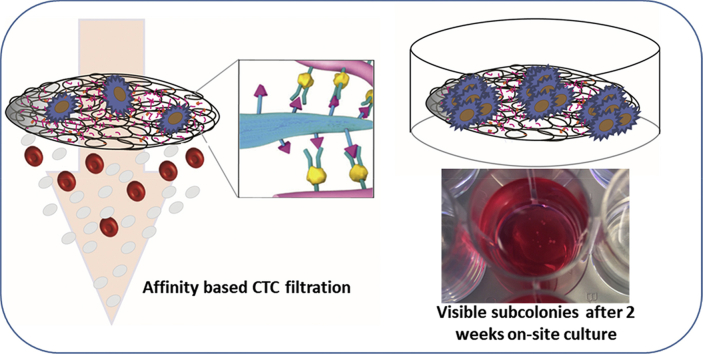
Abstract
Metastasis is the major cause of death in cancer patients accounting for about 90% of the mortality. The detection and analysis of the hallmark of metastasis, circulating tumor cells (CTCs), have significant impact in cancer biology and clinical practice. However, the scarcity of CTCs in blood, particularly in that of colorectal cancer patients, is a serious bottleneck in the development of CTC-based precision medicine. Herein, the melt electrowriting (MEW) technology was used for reproductive fabrication of a biocompatible antibody-presenting polycaprolactone filter with tailored porous structure. It is demonstrated, for the first time, that such filter can be used not only to catch cancer cells spiked in whole blood but also to culture the cancer cells directly on site. Specifically, HT29 colon cancer cells can be captured with an efficiency of 85%, and when spiked into 4 mL of whole blood, 47% were captured on one Ø12mm filter. Furthermore, repeated capture and culture experiments have shown that as few as 20 HT29 colon cancer cells spiked into 4 mL of whole blood can be captured on the filter and within 2 weeks be expanded on site to become tumor bodies that are visible to the untrained eye. This filter allows for downstream analysis, such as flow cytometry, immunocytochemistry, Western blotting, and rt-qPCR. This technology represents a simple and cost-effective platform that potentially enables fast and efficient culture of rare CTCs from patients’ blood. This provides non-invasive alternatives for solid biopsy tumor materials for treatment screening, with great potential to realize precision medicine for cancer treatment.
Keywords: Circulating tumor cells, Melt electrospinning writing, On-site culture, Polycaprolactone, Bioconjugation, Colorectal cancer
Graphical abstract
1. Introduction
At present, late stage colorectal cancer is associated with poor survival rates, and colorectal cancer is one of the leading causes of cancer-related deaths [1]. When colorectal tumors are diagnosed at a late stage, there are often limited options in terms of curative surgery and chemotherapy. The tumor heterogeneity leads to complexity and significant patient-to-patient differences in treatment response is a pivotal challenge [2]. Circulating tumor cells (CTCs) might be one of the avenues to overcome these challenges [3,4]. Several studies have shown that stable CTC cell lines can be established as 2D cultures, and such cell lines might extend our understanding of the CTC biology and the underlying mechanisms that drive cancer dissemination [5]. However, it can be difficult to establish CTC-derived cell lines of colorectal cancer patients because of their extreme rareness, one CTC among a billion hematopoietic cells [6].
Tremendous effort has been directed toward exploring the diagnostic and prognostic potential of CTCs. To facilitate their isolation and expansion, Zhang et al. recently demonstrated efficient chip-based CTC capture and subsequent fast expansion in three-dimensional (3D) co-cultures that can be used for downstream analysis and drug testing [7]. Other groups have followed, and it appears that CTC capture and culture via microtechnologies and nanotechnologies have gained momentum [3,8]. A wide range of CTC isolation technologies have been developed, using different principles such as immunoaffinity, size-based capture, and inertial focusing for CTC enrichment [[9], [10], [11]].
Electrospinning is a robust nanofiber-producing technique, where viscous liquids made of virtually any polymers, supramolecules, or composites can be extruded and elongated as submicron fibers under an electric field. Fibers in the nanometer and submicron scale have an enormous variety of applications, which is rooted in their ultrahigh surface area. Only to name a few, applications of such fibers include catalysis, sensors, and filtration [12]. Their overall extracellular matrix-mimicking structure together with their capacity for encapsulation or bioconjugation of biomolecules has within decades made them attractive in both tissue engineering and cancer modeling [13]. It has previously been demonstrated that electrospun scaffolds can be utilized for isolation of CTCs. One example can be seen in the work by Zhang et al. in which spiked gastric and colon cancer cells were captured on anti-EpCAM–conjugated TiO2 electrospun scaffolds incubated with a 1 mL target cell suspension. They furthermore demonstrated a few examples of CTC capture from cancer patient blood [14]. Ma et al. reported the use of 3D polystyrene (PS) fibrous scaffolds to capture spiked MCF-7 breast cancer cells by incubating anti-EpCAM–conjugated PS scaffolds with the cells for 30 min [15]. In 2017, Xu et al. published their study on the use of 2D hyaluronic acid–functionalized poly lactic-co-glycolic acid (PLGA) nanofibers embedded in a microfluidic system for the capture a variety of cancer cells [16]. Additionally, a binary blend of polyethylene oxid (PEO) and nylon-6 has been utilized by Lee et al. for the capture of CTCs from colorectal cancer patients by incubating drops of samples with their binary blend fibers [17]. However, none of those studies has used the fibers as a filtration device nor demonstrated the on-site culture capacity.
The above-mentioned studies all used conventional solution electrospinning, where the stacking of fibers is intrinsically random because of the relatively high conductivity of polymer solutions. Comparing with solution electrospinning, melt electrowriting (MEW) of polymer melts with lower conductivity is distinguished by the possibility to produce highly defined tailored architectures using computer-aided programming [18]. This improves the reproducibility of the filter with both tailored porosity and transparency which facilitate microscopy analysis at clinics [3,5,19].
Here, we present a novel MEW microfibrous filter made of polycaprolactone (PCL) that enables size- and immunoaffinity-based capture and on-site culture of EpCAM-positive cancer cells. Scanning electron microscopy (SEM) was used to characterize the filter structure. Fluorescent imaging and enzyme-linked immunosorbent assay (ELISA) were used to analyze anti-EpCAM antibody bioconjugation. The capture efficiency of HT29 colon cancer cells spiked in milliliters of whole blood was assessed at different flow rates using immunocytochemistry staining. Captured cells were subsequently cultured in situ on the filter, where a few cells can be expanded within 2 weeks to subcolonies that are visible to the naked eye. Further down-stream analysis, such as Western blot, flow cytometry, qPCR, and fluorescence microscopy was also demonstrated.
2. Materials and methods
2.1. Culture of cells
HT29 and NIH3T3 were maintained in complete growth medium, Dulbecco's Modified Eagle's Medium (DMEM, Sigma, cat no. D6046) holding 10% fetal bovine serum (FBS, Gibco, cat no. 10082-147) and 1:100 diluted Pen-strep (LONZA, cat no. 17-602E). The cells were passaged when 80–100% confluent and cultured at 37°C, 95% humidity, and 5% CO2. Detachment of cells was done by trypsination as outlined by manufacturer (LONZA, cat no. BE17-161E). Dulbecco's phospate buffered saline (DPBS, Sigma, D8537) was used for washing cells in between the steps of passaging the cells.
