dye (DiI, red), extracellular DNA dye (SYTOX Green, green) and another DNA dye (Hochest 33258, blue); Scale bar: 50 μm (C). (D and E) Immunofluorescence staining of LPS-stimulated PMNs characterized by extrusion of DNA (DAPI, blue) and NE (green); Scale bar: 50 μm. PMNs were treated with the same concentration (1 μg/mL) of LPS for 0, 2, and 4 h (D); or treated with different concentrations (0, 0.1, 1.0, and 10 μg/mL) of LPS for 2 h (E). (F, G, and H) PMNs were treated with sequential concentrations (0. 0.1, 1.0, 10 μg/mL) of LPS for a series of times (0, 1, 2, 3, and 4 h). Fluorescent values of extracellular DNA stained with SYTOX Green (F). Western blotting analysis of HiCt3 in PMNs, and β-Actin used as a protein loading control (G and H). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. NETs formation is the major cytotoxic factor causing lung epithelial apoptosis and cytoskeletal destruction
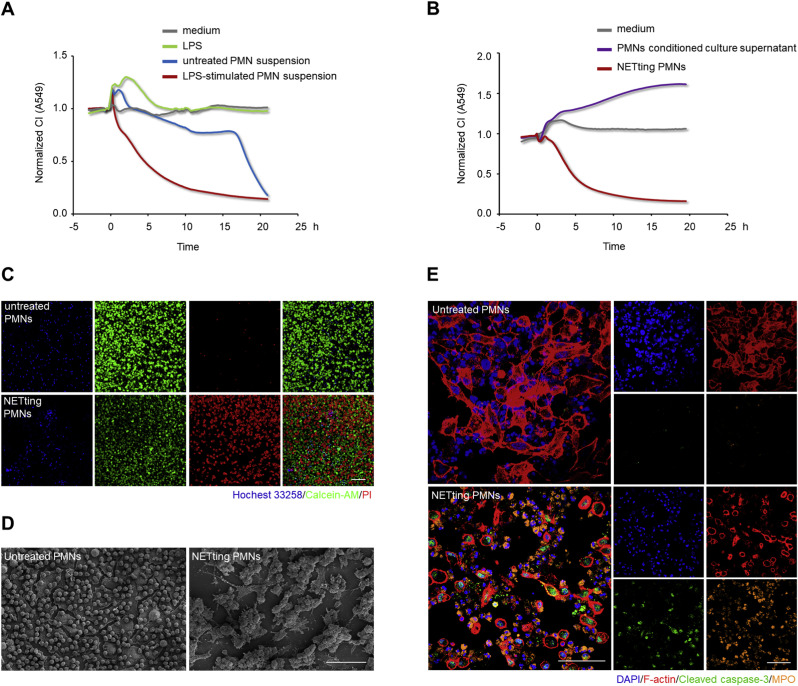
To further understand the destructive mechanism of LPS-activated PMNs on lung epithelial injuries, A549 lung epithelial cells were incubated in media with and without LPS, and co-cultured with equal numbers of untreated or LPS-treated PMN suspensions. CI values obtained from the xCELLigence system identified that stimulus LPS and untreated PMNs were non-cytotoxic to lung epithelial cells during the early co-culture period (Fig. 3 A). Since NETs generation occurs with LPS stimulation, LPS-activated PMNs were regarded as NETting PMNs. However, PMNs stimulated by LPS also secrete a large number of cytokines regulating epithelial function [77,43]. In view of this, PMNs conditioned culture supernatant and NETting PMN pellets, separated from LPS-stimulated PMN suspensions, were incubated with A549 cells individually to preclude the disturbance of other cytotoxic secretions. As shown in Fig. 3B, a sharp lung epithelial CI decline was exclusively caused by the NETting PMNs. We further analyzed the damage accounting for lung epithelial CI decline by co-culturing Hochest 33258 labeled NETting PMNs or untreated PMNs with A549 cells. Compared with untreated PMNs, NETting PMNs promoted lung epithelial cell apoptosis, as indicated by an increase in PI-positive cells and a decrease in Calcein AM-positive cells (Fig. 3C). Meanwhile, high-resolution SEM revealed that damaged lung epithelial cells were wrapped with NETs fibrous structures and gathered into cell clusters (Fig. 3D). Antibody-based immunofluorescence led to deeper insights into the destructive roles of NETting PMNs. Apart from dramatic cytoskeletal damages, cell apoptosis also occurred, as demonstrated by a significant increase in cleaved caspase-3 in epithelial cells after co-culture (Fig. 3E). Collectively, NETs generation in LPS-activated PMNs is the predominant factor causing lung epithelial injuries, including cell apoptosis and cytoskeletal destruction.
Fig. 3.