2.2. Melt electrospinning writing and preparation of filter
Melt electrospinning writing (CAT000111, Spraybase) was performed to print the filters. PCL (Mw = 45 kDa, Sigma-Aldrich) was loaded in a grounded stainless steel syringe and heated to 80°C. Then a gas pump was used to supply air pressure of 0.5 bar for extruding the melted PCL polymer out from the syringe connected with a blunt-end stainless steel needle. An automated plate collector was placed under the needle and connected to a high voltage of 3.5 kV. Movement of the collector was precisely controlled by UCCNC software. The distance between the needle and collector was 4 mm. Coiled fibers were first printed as the bottom layer at 400 mm/min using a 0.3 mm in diameter needle. The relatively low speed facilitated coiling of the fibers. The spacing was 100 μm. The coiled fibers were printed in two perpendicular directions and in their two diagonal directions with the same spacing distance. This formed a porous monolayer membrane of coiled fibers. Afterward, a grid frame composed of bigger straight fibers for stabilizing the underneath membrane was printed on top of the bottom layer at a 2,000 mm/min. The top layer needle had a diameter of 0.55 mm, and the spacing was 300 μm. The top grid frame was prepared by printing straight fibers in two perpendicular directions.
After electrospinning, filters were transferred to a 70% EtOH bath from where they were mounted to glass slides and with a scalpel cut to fit the filter holders (Millipore, cat no. SX0001300).
2.3. Scanning electron microscopy
The structure and surface morphology of the PCL fibrous scaffolds were characterized by scanning electron microscope (Hitachi TM3030).
2.4. Bioconjugation of a biotinylated anti-human EpCAM antibody to the filters
If the filters were used for culture, they were placed in a drop of 70% ethanol before bioconjugation, and the ethanol was evaporated in a sterile laminar flow bench. A UV source (around 260 nm) of a sterile bench was then used for additional sterilization for more than 1 h. All buffers were autoclaved or sterile filtered (0.2 μm).
All conjugation steps were performed at room temperature. The anti-EpCAM antibody was conjugated to a filter via polydopamine coating [23]. In short, the filter was kept on glass slides (VWR, cat no 631-1551) within an area encircled using a liquid blocker super PAP pen (Fisher scientific, cat no. NC 9827128). Dopamin hydrochloride (Sigma, cat no. H8502) was added (2 mg/mL in 500 μL 10 mM Tris-HCL, pH 9) to the filter for minimum 30 min, making sure that the filter was soaked with the dopamine solution hereby creating a polydopamine surface. Next, the filter was washed four times in 400 μL sodium phosphate buffer (50 mM, pH 7.8). Streptavidin (15 μg/mL) was then conjugated to the polydopamine layer for minimum 45 min in sodium phosphate buffer. The filter was washed four times in phosphate buffered saline (PBS, Sigma, cat no. P4417), before transferring the filter to a fresh glass slide. Here, the filter was incubated with 5 μg/mL of biotinylated anti-EpCAM antibody (RnD Systems, BAF960) in 1% bovine serum albumin in PBS for more than an 1 h. Finally, the filter was washed in PBS four times.
2.5. Immunocytochemistry
In general, scaffolds and cells were stained on VWR glass slides (cat no. 631-1551) or on cover slides (VWR, cat no. 631-0138). The cells or scaffolds were placed in an encircled area by using the aforementioned PAP pen to create a hydrophobic barrier. Next, samples were washed in DPBS or PBS and fixed in 4% formaldehyde (AppliChem, cat no. A3813), followed by four times rinsing in PBS. The cells were then permeabilized and blocked in BD Perm/Wash buffer (BD Biosciences, cat.554723) for 10 min, before adding primary antibodies or phalloidin alexa fluor 488 (ThermoFisher Scientific, cat 12379) in Perm/Wash buffer. The primary antibodies mouse monoclonal anti-pan cytokeratin (ab86734, Abcam) and rabbit recombinant anti-CD 45 (ab40763, Abcam) were used in 1:200 and 1:100 ratios, respectively, and incubated 2 h at room temperature or overnight at 4°C. Phalloidin alexa fluor 488 was used in 1:100 ratio for 1 h staining and 1:400 for overnight staining. Next, washing was performed three times, 3 min each, in 400 μL Perm/wash buffer. Finally, the secondary antibodies anti-mouse and anti-rabbit alexa fluor 488/594 (Abcam, cat no. ab150105, ab150076) was used 1:400 in Perm/Wash buffer for 1 h at room temperature. The washing was repeated. Hoechst 33342 (Life Technologies, cat no. H3570) was then used 1:10,000 in PBS for 5 min, the sample was rinsed, gently desiccated, and mounted with aqueous mounting media (Sigma, cat no. F4680). Staining was analyzed in EVOS FL Auto Imaging system or Zeiss confocal laser scanning microscopy (LSM 700 and 780).
2.6. Cell seeding on scaffolds and cell counting kit-8 measurements
Before cell culture, all filter scaffolds were conjugated to the anti-EpCAM antibody under sterile conditions. HT29 cells were seeded to the scaffolds in 20 μL media in a TC 96-well culture plate (SARSTEDT, cat no. 83.3925.500) in different densities (100, 200, and 400 cells per scaffold, four replicate for each seeding density). After 1 h, additional 180 μL media were added to the scaffolds. The next day (day 1), each scaffold was transferred to new wells, fresh media was added, and the scaffolds were left there until day 5 where the cell viability was assessed by using the cell counting kit-8 as outlined by the manufacturer (Dojindo Molecular Technologies). The cell counting kit-8 was applied again on day 15, and each time the measurements were performed in new wells to rule out any signal from cells not growing directly on the scaffolds. After finishing the cell counting on day 15, the scaffolds were rinsed in PBS, and the presence of cells was confirmed by immunocytochemistry using the anti-pan cytokeratin antibody and Hoechst stain as outlined above.
2.7. Prestaining HT29 with cell tracker dye
Approximately 5 million HT29 cells were washed twice in DPBS. Cells were centrifuged at 300 g for 3 min each time. The cells were re-suspended in DMEM without fetal bovine serum but holding 1 μM CellTracker Red CMPTX dye (ThermoFisher, cat no. C34552). After 30 min in the CellTracker solution at 37°C, the cells were washed twice in complete growth media and kept in media until further use.
2.8. Processing blood samples
Eight milliliters of blood from volunteers from our research group were drawn into EDTA-coated tubes. The blood was used according to the National Ethical Guidelines for quality control and quality assurance (guideline no. 11052, July 2, 1999). The blood was processed within 6 h. Lysis of red blood cells (RBCs) was performed by mixing 4 mL blood with 40 mL lysis buffer (155 mM NH4Cl, 12 mM NaHCOO3, 0.1 mM EDTA), it was incubated for 10 min at room temperature followed by centrifugation (5 min, 1,000 g). If necessary, the cells were resuspended, and the centrifugation was repeated. Without disturbing the pellet, the supernatant was gently aspirated, and PBS was added to total a volume of 1.5 mL.
2.9. Spike-in experiments
For whole-blood samples with non-lysed RBCs, 200 prestained HT29 were added to 1 mL whole blood and filtrated. For samples where RBCs were lysed, 200 prestained HT29 cells were added to 4 mL whole blood, and thereafter, the RBCs were lysed as described above. The flow rate was 0.5 mL/h. For spike-in experiments concerning culture after capture, 20 or 200 HT29 cells were spiked into 4 mL whole blood. The blood was subsequently treated with RBC lysis buffer, and the nucleated cells were filtered.
2.10. The filtrations
A filter was transferred to a sterile (if needed for culture) water bath, and the floating filter could be mounted from underneath directly to the bottom part of a filter holder. The top part of the filter holder was gently attached to the bottom part and through a tubing connected to a 20 mL syringe placed in a syringe pump (World Precision Instruments, cat no. 941-371-10003) (refer to figure 1). Next, the filter holder was carefully filled with 10% FBS in PBS, taking care that air was not trapped above the filter. The filter was left like this for 10 min before commencing the filtration. A 3 mL syringe was then connected to the top of the filter holder, and the sample was gently loaded to this syringe. The syringe pump was set to withdraw mode. Just before the entire sample had run through the filter, additional 300 μL PBS were added for washing. The immunocytochemistry was performed directly on the filter holder, and the cells could be visualized while still being placed on the filter holder by gently placing the bottom part of the filter holder directly on top of a cover slide.
2.11. Cell lysing and Western blotting
Four hundred HT29 cells per scaffold were seeded. Each scaffold was conjugated the anti-EpCAM antibody. The cells were cultured for 28 days, where after the cells, still on the scaffolds, were washed in DPBS 2–3 times and could be kept at −20°C until further use. To lyse the cells on the scaffolds, 100 μL per scaffold of M-PER solution (Thermo Scientific, cat no. 78503) were used. After 10 min incubation at room temperature, the lysates were centrifuged 15 min at 14,000 g. The supernatant was harvested. To prepare samples for sodium dodecyl sulfate-polyacryl amide gel electrophoresis (SDS-PAGE), 80 μL of each sample were mixed carefully with 20 μL XT sample buffer (BIORAD, cat no. 161-0791) and 6 μL XT reducing agent (BIORAD, cat no. 161-0792). The samples were left at 90°C for 5 min. Next, 20 μL of each sample were loaded to a 12% XT Bis-tris gel (BIORAD, cat no. 3450118) and run at 200 V for 50 min. Proteins were blotted to a nitrocellulose membrane by using the turbo-blotting system of BIORAD (Trans-Blot Turbo). The membrane was blocked 45 min in 3% bovine serum albumine in PBS (BSA-PBS) at room temperature on a rolling table. The membrane was then probed with anti-beta actin antibody (cat no. ab8227, Abcam) or the anti-EpCAM antibody diluted 1:1000 in 7 mL 2% BSA-PBS and incubated overnight at 4°C. In the first and subsequent washing steps, the volume used was 10–15 mL per wash. The membrane was rinsed twice and washed in 3 × 5 min in PBS. The anti-beta actin antibody was detected by using anti-rabbit IgG Cy3 conjugate (Sigma Aldrich, cat no. C2306) 1:1000 in 7 mL 2% BSA-PBS for 2 h at room temperature. The anti-EpCAM antibody was detected with streptavidin alexa fluor 488 1:1000 in 7 mL 2% BSA-PBS for 2 h at room temperature. The membrane was then rinsed once in PBS, washed 4 × 5 min in PBS-tween20 (0.05%), washed 2 × 5 min in PBS, and finally rinsed in PBS. To detect the signal, we used a typhoon scanner (Amersham), which was set for 200 μm resolution and PMT voltage to 500.
2.12. Flow cytometry
HT29 cells were seeded on anti-EpCAM conjugated scaffolds (1,000 cells/filter) and cultured for 10 days in a 96-well culture plate before harvesting the cells from the scaffolds by using trypsin. Cells from one scaffold were then incubated with 10 μg/mL of the anti-EpCAM antibody in PBS for 30 min at room temperature, washed once in PBS, and centrifuged at 400 g for 5 min. The anti-EpCAM antibody was detected by using 1:100 diluted streptavidin alexa fluor 488(Thermofisher, cat no. S11223) for 25 min at room temperature. Washing was repeated, the cells were resuspended in 300 μL PBS, and the flow cytometry was performed on a SONY SH800 Cell Sorter. Non-stained cells detached from a separate scaffold was used as a negative control.
2.13. RNA isolation and cDNA synthesis
Total RNA was extracted from each individual filter using the miRCURY RNA isolation Kit—Tissue (Exiqon) according to the manufacturer's protocol (Proteinase K digestion was omitted). The RNA concentration, including the A260/280 and A260/230 ratios, were determined using a Nanodrop 2000 (ThermoFisher Scientific). The isolated RNA from filter 1 (10.5 ng/μl) and filter 2 (15.2 ng/μl) were then reverse transcribed with the ExiLERATE LNA qPCR—cDNA synthesis kit (Exiqon), according to the manufacturer's instructions.
2.14. Quantitative real-time PCR
The qPCR reactions were prepared using the ExiLERATE LNA qPCR—SYBR Green master mix (Exiqon), cDNA template, and LNA primer sets against human phosphoglycerate kinase 1 (PGK1) and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Exiqon) according to the manufacturer's instructions. Each reaction was performed at a final volume of 10 μL. The reactions were run on a Stratagene Mx3000P (Agilent Technologies) with the following thermal profile: 95°C for 30 s; 40 cycles at 95°C for 10 s and 60°C for 1 min; and 55°C for 30 s and 95°C for 30 s. Dissociation curves and amplification plots were analyzed with the MxPro qPCR software (Agilent). No amplification was observed for no template controls and no primer controls confirming primer and target amplification specificity.
2.15. Culture after capture
Three filtrations were performed to capture 200 HT29 cells: two non-spiked samples (200 HT29 cells each) and one spiked sample (200 HT29 in 4 mL blood). Moreover, additional filtrations were independently performed on three spike-in samples, each having 20 HT29 cells spiked into 4 mL blood. After filtration, the filters were gently rinsed in PBS and transferred to complete growth medium and cultured in a 24-well TC plate (SARSTEDT, cat no. 83.3922) at 37°C, 95% humidity and 5% CO2, this was considered as day 1.
The next day, the filters were gently rinsed in DPBS and transferred to new wells holding fresh medium. The transfer step was repeated on day 5 and 12. At day 21, the culture was ended, and the filters were rinsed in DPBS and fixed in 4% formaldehyde for 10 min. Finally, the filters were stained for CD45 and pan-cytokeratin and evaluated by fluorescence microscopy.