NETs formation is the main cytotoxicity causing lung epithelial apoptosis and cytoskeletal destruction. (A and B) CI response curves of A549 lung epithelial cells in the xCELLigence system. A549 cells were incubated with media containing LPS (1 μg/mL) or not, and co-cultured with equal numbers (2.4 × 104/well) of untreated PMN or LPS (1 μg/mL)-treated PMN suspensions (A). A549 cells were incubated with conditioned culture supernatant or NETting PMNs (2.4 × 104/well), separated from LPS-stimulated PMN suspensions (B). (C, D, and E) A549 lung epithelial cells were co-cultured with equal numbers of untreated PMNs or NETting PMNs at 1:8 number ratio for 10 h. Fluorescence staining of A549 cells co-stained with Calcein-AM (green)/PI (red) fluorescein after co-culturing with Hoechst 33258 (blue) labeled PMNs; Scale bar: 100 μm (C). High-resolution SEM images of the co-culture system; Scale bar: 500 μm (D). Immunofluorescence staining of A549 cells co-dyed with F-actin (phalloidin, red) and cleaved caspase-3 (green), as well as PMNs co-stained with DNA (DAPI, blue) and MPO (orange); Scale bar: 100 μm (E). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Inhibition of the generation of NETs relieves lung epithelial injuries
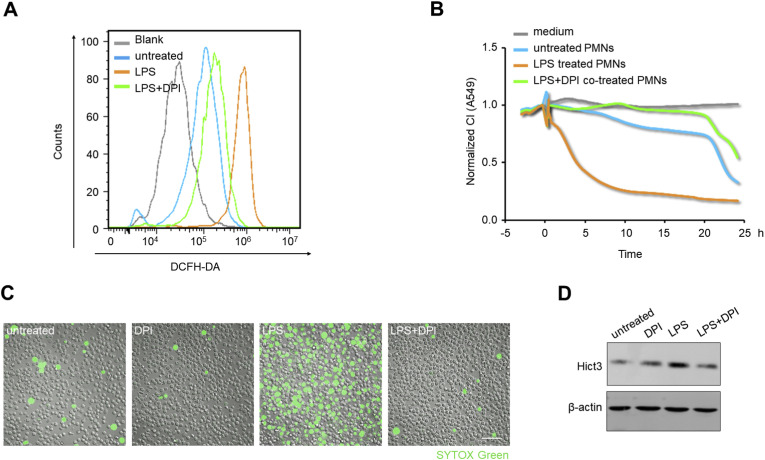
In order to explore whether inhibiting NETs formation could ease lung epithelial injuries, we used diphenyleneiodonium chloride ([DPI], NADPH oxidase inhibitor) [38,44] to inhibit NETosis. Using flow cytometry with DCFH-DA fluorescein, we confirmed that LPS stimulation enhanced the intracellular ROS level of PMNs, while DPI co-treatment reversed ROS increase (Fig. 4 A). To explore the protective role of DPI during the interaction of lung epithelial cells and PMNs, equal numbers of PMNs were co-cultured with A549 lung epithelial cells in the xCELLigence system, after the various treatments mentioned above. In this experiment, PMNs after LPS treatment prominently decreased lung epithelial CI values, whereas co-treatment with DPI significantly relieved the CI decline (Fig. 4B). Subsequently, the inhibitory effect of DPI on NETs formation was verified by detecting the amount of released extracellular DNA, and the extent of histone 3 citrullination. As shown in Fig. 4C and Supplemental Videos, DPI blocked spontaneous NETs generation, and significantly suppressed extracellular DNA production from LPS-stimulated PMNs. Meanwhile, LPS significantly increased the level of HiCt3 protein, whereas DPI co-treatment attenuated this increase (Fig. 4D). These led to the conclusion that the inhibition of NETosis can alleviate activated PMNs-induced lung epithelial damage.
Fig. 4.
Inhibition of the formation of NETs relieves lung epithelial injuries. (A) The intracellular ROS levels in PMNs detected by flow cytometry with DCFH-DA fluorescein after 1 h of no/different treatments (LPS (1 μg/mL), LPS (1 μg/mL) + DPI (1 μM)). (B, C, and D) PMNs were untreated or treated with LPS (1 μg/mL), DPI (1 μM), LPS (1 μg/mL) + DPI (1 μM) for 4 h, respectively. CI response curves of A549 cells co-cultured with equal numbers (2.4 × 104/well) of above differently treated PMNs (B). Live-imaging of SYTOX Green-stained extracellular DNA released from the PMNs; Scale bar: 100 μm (C). Western blotting analysis of HiCt3 levels in the PMNs (D). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Supplementary video related to this article can be found at https://doi.org/10.1016/j.yexcr.2020.112101
The following are the supplementary data related to this article:
3.5. NETs mediate lung epithelial injuries
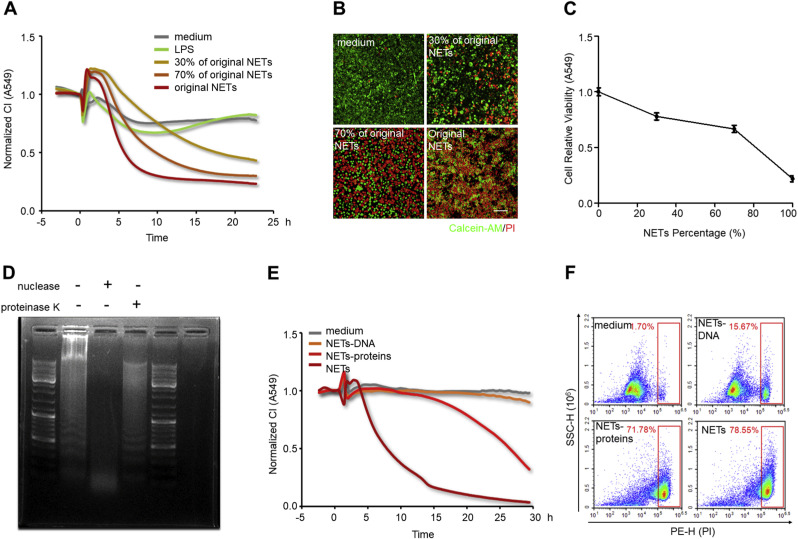
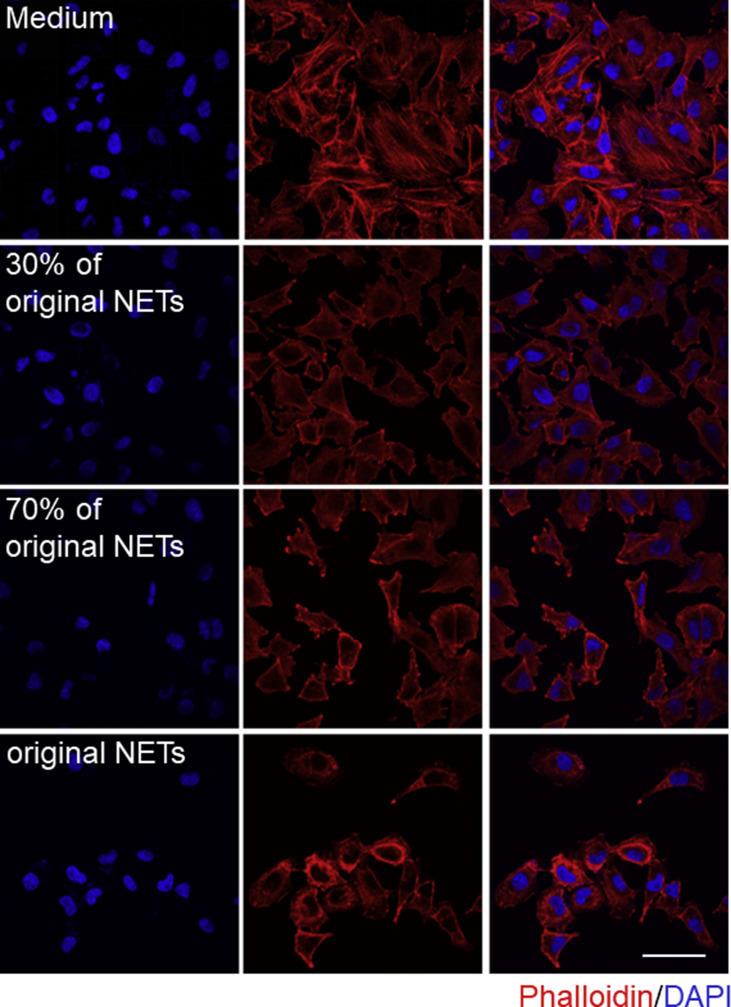
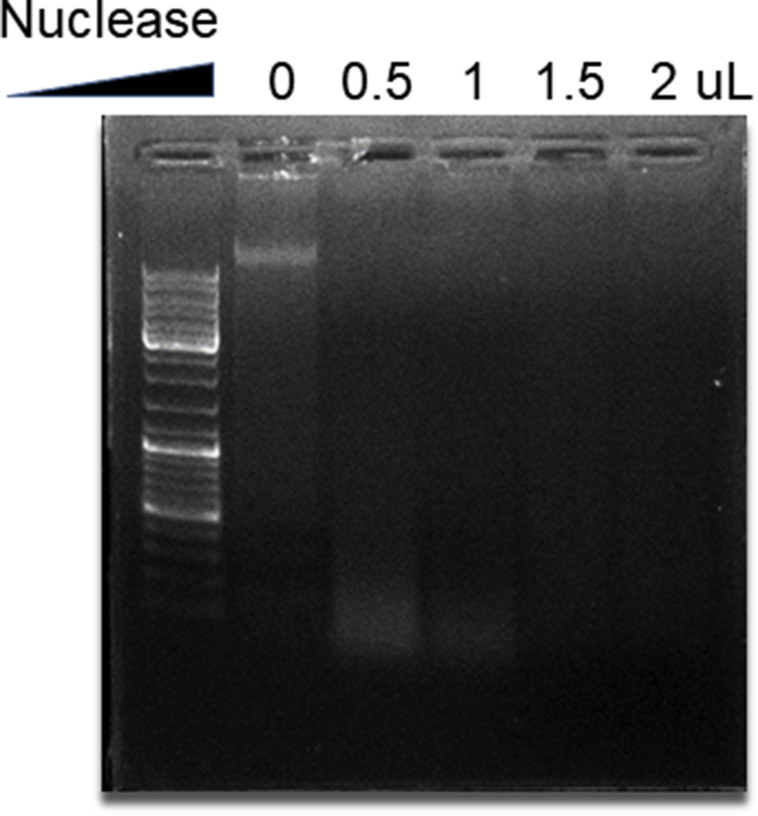