2.16. Analysis of cancer cell cluster thickness, three-dimensional growth, and area analysis
Confocal imaging was performed using Zeiss LSM780. The following excitation/emission parameters were used for generating composite images: (i) Hoechst 33342 - 405/415-502 nm, (ii) Alexa Fluor 488 - 488/500-571 nm, (iii) Alexa Fluor 594 - 561/615-696. Fluorescence of Hoechst and Alexa Fluor 594 was acquired simultaneously. Maximal intensity projections and color-coded depth projections were generated using Zeiss ZenBlue software. In the color-coded projection, point 0 refers to the most shallow focal point. 3D projections were generated using Fiji ImageJ [43] using attached macro (supplementary methods). Resulting animation frames were then rearranged using Adobe Photoshop CC 2015 into final projections (supplementary animation). The determination of the area of HT29 subcolonies was performed using ImageJ. Stitched 8-bit images were first cropped and converted to binary images by adjusting the threshold. As a scale reference, the stabilization frame of the filter with dimensions of 300 × 300 μm was used. Subsequently, the analyze particle tool in ImageJ was applied to determine the area of particles (cells). In the co-culture study (Fig. s5), particles with an area less than 36 μm2 were not analyzed to avoid measurement of visual artefacts. The analysis was blinded.
2.17. Statistics
Students t-test, two-tailed and equal variance. Groups were considered different for p-values < 0.05.
3. Results
3.1. The filter fabrication and bioconjugation
The MEW technique was used to print defined patterns of a microcoiled MEW filter. The MEW filter holds 30 fiber coils per mm, which were written in the x and y directions and in two diagonal directions with 100 μm spacing.
Previously, PCL dissolved in chloroform (CHCl3) has been electrospun into biocompatible microfibers by electrospinning [20]. On the other hand, PCL dissolved in a mixture of dichloromethane (DCM) and N,N-dimethylformamide (DMF) has been extruded to nanofibers by electrospinning [21]. In parallel, filters using similar approaches (Supporting method) were produced for comparison, namely a microfibrous CHCl3 filter and a nanofibrous DCM/DMF filter. As a final step, a frame for stabilization was printed on top of all three types of filters in the x and y directions with 300 μm spacing (Fig. 1c).
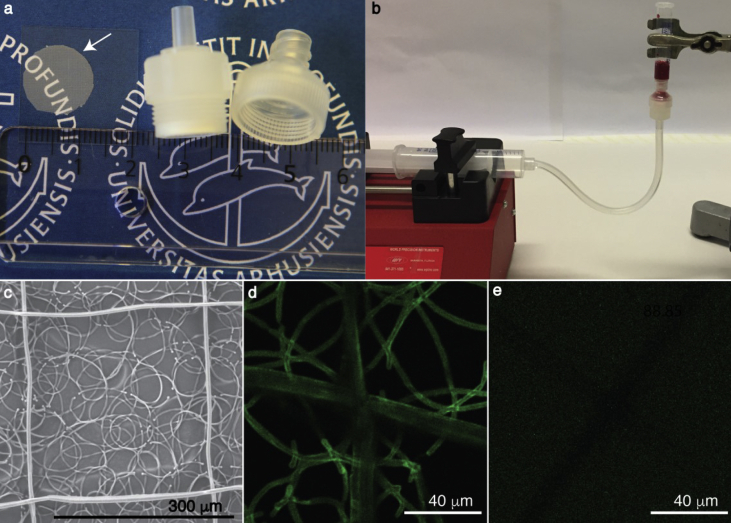
Fig. 1.
The filtration setup, MEW filter structure, and bioconjugation. (a) The MEW filter, marked with a white arrow, with diameter of Ø12 mm is transparent and can readily be mounted to standard filter holders. (b) The filtration setup consists of a syringe pump connected through a tubing to the filter holder and of a syringe attached on top of the filter holder, where the sample is loaded. (c) Scanning electron microscopy (SEM) image of the MEW filter. Scalebar, 300 μm. (d) The confocal microscopy image of MEW filter conjugated with a biotinylated anti-EpCAM antibody, which was detected by streptavidin alexa fluor 488 (green). The image was acquired, at 10% laser power. (e) A non-conjugated MEW filter was incubated with streptavidin alexa fluor 488 as a negative control. The laser power was set to 100%. MEW, melt electrowriting.
Characteristics of the three filter types were determined by using the DiameterJ plugin for imageJ (Supplementary Table s1 and Fig. s1). The average diameters and standard deviations of the DCM/DMF filter, CHCl3 filter, and MEW filter were 0.9 ± 0.2 μm, 7.7 ± 2.5 μm, and 3.3 ± 0.45 μm, respectively. The average pore sizes and standard deviations of the DCM/DMF, CHCl3, and MEW filters were 7.4 ± 6.3 μm2, 112.8 ± 118.9 μm2, and 141.5 ± 146.8 μm2, respectively.
As there are billions of blood cells and only a few CTCs in the patient blood, it is beneficial to decrease the amount of captured background blood cells to ease downstream culture and analysis. Initial pore size-based filtrations were performed on white blood cells using the three different filters, where the MEW filter captured fewer white blood cells compared with the CHCl3 filter, and much fewer than that on the DCM/DMF filter (Fig. s1), because of the significantly smaller pore sizes of the CHCl3 and DCM/DMF filters.
Although solution electrospinning offers faster fiber production, challenges with reproducibility of the DCM/DMF filters were encountered, because of the intrinsic chaotic nature of solution electrospinning. The MEW filter was thus chosen to test CTC capture and in situ culture. This filter is transparent, and it can be used with standard filter holders (Fig. 1a), which can be easily connected to syringes and pumps for flow-rate controlled filtration (Fig. 1b).
In terms of isolating CTCs of an epithelial origin, the EpCAM surface molecule has become a golden standard [22]. Accordingly, anti-EpCAM antibody bioconjugation on the MEW filter was performed via a polydopamine-based conjugation of streptavidin, followed by a biotinylated anti-EpCAM antibody to coat each single MEW fiber [23]. The conjugation was successfully performed in 3–4 hours, where the biotinylated antibody was readily detected with streptavidin alexa fluor 488 (Fig. 1d), as opposed to a non-conjugated filter, where no signal of streptavidin alexa fluor 488 was detected (Fig. 1e).
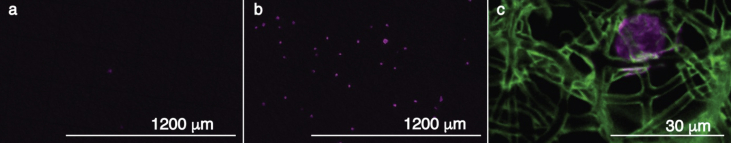
To assess the function of the anti-EpCAM bioconjugation, a non-conjugated MEW filter was compared with an anti-EpCAM–conjugated MEW filter, through which 2,000 prestained HT29 cells were filtrated at 2 mL/h. Clearly, the conjugation of the anti-EpCAM antibody improved capture efficiency of the HT29 cells (Fig. 2).
Fig. 2.
Filtration of 2,000 HT29 cells on filters with and without anti-EpCAM bioconjugation. (a) A representative image of a non-conjugated filter after filtration. (b) A representative image of an anti-EpCAM conjugated filter after filtration. Prestained HT29 cells are shown in magenta. (c) A zoom-in image of HT29 cells (magenta, stained for cytokeratin) captured on the conjugated filter (green, stained with streptavidin AF488).
The conjugation efficiency relies on the formation of a polydopamine layer, and the conjugation of the streptavidin to the formed quinone groups of the polydopamine [23]. To make a rough assessment of the amount of antibody bound to the filter surface, ELISA was used to detect any unbound antibody after performing the conjugation (Fig. s2). By calculation, approximately 11,200 anti-EpCAM molecules were bound for every squared micrometer.
3.2. The filter biocompatibility and down-stream analysis
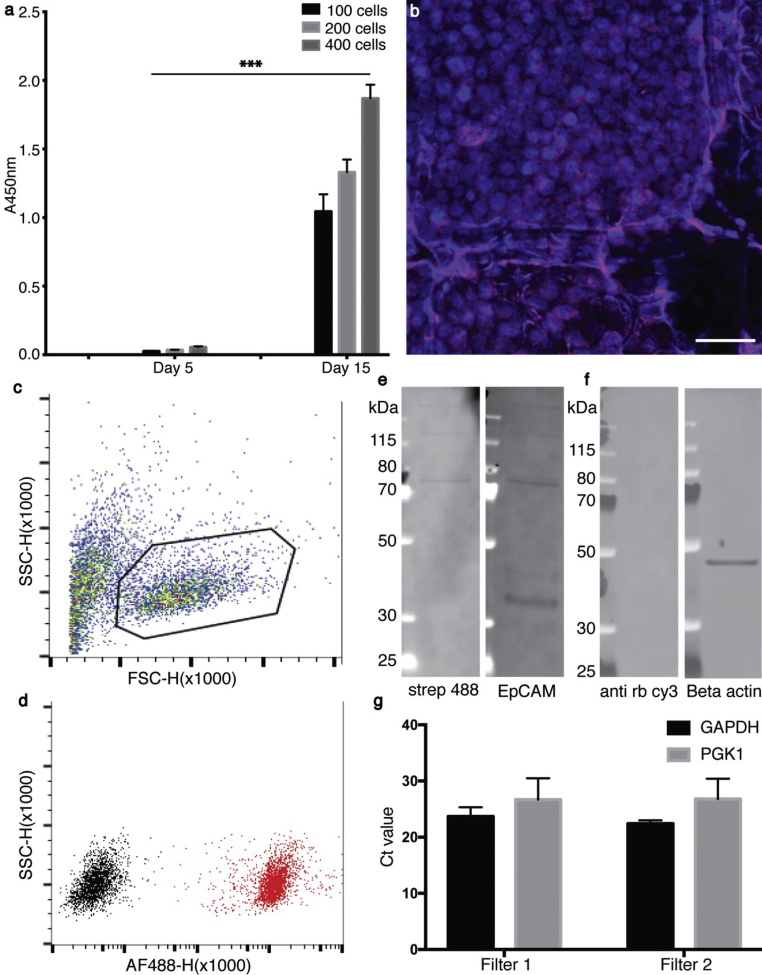
As CTCs are rare in blood and as we aimed for a scaffold that enables culture of these rare events, we assessed whether a few cells could be expanded on the anti-EpCAM–conjugated MEW filter by seeding 100, 200, and 400 HT29 cells on the MEW filter of Ø = 6 mm. The cells were successfully expanded on the MEW filters within 2 weeks, as shown by CCK-8 assay measurements, immunochemical staining, and imaging (Fig. 3a and b). While it has been shown that nucleic acid–based analysis can be performed on breast cancer cells grown on PCL scaffolds [24], it could also be relevant to analyze cells from PCL-based scaffolds by common proteomic methods, such as flow cytometry and Western blotting. The expanded cells were therefore detached from the filter, and flow cytometry was performed on the cells. The cells were gated according to their forward vs side scatter (Fig. 3c). When comparing the signal of non-stained cells from one scaffold with the signal of anti-EpCAM stained cells from another scaffold, a clear shift was observed (Fig. 3d). Additionally, cell lysates were obtained from cultured scaffolds, on which EpCAM (Fig. 3e) and beta actin (Fig. 3f) could be detected by Western blotting. To demonstrate the feasibility of rt-qPCR analysis of cells grown on the filters, the house-hold genes GAPDH and PGK1 were analyzed on two randomly chosen filter samples and successfully detected (Fig. 3g).
Fig. 3.
Biocompatibility test and downstream analysis. (a) CCK-8 viability assay: HT29 cells were seeded as 100, 200, and 400 cells per scaffold and followed for 15 days. ∗∗∗ p-value < 0.0005 (b) The proliferation/expansion of the HT29 cells on a ‘100 cell seeding’ scaffold was confirmed by immunochemistry and confocal microscopy on day 15. The cells were stained for cytokeratin (magenta) and their nuclei (blue). Scalebar, 40 μm. (c) Flow cytometry on HT29 cells, detached from the scaffolds, were gated according to their SSC-H x FSC-H, and (d) cells within this gate were stained for EpCAM using biotin anti-EpCAM and streptavidin AF488 for detection. Western blot (e), on cell lysates from cells grown on the scaffolds were analyzed for EpCAM, using biotin anti-EpCAM and streptavidin AF488 for detection. A band around 40 kDa was observed corresponding to the apparent size of EpCAM. Additional bands deriving from the streptavidin AF488 only were also observed. (f) Beta actin was also detected in Western blot using rabbit anti-actin and anti-rabbit cy3. An apparent band for beta actin around 45 kDa was observed. (g) The rt-qPCR analysis of GAPDH and PGK1 in two randomly chosen filter samples after filtration and culture, standard deviations are shown with error bars. PGK1, phosphoglycerate kinase 1; GAPDH, human glyceraldehyde-3-phosphate dehydrogenase.
3.3. Capture and culture
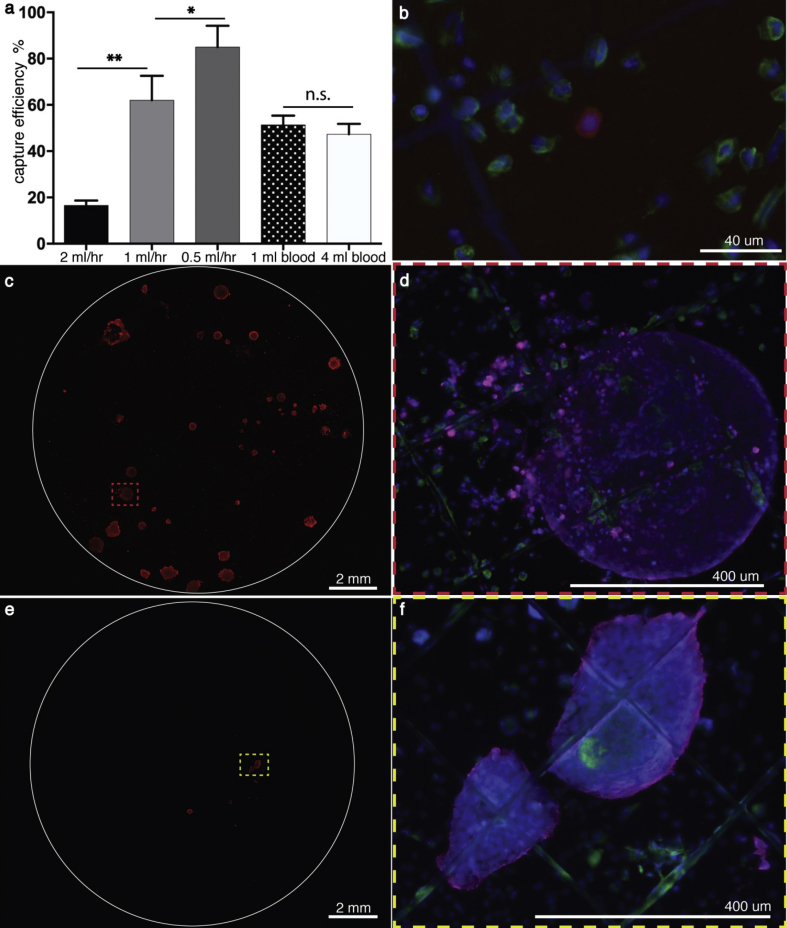
As flow rate influences the capture efficiency, different flow rates were first assayed using 200 prestained HT29 cells through anti-EpCAM–conjugated MEW filters (Ø 12 mm). It was shown that a capture of 85.0 ± 9.2% could be achieved at 0.5 mL/h, which was significantly higher than 62.0 ± 10.6% at 1 mL/h, and 16.5 ± 2.2% at 2 mL/h (Fig. 4a).