To ascertain the cytotoxic roles of NETs implicated in lung epithelial injuries, freshly isolated NETs were used to treat lung epithelial cells. A549 cells were incubated in media containing different proportions of NETs in the xCELLigence system. Epithelial CI values varied with NETs concentration; the higher the concentration of added NETs, the faster the lung epithelial CI decrease (Fig. 5 A). This result was also validated by immunofluorescence and Calcein-AM/PI double staining. After incubation with different concentrations of NETs, A549 cells exhibited progressive damage to their morphology (Fig. Supplemental Fig. S4) and an increased number of PI-positive cells and a decrease number of Calcein-AM-positive cells (Fig. 5B). Similarly, a Cell Counting Kit-8 assay used simultaneously consistently showed that NETs promoted lung epithelial apoptosis in a concentration-dependent manner (Fig. 5C). These results reinforce the fact that the cytotoxicity of NETting PMNs is primarily due to NETs formation. NETs are complicated structures of chromatin filaments studded with granular proteases [69]. To further pinpoint the prevailing cytotoxic components in NETs, freshly isolated NETs were dissociated into NETs-DNA and NETs-proteins after digestion with nuclease or proteinase K. The nucleic acids changed from heterogeneous fragments into small fragments after DNA hydrolyzation by nuclease, as shown in Fig. Supplemental Fig. S5. The minimum volume (1.5 μL) of nuclease completely removing nucleic acid was determined for follow-up experiments. After the protein components of NETs were obliterated by proteinase K, the smearing (negatively charged proteins impeding the movement of nucleic acids) around the sample hole disappeared (Fig. 5D). Subsequently, using the xCELLigence system, lung epithelial cells were incubated with different NETs components. CI decline was significantly induced by NETs-proteins, close to the effect of whole NETs (Fig. 5E). Meanwhile, flow cytometry revealed the same result: lung epithelial cell apoptosis was significantly enhanced when cells were treated by NETs-proteins compared to NETs-DNA, demonstrated by an increased percentage of PI-positive cells (Fig. 5F). In short, isolated NETs can notably cause lung epithelial injuries in a time and concentration-dependent manner, and protein components in NETs are the prevailing cytotoxic mediators.
Fig. 5.
NETs mediate lung epithelial injuries. (A, B, and C) A549 lung epithelial cells were exposed to NETs at 30%, 70% and 100% of the original concentration. CI response curves of A549 cells in xCELLigence system (A). Fluorescence staining of A549 cells co-dyed with Calcein-AM (green)/PI (red) after exposure to NETs for 10 h; Scale bar: 100 μm (B). The cell viability of A549 determined by CCK-8 after exposure to NETs for 10 h (C). (D) Representative analysis of NETs digestion by gel electrophoresis. From left to right: untreated NETs, NETs treated with nuclease, and NETs treated with proteinase K. (E and F) A549 cells were incubated with the medium, NETs-proteins, NETs-DNA, and NETs, respectively. CI response curves of A549 cells in xCELLigence system (E). The percentages of PI-positive A549 cells measured by flow cytometry after incubation with different NETs components for 24 h (F). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Gradient concentrations of NETs detected by the xCELLigence system correspond with NETosis Assay Kit measurement
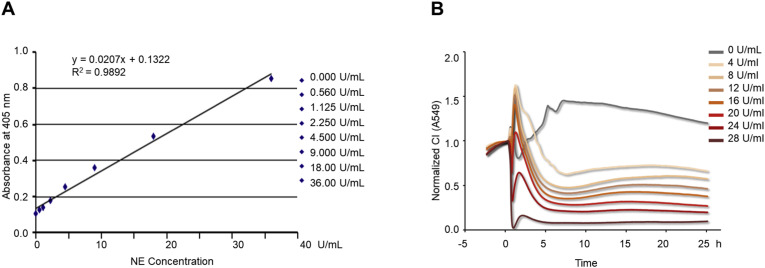
To further confirm the equivalence of the xCELLigence system with other NETs detecting methods, we compared our assay with the commercially available NETosis Assay Kit. The NE concentration in our isolated NETs was measured as 28 U/mL using the NETs standard curve (Fig. 6 A) plotted according to the manufacturer’s instructions. A series of NETs samples were then prepared by diluting the original NETs with RPMI 1640 medium, and the NE concentrations of these dilutions were measured as 24, 20, 16, 12, 8, and 4 U/mL using the same NETs standard curve. Subsequently, A549 lung epithelial cells were incubated with these different concentrations of NETs in the xCELLigence system. The lung epithelial CI values decreased in proportion to the NE concentrations in the NETs (Fig. 6B). This result suggests that the ability of the xCELLigence system to detect NETs by evaluating the decrease in CI corresponds well with the commercial NETosis Assay Kit measuring NETs by NE concentrations.