Fig. 4.
Capture efficiency and on-site culture. (a) Different flow rates were used to assess the impact of the flow on the capture. The measurements were performed in triplicates, ∗ p-value < 0.05, ∗∗ p-value < 0.005. Thereafter, 200 HT29 cells were spiked into 1 mL blood and 4 mL blood, respectively. (b) An image of a captured HT29 cell (red, prestained with CellTracker Red CMPTX dye) after filtration, corresponding to day 1. The cancer cell is surrounded by white blood cells (green, stained with CD45). The nuclei are shown in blue. (c) Additionally, 200 HT29 cells were spiked into 4 mL blood, filtrated and also cultured for 21 days. A stitched image of the entire filter is shown here, where clusters of cancer cells are shown in red, stained for pan-cytokeratin. One of the clusters is seen at a higher magnification in (d). (e) A stitched image of the filter after filtration of 20 HT29 cells spike-in-sample and 2 weeks on-site culture. (f) A higher magnification image of one cluster in (e). The white blood cells were stained for CD45 (green), the cancer cells for cytokeratin (magenta), and their nuclei were visualized by Hoechst (blue).
Next, 0.5 mL/h was used to investigate the capacity of the MEW filter on clinical relevant spiked-in-samples, where 200 HT29 colon cancer cells were spiked into 1 mL and 4 mL whole blood, respectively.
The ‘1 mL blood’ spike-in-sample (whole blood sample) was filtered without lysing the RBCs. Here, 51.3 ± 4.0% of the cancer cells were captured. Owing to the long (0.5 mL/h, 8 h) filtration process, the ‘4 mL blood’ sample was treated with RBC lysing buffer after spiking in HT29 cells to avoid coagulation. The nucleated cells were then filtered, and 47.2 ± 4.7% of the cancer cells were captured (Fig. 4a). Subsequent to these filtrations, it was seen that the captured HT29 were dispersed on the filters as single cells and surrounded by white blood cells (Fig. 4b).
It was then assessed whether 200 colon cancer cells, spiked into 4 mL whole blood, could be captured and cultured on site. Although it was not possible to distinguish between cancer cells and surrounding blood cells on day 7, distinct tumor cell clusters were visible to the naked eye on day 14, with diameters up to approximately 300 μm (Fig. s3). After 21 days of culture, several cancer clusters had emerged on the filter (Fig. 4c and d). The size of the clusters varied substantially, ranging from 6.7 × 103 to 3.7 × 105 μm2 with an average of 1 × 105 μm2 ± 1.1 × 105 μm2 standard deviation. The clusters were surrounded by residual blood cells that appeared to have attached to the filter and survived there (Fig. 4d and f).
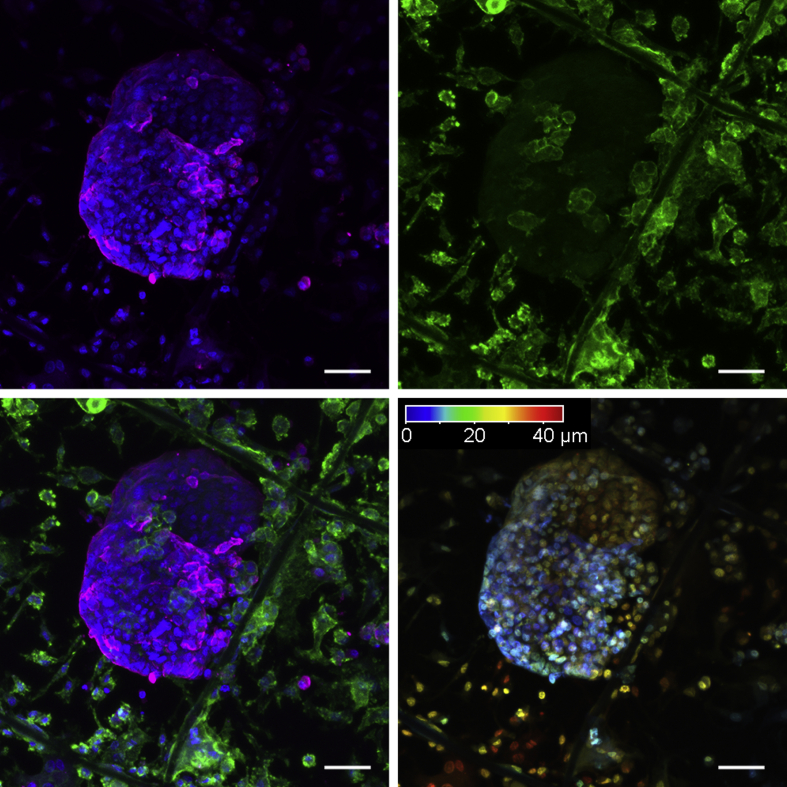
The CTCs are extremely rare, roughly one CTC among one billion blood cells. To get even closer to clinical relevance, the filter was tested further by performing three additional filtrations. In these independently performed filtrations, 20 HT29 colon cancer cells were spiked into 4 mL whole blood, and the nucleated cells were filtered. In all three filtrations, clusters of cancer cells emerged after 3 weeks of culture (Fig. 4e–f, and Supplementary Fig. s4). Again, the clusters displayed a considerably high size variation in which the average cancer cluster area of the three experiments was 6.4 × 104 μm2, and the standard error of the mean was 4.9 × 104 μm2. In addition, one of the ‘20 spike’ samples was stained positive for CD68, indicating that the surrounding cells were likely to be macrophages (Supplementary Fig. s4d). Analysis of this filter by confocal microscopy further indicated that some of the macrophages infiltrated the tumor clusters (Supplementary Animation 1). Finally, it was observed that cells were growing in several layers to a thickness similar to that of the filter, which is approximately 50 μm (Fig. 5 and Supplementary Animation 2).
Fig. 5.
Three-dimensional growth. Upper left, a cancer cluster stained for cytokeratin (magenta) and DNA (blue). Upper right, showing the green channel for CD45 staining only. Bottom left, all channels. Bottom right, a color-coded depth analysis of the cancer cluster, where point 0 (blue) refers to the closest focal point. Scale bar, 50 μm.
The following are the supplementary data related to this article:
4. Discussions
With the increasing amount of available targeted cancer therapies and with the often seen dynamically changing drug resistance, there is a clinical need for tools that enable tailoring of targeted cancer therapy programs [25]. CTCs might play an essential role as biological sources for creating relevant tumor models holding the subsets of cells that cause disease relapse, dissemination, and drug resistance [26,27]. Although the use of the conventional solution electrospinning technique for developing CTC capture microfluidic devices has previously been demonstrated [14,15], this is, to our knowledge, the first study to demonstrate the use of the MEW technique for tailoring a filter that enables both capture and in situ culture of cancer cells in a background of blood. Conjugating antibodies to the filter can be performed within 3–4 hours without any cumbersome steps, offering the possibility to readily target several CTC relevant markers, such as surface vimentin, CD133, N-cadherin, and epidermal growth factor receptor (EGFR). These markers are valuable for capturing CTCs with little or no EpCAM expression [28].
Furthermore, the filter can in principal be applied for any rare cell type in the blood that can be distinguished from its surroundings through surface marker recognition, for example circulating fibrocytes [29] and CD133 positive progenitor cells [30]. Isolating and culturing fibrocytes on the filter might enable and generalize the access to fibrocytes, which are involved in fibrotic disorders such as pulmonary fibrosis and asthma [31].
The MEW filter can be straightforwardly implemented in cell culture and subsequently be analyzed by Western blotting, flow cytometry, and rt-qPCR. In addition, the filter is transparent and is thus ideal for microscopy-based analysis. PCL- scaffolds [32,33] are well established to be compatible with additional down-stream applications, such as analysis of nucleic acid composition and down-stream drug screening, which will be initiated in our further studies.
Co-culture might facilitate the expansion of some CTC clusters as demonstrated by Zhang et al. [7]. Motivated by this work, it was tested in a preliminary study whether the expansion of cancer cells on the filter can be sped up by co-culturing with fibroblasts. In this preliminary study, NIH3T3 fibroblasts were cultured on tissue culture plastic, and filters seeded with 200 HT29 cells were placed on top of the fibroblasts. After 7 days of culture, the filters were infiltrated by both types of cells (Fig. s5). Although these preliminary data are inconclusive, they indicate that the cancer cells grow differently in co-culture compared with monoculture, which can be seen by the difference in mean area values of the cancer islets. The observed differences might be explained by spatial competition between the cells, in which the fast growing fibroblasts possibly force the HT29 cancer cells to grow in a different manner compared with the monoculture, simply because of limited space. In addition, paracrine signaling between cancer cells and fibroblast most likely affect the growth of both cell types [34]. No significant difference in total area coverage was observed between the monoculture and co-culture (Table s2).
We have demonstrated that cancer cells at clinical relevant concentrations can be isolated from a background of billions of blood cells and in 2 weeks be grown in situ to sizes that are visible to the naked eye. The ability of single dispersed cancer cells to form colonies has been demonstrated previously. Meng et al. demonstrates that CD133, a molecule proposed to mark cells with stemness, does not suffice to select those cells with the capacity of forming colonies. They do however suggest that the colony formation feature connects to properties of cancer stem cells [35]. In the work by Held et al., it is shown that the colony forming cells of melanomas can form tumors in mice from single cells, which are more resistant to chemotherapy [36]. In both studies, the mechanisms by which single cells can form colonies are largely undescribed, but it is suggested to be related to the stemness of the cancer cells.
Intriguingly, some blood cells could apparently attach to the filter and survive, as seen in Fig. 4f. Immunocytochemistry against CD68 (Supplementary Fig. s4d and Animation s1) suggests that these cells most likely are macrophages. It is well-known that some blood cells facilitate both dissemination and growth of cancer [37,38]. Even though the mechanisms remain elusive, it has furthermore been suggested that the interaction between white blood cells and CTCs might facilitate the CTC cluster formation in vitro [8,39]. It has been proposed that co-isolated monocytes can differentiate into tumor-associated macrophages (TAMs) by paracrine manipulation enforced by the cancer cells [34]. TAMs are identified to express cytokines and chemokines that can suppress the immune system and promote tumor progression. Therefore, we can only speculate that patient-derived macrophages, captured alongside with CTCs, might facilitate the cancer growth on the filter. However, there is a risk that the background of blood cells might mask the conjugated antibodies and hereby decrease the capture efficiency. Consequently, future work could include efforts to decrease the capture of blood cells by improving the homogeneity of filter pore size. Importantly, future steps of this work should include thorough validation on patient blood to test the clinical applicability of the filter. In such a study, it will be of key importance to analyze the CTC tumor bodies for well-known markers of colorectal cancer. This includes phenotyping the CTC tumors by immunochemistry against markers such as EpCAM, cytokeratin, MUC1, and EGFR, but more specifically, targeted genotyping of the BRAF and KRAS genes should be performed [40]. Finally, these characterizations should be compared with the molecular characteristics of circulating tumor DNA and archival tumor tissue of the same patient.
Timely treatment is crucial for cancer patients. Our work has therefore focused on establishing a fast expansion of CTCs for clinical relevant down-stream analysis and treatment screening. This is in line with recent work by Khoo et al. [41,42] and Zhang et al. [7], in which platforms have been built to assist the development of point of care treatments of individual patients.
5. Conclusions
In conclusion, we have developed a scaffold to potentially capture and culture CTCs on site, where cancer cell clusters form within 2 weeks. To our knowledge, this is the first time that captured cancer cells are able to be cultured on site on a MEW scaffold. A key element of our capture and culture device is that the CTCs are dispersed on the filter as single cells or clusters of few cells from where they expand to big cancer clusters. Consequently, each of the expanded clusters might represent one or a few clones of the tumor, hereby facilitating the identification of important clonal subsets of the cancer. These cluster subsets of patient CTCs might hold valuable information such as drug resistance and DNA mutations and will thus bring the clinicians closer toward personalized medicine management.