Fig. 6.
Gradient concentrations of NETs detected by xCELLigence system correspond with NETosis assay Kit measurement. (A) The plotted NETs quantitative standard curve instructed by the NETosis assay kit; OD at 405 nm (linear y-axis) versus NE concentration (linear x-axis) for standards. (B) CI response curves of A549 cells exposed to a series of NETs containing sequential NE concentrations in the xCELLigence system.
3.7. The xCELLigence system distinguishes NETs formation differences in PMNs between pneumonia patients and healthy controls
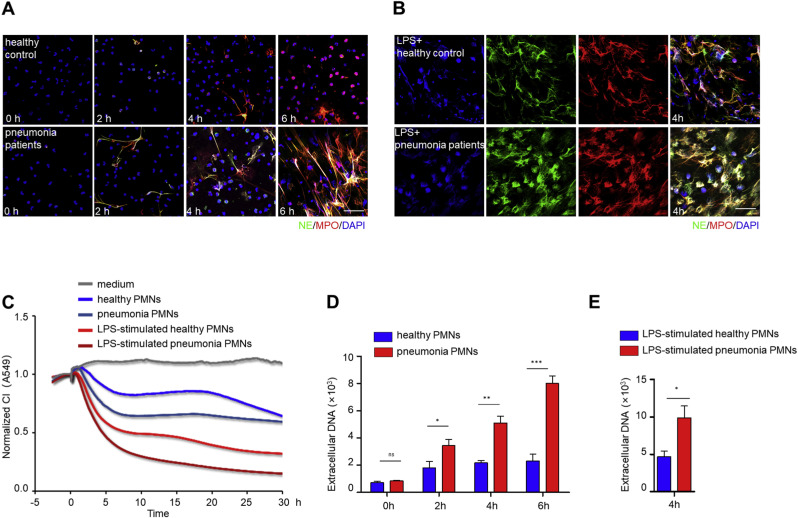
Accumulating evidence underscores the fact that most patients with severe pneumonia suffer from complications of ALI/ARDS [5,66,45]. In our experiments, PMNs were isolated from the peripheral blood of severe pneumonia patients and healthy controls. PMNs derived from pneumonia patients were more prone to generate NETs either spontaneously (Fig. 7 A) or by LPS stimulation (Fig. 7B), as determined by the co-localization of DNA with NE and MPO by immunofluorescence. Subsequently, these PMNs, either untreated or further activated by LPS stimulation, were added to A549 lung epithelial cells in the xCELLigence system. The resultant patterns revealed that the PMNs derived from pneumonia patients had a higher damaging capacity than those from healthy controls. When further stimulated by LPS, both kinds of PMNs were able to aggravate the lung epithelial CI declines in an additive manner (Fig. 7C). As an alternative validation, the fluorescent values of SYTOX Green-stained extracellular DNA were higher in pneumonia PMNs than in healthy control PMNs, whether incubated in the absence of LPS (Fig. 7D) or in the presence of LPS (Fig. 7E). These results indicate that the xCELLigence system is able to identify the potential spontaneity and high sensitivity of pneumonia PMNs to form NETs.
Fig. 7.
The xCELLigence system distinguishes NETs formation differences in PMNs from pneumonia patients and healthy control. (A and B) PMNs from severe pneumonia patients and healthy control were untreated or further stimulated with LPS (1 μg/mL), respectively. Immunofluorescence staining of NETs, visualized as co-localized extracellular DNA (blue), NE (green) and MPO (red); Scale bar: 50 μm. Spontaneous NETs formation captured at 0, 2, 4, 6 h (A). NETs formation after 4 h of LPS stimulation (B). (C) CI response curves of A549 cells co-cultured with equal numbers (1.2 × 104/well) of PMNs from severe pneumonia patients or healthy controls with or without LPS (1 μg/mL) stimulation in the xCELLigence system. (D and E) Fluorescent values of extracellular DNA stained with SYTOX Green. Time-course production of extracellular DNA without LPS stimulation (D). The production of extracellular DNA after 4 h of LPS (1 μg/mL) stimulation (E). The experiment was performed thrice, and the results are shown as mean ± SEM. **p < 0.01; *p < 0.05; ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.8. The xCELLigence system detects the alleviative effects of ASA and APAP on NETs formation
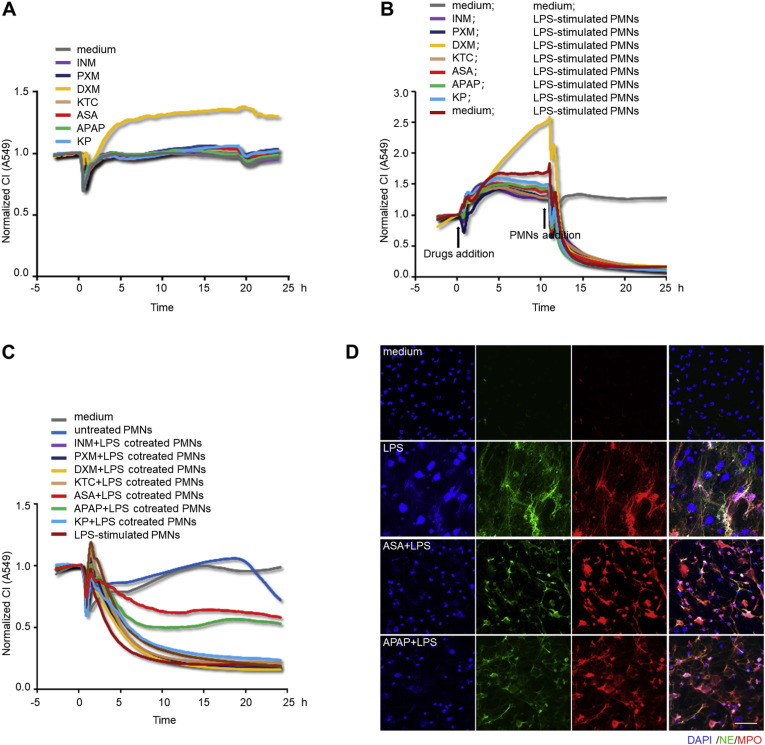
Since NETosis usually occurs in an inflammatory microenvironment, inflammatory stimuli are responsible for the formation of NETs [10,70,41,78,79], but in clinical practice, the application of anti-inflammatory drugs, especially dexamethasone (DXM), for the therapy of ALI/ARDS is still disputed [[50], [80], [81]]. In this study, we attempted to use the xCELLigence system to investigate the effects of common anti-inflammatory drugs during lung epithelial cells-PMNs interaction. Although there was an increase in CI with DXM, no CI variation was observed when lung epithelial cells were incubated with the other 6 kinds of nonsteroidal anti-inflammatory drugs (NSAIDs) (Fig. 8 A). Next, the lung epithelial cells were pre-incubated with the drugs for 10 h prior to PMNs addition, but a sharp decrease in CI inevitably emerged once NETting PMNs were added (Fig. 8B), indicating that the anti-inflammatory treatments were not enough to combat the destructive power of the already formed NETs. Finally, PMNs were pretreated with anti-inflammatory drugs prior to LPS stimulation, and these differently treated PMNs were co-cultured with lung epithelial cells severally in the xCELLigence system. As shown in Fig. 8C, ASA pre-treatment was most able to rescue the decline in lung epithelial CI, closely followed by APAP. This is consistent with the findings of Caudrillier et al. [24], Looney et al. [51], Zawrotniak et al. [52], and Lapponi et al. [82] In agreement with these findings, immunofluorescence staining verified the mitigative effects of ASA and APAP on NETs formation, showing the reduced severity of distorted and bulgy nuclei as well as diffused antimicrobial enzymes, compared to PMNs with single LPS stimulation (Fig. 8D). Altogether, these results suggest that this cell-based assay has potential application in screening potential NETs remission drugs.
Fig. 8.
The xCELLigence system detects the alleviative effects of ASA and APAP on NETs formation. (A) CI response curves of A549 cells incubated with the media containing the same concentration (1 μM) of NSAIDs (INM, PXM, KTC, ASA, APAP, KP) and DXM in the xCELLigence system. (B) CI response curves of A549 cells co-cultured with NETting PMNs (2.4 × 104/well), after 10 h of pre-incubation with the aforementioned anti-inflammatory drugs. The arrows represent the time points of drugs or PMNs addition. (C) CI response curves of A549 cells co-cultured with PMNs (2.4 × 104/well) that had been pre-incubated with the different anti-inflammatory drugs (1 μM) for 1 h prior to 4 h of LPS (1 μg/mL) stimulation. (D) Immunofluorescence staining of PMNs with 1 h of ASA (1 μM) or APAP (1 μM) pre-treatment prior to 4h of LPS (1 μg/mL) stimulation; NETs were identified by the extrusion of MPO (red), NE (green) and DAPI (blue); Scale bar: 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
NETs, the lattices of DNA filaments decorated with toxic histones and enzymes, have been recognized for their function in immobilizing and neutralizing microbial pathogens including bacteria [69] and viruses [13,14] during infections. However, uncontrolled and persistent NETs generation from activated PMNs can cause tissue injury in a plethora of inflammatory disorders. Accumulating reports have claimed that NETs are implicated in ALI/ARDS in humans [68,10,13] as well as in animal models [24,83,84]. Lately, clinical statistics of NCIP have revealed that critically ill patients with comorbidities and ARDS are at an increased risk of death [[1], [2], [3], [4]]. Additionally, concluded from the case reports with autopsy findings, the prominent pathological features in lung specimens of NCIP patients are the thick mucus obstructing small airways and diffuse alveolar damage [85]. Since NETs are highly viscous and there is widespread existence of NETs in infection-associated pulmonary diseases [15], it is reasonable to presume that NETs might be involved in the pathogenesis of NCIP. Therefore, it is helpful and profound to directly visualize the crosstalk of lung epithelial cells and activated PMNs or NETs.
In this study, we established a novel cell-based NETs detection method by means of the xCELLigence system. Using this assay, the pathological roles of NETs or NETting PMNs on lung epithelial injuries were detectable, and this damage was further clarified in a time and number/concentration-dependent manner. Although existing methods [69,23,36,57] have long been applied for detecting NETs, each of them has its disadvantages. For example, immunofluorescent staining is labor-intensive and time-consuming, and cell-free DNA quantification may be affected by contaminated DNA derived from necrosis. Hence, there is still no single universally accepted gold-standard. Here, in order to acquire bias-free results, several methods were used together to determine multiple aspects of LPS-stimulated formation of NETs. For example, NE/MPO antibody-based immunofluorescence and high-resolution SEM showed the morphological features of reticular chromatins studded with granular proteins; membrane-impermeable SYTOX Green-based extracellular DNA staining indicated cytomembrane rupture along with the extrusion of nuclei, and HiCt3 protein-based western blotting revealed the reliance of NETosis on histone 3 citrullination. The combination of these approaches not only avoided the pitfalls of a single approach but also reached a consistent conclusion that LPS could induce NETs formation and DPI inhibited it.
The common limitation of the aforementioned traditional methods is their incapability of detecting the pathological roles exerted by NETs in disease conditions firsthand. Our established cell-based NETs monitoring methodology remedied this shortcoming: the CI curves presented in the xCELLigence system reflected lung epithelial cell changes in response to the co-cultured PMNs or NETs. Our results demonstrated that LPS-activated PMNs, namely, NETting PMNs, whether derived from humans or mice, caused homologous lung epithelial injuries. The damage was also confirmed by immunofluorescence with phalloidin staining F-actin and an antibody labeling cleaved caspase-3, in combination with flow cytometry using PI fluorescein probing necrotic or apoptotic cells, which revealed lung epithelial cytoskeletal destruction and apoptosis/necrosis induced by NETs. In contrast, our method could notably detect the destructive roles of NETs holistically, especially evaluate NETs-induced lung epithelial damage in a time and number/concentration-dependent manner. Moreover, compared to the above end-point methods requiring antibody or fluorescein labeling, this innovative cell-based technology precluded the need for cellular labeling with noninvasive measurement, and allowed synchronous detection of multiple samples in a high-throughput manner.
Besides NETs formation, ROS, protease, and cytokines are also reported to be generated from activated PMNs [41,78,86]. To avoid overestimating the NETs effects in this co-culture setting, we incubated lung epithelial cells with PMNs conditioned culture medium (which might contain ROS, protease, cytokines, and a small amount of cell-free DNA) or activated NETting PMNs, which were separated from LPS-stimulated PMN suspensions by centrifugation. The lung epithelial CI decline, due to cell death, and cytoskeletal destruction, was exclusively induced by NETting PMNs. The inhibition of NETs formation using DPI partially but significantly reduced the lung epithelial CI decline. These findings suggested that NETs generation was the major cytotoxic factor accounting for lung epithelial injuries caused by LPS-stimulated PMNs. Additionally, our assay detecting NETs by monitoring the extent of lung epithelial CI decline had been shown to be equivalent in quantification with the widely used NETosis kit based on measuring NETs-derived NE concentration. This comparability lended further credibility and accuracy to this methodology. In our work, high concentrations of granular protein components in NETs were proved as the prevailing detrimental mediators, and the DNA backbone of NETs was almost non-cytotoxic to lung epithelial cells. The former corresponded with the existing data [59,60], however, the latter was contrary to the findings that in vivo DNase administration degrading DNA could significantly reduce the tissue injury and mortality in the mouse ALI models [24,87]. This paradox may be ascribed to the discrepancies resulting from the in vitro and in vivo experiments. NETs play cytotoxic roles due to the web-like fibrous structure of the DNA serving as a surface for granular protease aggregation and entrapment, facilitating enzymolysis [69,79]. However, in in vitro situations, even after DNA degradation, unbound proteases that remain concentrated in culture wells can cause similar lung epithelial injuries with enough time and concentration.
Reviewing our methodology, unexpectedly, lung epithelial CI decline also emerged in the late stage of co-culture with untreated PMNs, around 18 h after PMNs addition. PMNs are terminally differentiated cells with a life span of 12–15 h [88]. After this period, they undergo apoptosis or other forms of death, including necrosis [88]. Their life span can be appropriately extended after exposure to several cytokines [78,89]. These unexpected CI declines in the late stage were possibly caused by enzymes passively leaked from dead PMNs. It might not be easy to distinguish NETosis from late apoptosis and necrosis, as it has been claimed that NETosis is a type of necroptosis [64,90]. However, in this assay, the CI changes during the early co-culture period are the key points, in which violent NETosis is the dominant event.
In this study, we presented the utility of the xCELLigence system to detect the destructive effects of NETs on lung epithelial cells. Furthermore, this cell-based assay showed great superiority in distinguishing the formative potential of NETs in PMNs from severe pneumonia patients, and discovering putative therapeutics for NETosis alleviation. It has been analyzed that CRS and elevated PMN counts are the key points contributing to NCIP transformation from mild to critical cases [1,2]. Therefore, this work not only presents a new idea and method to evaluate NETs as well as NETs-associated ALI/ARDS in a dynamic, real-time, and quantitative manner, but also builds a solid foundation for exploring therapeutic interventions in the current clinical setting.
Financial support
This work was supported by grants from National Natural Science Foundation of China (Grant number 81970049) and National Major Scientific Instruments Research Project of China (Grant number 31627801).
Author contribution
Dandan Lv performed the majority of the experiments, but also analyzed and interpreted data and wrote the initial manuscript together with Yiming Xu. Yiming Xu performed additional experiments. Dandan Lv and Yiming Xu were responsible for cell isolation. Dandan Lv and Kejing Ying were responsible for patient blood samples collection. Hongqiang Cheng was responsible for the study, design, writing, analysis and interpreting data. Kejing Ying, Yuehai Ke and Xue Zhang were responsible for experimental designs. All authors contributed to writing and revising the manuscript. Xue Zhang, Ph.D. and Kejing Ying, Ph.D. contributed equally to the manuscript.
Declaration of competing interest
There are no conflicts of interest.
Acknowledgments
We would like to thank Ms. Shuangshuang Liu for instructing the application of Nikon A1R confocal microscope throughout the study. Thanks to Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yexcr.2020.112101.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Videos. Equal numbers of PMNs were untreated (A) or respectively treated with LPS (1 μg/mL) (B), DPI (1 μM) (C), and LPS (1 μg/mL) +DPI (1 μM) (D).
Supplemental Fig. S1.
The isolated purity of human peripheral blood PMNs labeled with Ly6G by flow cytometry.
Supplemental Fig. S2.
The polymorphonuclear morphology of isolated PMNs by Giemsa stain.
Supplemental Fig. S3.
High-resolution SEM micrograph of monolayer A549 lung epithelial barrier; Scale bar: 500 μm.
Supplemental Fig. S4.
Immunofluorescence staining of A549 cells co-stained with F-actin (phalloidin, red) and DNA (DAPI, blue) after incubation with different concentrations of NETs for 10 h; Scale bar: 50 μm.
Supplemental Fig. S5.
Representative analysis of DNA degradation efficiency after the NETs were treated with different volumes of nuclease by gel electrophoresis.
References
- 1.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellani G., Laffey J.G., Pham T. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 6.Libby L.J., Gelbman B.D., Altorki N.K. Surgical lung biopsy in adult respiratory distress syndrome: a meta-analysis. Ann Thorac Surg. 2014;98(4):1254–1260. doi: 10.1016/j.athoracsur.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Mikacenic C., Moore R., Dmyterko V. Neutrophil extracellular traps (NETs) are increased in the alveolar spaces of patients with ventilator-associated pneumonia. Crit Care. 2018;22(1):358. doi: 10.1186/s13054-018-2290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L., Liu L., Zhang Y. High level of neutrophil extracellular traps correlates with poor prognosis of severe influenza. J Infect Dis. 2018;217(3):428–437. doi: 10.1093/infdis/jix475. [DOI] [PubMed] [Google Scholar]
- 14.Cortjens B., de Jong R., Bonsing J.G. Local dornase alfa treatment reduces NETs-induced airway obstruction during severe RSV infection. Thorax. 2018;73(6):578–580. doi: 10.1136/thoraxjnl-2017-210289. [DOI] [PubMed] [Google Scholar]
- 15.Porto B.N., Stein R.T. Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front Immunol. 2016;7:311. doi: 10.3389/fimmu.2016.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An Z., Li J., Yu J. Neutrophil extracellular traps induced by IL-8 aggravate atherosclerosis via activation NF-κB signaling in macrophages. Cell Cycle. 2019;18(21):2928–2938. doi: 10.1080/15384101.2019.1662678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Buhr N., von Köckritz-Blickwede M. How Neutrophil Extracellular Traps Become Visible. J Immunol Res. 2016;2016:4604713. doi: 10.1155/2016/4604713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruschi M., Petretto A., Santucci L. Neutrophil Extracellular Traps protein composition is specific for patients with Lupus nephritis and includes methyl-oxidized αenolase (methionine sulfoxide 93) Sci Rep. 2019;9(1):7934. doi: 10.1038/s41598-019-44379-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavillet M., Martinod K., Renella R. Flow cytometric assay for direct quantification of neutrophil extracellular traps in blood samples. Am J Hematol. 2015;90(12):1155–1158. doi: 10.1002/ajh.24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W., Fogg D.K., Kaplan M.J. A novel image-based quantitative method for the characterization of NETosis. J Immunol Methods. 2015;423:104–110. doi: 10.1016/j.jim.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caudrillier A., Kessenbrock K., Gilliss B.M. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J Clin Invest. 2012;122(7):2661–2671. doi: 10.1172/JCI61303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginley B.G., Emmons T., Lutnick B. Computational detection and quantification of human and mouse neutrophil extracellular traps in flow cytometry and confocal microscopy. Sci Rep. 2017;7(1):17755. doi: 10.1038/s41598-017-18099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraaij T., Tengström F.C., Kamerling S.W.A. A novel method for high-throughput detection and quantification of neutrophil extracellular traps reveals ROS-independent NET release with immune complexes. Autoimmun Rev. 2016;15(6):577–584. doi: 10.1016/j.autrev.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Yan G., Sun W., Pei Y. A novel release kinetics evaluation of Chinese compound medicine: application of the xCELLigence RTCA system to determine the release characteristics of Sedum sarmentosum compound sustained-release pellets. Saudi Pharm J. 2018;26(3):445–451. doi: 10.1016/j.jsps.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun M., Fu H., Cheng H. A dynamic real-time method for monitoring epithelial barrier function in vitro. Anal Biochem. 2012;425(2):96–103. doi: 10.1016/j.ab.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Guan N., Deng J., Li T. Label-free monitoring of T cell activation by the impedance-based xCELLigence system. Mol Biosyst. 2013;9(5):1035–1043. doi: 10.1039/c3mb25421f. [DOI] [PubMed] [Google Scholar]
- 33.Moodley K., Angel C.E., Glass M. Real-time profiling of NK cell killing of human astrocytes using xCELLigence technology. J Neurosci Methods. 2011;200(2):173–180. doi: 10.1016/j.jneumeth.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto Y., Fukada Y., Mori D. Prostaglandin E2 stimulates granulocyte colony-stimulating factor production via the prostanoid EP2 receptor in mouse peritoneal neutrophils. J Immunol. 2005;175(4):2606–2612. doi: 10.4049/jimmunol.175.4.2606. [DOI] [PubMed] [Google Scholar]
- 36.Grootjans S., Hassannia B., Delrue I. A real-time fluorometric method for the simultaneous detection of cell death type and rate. Nat Protoc. 2016;11(8):1444–1454. doi: 10.1038/nprot.2016.085. [DOI] [PubMed] [Google Scholar]
- 37.Lood C., Blanco L.P., Purmalek M.M. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadesalingam A., Chen J.H.K., Farahvash A. Hypertonic saline suppresses NADPH oxidase-dependent neutrophil extracellular trap formation and promotes apoptosis. Front Immunol. 2018;9:359. doi: 10.3389/fimmu.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrientos L., Marin-Esteban V., de Chaisemartin L. An improved strategy to recover large fragments of functional human neutrophil extracellular traps. Front Immunol. 2013;4:166. doi: 10.3389/fimmu.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelaseyed T., Bretscher A. Regulation of actin-based apical structures on epithelial cells. J Cell Sci. 2018;131(20):jcs221853. doi: 10.1242/jcs.221853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weindl G., Naglik J.R., Kaesler S. Human epithelial cells establish direct antifungal defense through TLR4-mediated signaling. J Clin Invest. 2007;117(12):3664–3672. doi: 10.1172/JCI28115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck A., Sanchez Klose F.P., Venkatakrishnan V. DPI selectively inhibits intracellular NADPH oxidase activity in human neutrophils. Immunohorizons. 2019;3(10):488–497. doi: 10.4049/immunohorizons.1900062. [DOI] [PubMed] [Google Scholar]
- 45.De Freitas Caires N., Gaudet A., Portier L. Endocan, sepsis, pneumonia, and acute respiratory distress syndrome. Crit Care. 2018;22(1):280. doi: 10.1186/s13054-018-2222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raghavendran K., Pryhuber G.S., Chess P.R. Pharmacotherapy of acute lung injury and acute respiratory distress syndrome. Curr Med Chem. 2008;15(19):1911–1924. doi: 10.2174/092986708785132942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Looney M.R., Nguyen J.X., Hu Y. Platelet depletion and aspirin treatment protect mice in a two-event model of transfusion-related acute lung injury. J Clin Invest. 2009;119(11):3450–3461. doi: 10.1172/JCI38432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zawrotniak M., Kozik A., Rapala-Kozik M. Selected mucolytic, anti-inflammatory and cardiovascular drugs change the ability of neutrophils to form extracellular traps (NETs) Acta Biochim Pol. 2015;62(3):465–473. doi: 10.18388/abp.2015_1055. [DOI] [PubMed] [Google Scholar]
- 57.Rebernick R., Fahmy L., Glover C. DNA area and NETosis analysis (DANA): a high-throughput method to quantify neutrophil extracellular traps in fluorescent microscope images. Biol Proced Online. 2018;20:7. doi: 10.1186/s12575-018-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosmann M., Ward P.A. Protein-based therapies for acute lung injury: targeting neutrophil extracellular traps. Expert Opin Ther Targets. 2014;18(6):703–714. doi: 10.1517/14728222.2014.902938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saffarzadeh M., Juenemann C., Queisser M.A. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desai J., Mulay S.R., Nakazawa D. Matters of life and death. How neutrophils die or survive along NET release and is “NETosis” = necroptosis? Cell Mol Life Sci. 2016;73(11–12):2211–2219. doi: 10.1007/s00018-016-2195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes K.T., Beasley M.B. Pulmonary Manifestations of Acute Lung Injury: More Than Just Diffuse Alveolar Damage. Arch Pathol Lab Med. 2017;141(7):916–922. doi: 10.5858/arpa.2016-0342-RA. [DOI] [PubMed] [Google Scholar]
- 67.Shimabukuro-Vornhagen A., Gödel P., Subklewe M. Cytokine release syndrome. J Immunother Cancer. 2018;6(1):56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grégoire M., Uhel F., Lesouhaitier M. Impaired efferocytosis and neutrophil extracellular trap clearance by macrophages in ARDS. Eur Respir J. 2018;52(2):1702590. doi: 10.1183/13993003.02590-2017. [DOI] [PubMed] [Google Scholar]
- 69.Brinkmann V., Reichard U., Goosmann C. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 70.Barbu E.A., Mendelsohn L., Samsel L. Pro-inflammatory cytokines associate with NETosis during sickle cell vaso-occlusive crises. Cytokine. 2020;127:154933. doi: 10.1016/j.cyto.2019.154933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L., Mao Y., Xu B. Induction of neutrophil extracellular traps during tissue injury: Involvement of STING and Toll-like receptor 9 pathways. Cell Prolif. 2019;52(3):e12579. doi: 10.1111/cpr.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li R.H.L., Ng G, Tablin F. Lipopolysaccharide-induced neutrophil extracellular trap formation in canine neutrophils is dependent on histone H3 citrullination by peptidylarginine deiminase. Vet Immunol Immunopathol. 2017;193–194:29–37. doi: 10.1016/j.vetimm.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Wright T.K., Gibson P.G., Simpson J.L. Neutrophil extracellular traps are associated with inflammation in chronic airway disease. Respirology. 2016;21(3):467–475. doi: 10.1111/resp.12730. [DOI] [PubMed] [Google Scholar]
- 74.Yan G., Du Q., Wei X. Application of Real-Time Cell Electronic Analysis System in Modern Pharmaceutical Evaluation and Analysis. Molecules. 2018;23(12):3280. doi: 10.3390/molecules23123280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin Y., Tang X., Cao X. 4-((5-(Tert-butyl)-3-chloro-2-hydroxybenzyl) amino)-2-hydroxybenzoic acid protects against oxygen-glucose deprivation/reperfusion injury. Life Sci. 2018;204:46–54. doi: 10.1016/j.lfs.2018.04.056. [DOI] [PubMed] [Google Scholar]
- 76.Najmeh S., Cools-Lartigue J., Giannias B. Simplified human neutrophil extracellular traps (NETs) isolation and handling. J Vis Exp. 2015;98:52687. doi: 10.3791/52687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.El-Benna J., Hurtado-Nedelec M., Marzaioli V. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunol Rev. 2016;273(1):180–193. doi: 10.1111/imr.12447. [DOI] [PubMed] [Google Scholar]
- 78.Ley K., Hoffman H.M., Kubes P., Ley K, Hoffman HM, Kubes P Neutrophils: New insights and open questions. Sci Immunol. 2018;3(30):eaat4579. doi: 10.1126/sciimmunol.aat4579. [DOI] [PubMed] [Google Scholar]
- 79.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 80.Azoulay É, Canet E, Raffoux E. Dexamethasone in patients with acute lung injury from acute monocytic leukaemia. Eur Respir J. 2012;39(3):648–653. doi: 10.1183/09031936.00057711. [DOI] [PubMed] [Google Scholar]
- 81.Standiford T.J., Ward P.A. Therapeutic targeting of acute lung injury and acute respiratory distress syndrome. Transl Res. 2016;167(1):183–191. doi: 10.1016/j.trsl.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lapponi M.J., Carestia A., Landoni V.I. Regulation of neutrophil extracellular trap formation by anti-inflammatory drugs. J Pharmacol Exp Ther. 2013;345(3):430–437. doi: 10.1124/jpet.112.202879. [DOI] [PubMed] [Google Scholar]
- 83.Gan T., Yang Y., Hu F. TLR3 Regulated Poly I:C-Induced Neutrophil Extracellular Traps and Acute Lung Injury Partly Through p38 MAP Kinase. Front Microbiol. 2018;9:3174. doi: 10.3389/fmicb.2018.03174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sercundes M.K., Ortolan L.S., Debone D. Targeting neutrophils to prevent malaria-associated acute lung injury/acute respiratory distress syndrome in mice. PLoS Pathog. 2016;12(12):e1006054. doi: 10.1371/journal.ppat.1006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mantovani A., Cassatella M.A., Costantini C. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 87.Lefrançais E., Mallavia B., Zhuo H. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight. 2018;3(3):e98178. doi: 10.1172/jci.insight.98178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cabrini M., Nahmod K., Geffner J. New insights into the mechanisms controlling neutrophil survival. Curr Opin Hematol. 2010;17(1):31–35. doi: 10.1097/MOH.0b013e3283333b29. [DOI] [PubMed] [Google Scholar]
- 89.Colotta F., Re F., Polentarutti N. Mantovani A. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood. 1992;80(8):2012–2020. [PubMed] [Google Scholar]
- 90.Schreiber A., Rousselle A., Becker J.U. Necroptosis controls NET generation and mediates complement activation, endothelial damage, and autoimmune vasculitis. Proc Natl Acad Sci U S A. 2017;114(45):E9618–E9625. doi: 10.1073/pnas.1708247114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.