6. Authors contributions
Mathias L. Jørgensen: Conceptualization, Methodology, Software, Writing - original draft preparation. Christoph Müller: Data curation. Mille Sikkesoq: Data curation. Marcin Nadzieja: Data curation. Zhongyang Zhang: Data curation. Jesper Just: Data curation. Karen-Lise Garm Spindler: Supervision. Menglin Chen: Conceptualization, Methodology, Supervision, Writing- Reviewing and Editing, Project administration
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the funding from Independent Research Fund Denmark (DFF-7017-00185) and Aarhus University Research Foundation (AUFF-E-2015-FLS-7-27) for support of the research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2020.100052.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bhandari A., Woodhouse M., Gupta S. Colorectal cancer is a leading cause of cancer incidence and mortality among adults younger than 50 years in the USA: a SEER-based analysis with comparison to other young-onset cancers. J. Invest. Med. 2017;65(2):311–315. doi: 10.1136/jim-2016-000229. Epub 2016/11/18. PubMed PMID: 27864324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard P.L., Hansen A.R., Ratain M.J., Siu L.L. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. PubMed PMID: 24048068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Praharaj P.P., Bhutia S.K., Nagrath S., Bitting R.L., Deep G. Circulating tumor cell-derived organoids: current challenges and promises in medical research and precision medicine. Biochim. Biophys. Acta Rev. Canc. 2018;1869(2):117–127. doi: 10.1016/j.bbcan.2017.12.005. Epub 2018/01/31. PubMed PMID: 29360544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K., Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 2010;16(9):398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K., Alix-Panabières C. Cell lines from circulating tumor cells. Oncoscience. 2015;2(10):815–816. doi: 10.18632/oncoscience.195. PubMed PMID: 26682259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cayrefourcq L., Mazard T., Joosse S., Solassol J., Ramos J., Assenat E. Establishment and characterization of a cell line from human circulating colon cancer cells. Canc. Res. 2015;75(5):892. doi: 10.1158/0008-5472.CAN-14-2613. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z., Shiratsuchi H., Lin J., Chen G., Reddy R.M., Azizi E. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014;5(23):12383–12397. doi: 10.18632/oncotarget.2592. PubMed PMID: 25474037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoo B.L., Grenci G., Lim Y.B., Lee S.C., Han J., Lim C.T. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 2017;13:34. doi: 10.1038/nprot.2017.125. https://www.nature.com/articles/nprot.2017.125#supplementary-information [DOI] [PubMed] [Google Scholar]
- 9.Joosse S.A., Gorges T.M., Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 2015;7(1):1–11. doi: 10.15252/emmm.201303698. Epub 2014/11/14. PubMed PMID: 25398926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozkumur E., Shah A.M., Ciciliano J.C., Emmink B.L., Miyamoto D.T., Brachtel E. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013;5(179) doi: 10.1126/scitranslmed.3005616. PubMed PMID: 23552373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riahi R., Gogoi P., Sepehri S., Zhou Y., Handique K., Godsey J. A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer. Int. J. Oncol. 2014;44(6):1870–1878. doi: 10.3892/ijo.2014.2353. PubMed PMID: 24676558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persano L., Camposeo A., Pisignano D. Active polymer nanofibers for photonics, electronics, energy generation and micromechanics. Prog. Polym. Sci. 2015;43:48–95. doi: 10.1016/j.progpolymsci.2014.10.001. [DOI] [Google Scholar]
- 13.Dvir T., Timko B.P., Kohane D.S., Langer R. Nanotechnological strategies for engineering complex tissues. Nat. Nanotechnol. 2010;6:13. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang N., Deng Y., Tai Q., Cheng B., Zhao L., Shen Q. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv. Mater. 2012;24(20):2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 15.Ma L., Yang G., Wang N., Zhang P., Guo F., Meng J. Trap effect of three-dimensional fibers network for high efficient cancer-cell capture. Adv. Healthc. Mater. 2015;4(6):838–843. doi: 10.1002/adhm.201400650. [DOI] [PubMed] [Google Scholar]
- 16.Xu G., Tan Y., Xu T., Yin D., Wang M., Shen M. Hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for cancer cell capture and culture. Biomater. Sci. 2017;5(4):752–761. doi: 10.1039/C6BM00933F. [DOI] [PubMed] [Google Scholar]
- 17.Lee A.-W., Lin F.-X., Wei P.-L., Jian-Wei G., Chen J.-K. Binary-blend fibber-based capture assay of circulating tumor cells for clinical diagnosis of colorectal cancer. J. Nanobiotechnol. 2018;16(1):4. doi: 10.1186/s12951-017-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown T.D., Dalton P.D., Hutmacher D.W. Direct writing by way of melt electrospinning. Adv. Mater. 2011;23(47):5651–5657. doi: 10.1002/adma.201103482. [DOI] [PubMed] [Google Scholar]
- 19.Yu M., Bardia A., Aceto N., Bersani F., Madden M.W., Donaldson M.C. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taskin M.B., Xu R., Gregersen H., Nygaard J.V., Besenbacher F., Chen M. Three-dimensional polydopamine functionalized coiled microfibrous scaffolds enhance human mesenchymal stem cells colonization and mild myofibroblastic differentiation. ACS Appl. Mater. Interfaces. 2016;8(25):15864–15873. doi: 10.1021/acsami.6b02994. [DOI] [PubMed] [Google Scholar]
- 21.Li Y.-F., Rubert M., Aslan H., Yu Y., Howard K.A., Dong M. Ultraporous interweaving electrospun microfibers from PCL–PEO binary blends and their inflammatory responses. Nanoscale. 2014;6(6):3392–3402. doi: 10.1039/C3NR06197C. [DOI] [PubMed] [Google Scholar]
- 22.Cohen S.J., Punt C.J.A., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26(19):3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 23.Lee H., Rho J., Messersmith P.B. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Adv. Mater. 2009;21(4):431–434. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balachander G.M., Talukdar P.M., Debnath M., Rangarajan A., Chatterjee K. Inflammatory role of cancer-associated fibroblasts in invasive breast tumors revealed using a fibrous polymer scaffold. ACS Appl. Mater. Interfaces. 2018;10(40):33814–33826. doi: 10.1021/acsami.8b07609. [DOI] [PubMed] [Google Scholar]
- 25.Maheswaran S., Haber D.A. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Canc. Res. 2015;75(12):2411. doi: 10.1158/0008-5472.CAN-15-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maheswaran S., Haber D.A. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Canc. Res. 2015 doi: 10.1158/0008-5472.CAN-15-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoma C.R., Zimmermann M., Agarkova I., Kelm J.M., Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014;69–70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Jie X.-X., Zhang X.-Y., Xu C.-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: mechanisms and clinical applications. Oncotarget. 2017;8(46):81558–81571. doi: 10.18632/oncotarget.18277. PubMed PMID: 29113414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan T.E., Cowper S., Wu S.-P., Bockenstedt L.K., Bucala R. Circulating fibrocytes: collagen-secreting cells of the peripheral blood. Int. J. Biochem. Cell Biol. 2004;36(4):598–606. doi: 10.1016/j.biocel.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Tondreau T., Meuleman N., Delforge A., Dejeneffe M., Leroy R., Massy M. Mesenchymal stem cells derived from CD133-positive cells in mobilized peripheral blood and cord blood: proliferation, Oct4 expression, and plasticity. Stem Cell. 2005;23(8):1105–1112. doi: 10.1634/stemcells.2004-0330. [DOI] [PubMed] [Google Scholar]
- 31.Gomperts B.N., Strieter R.M. Fibrocytes in lung disease. J. Leukoc. Biol. 2007;82(3):449–456. doi: 10.1189/jlb.0906587. [DOI] [PubMed] [Google Scholar]
- 32.Mitsak A.G., Kemppainen J.M., Harris M.T., Hollister S.J. Effect of polycaprolactone scaffold permeability on bone regeneration in vivo. Tissue Eng. Part A. 2011;17(13–14):1831–1839. doi: 10.1089/ten.TEA.2010.0560. Epub 2011/04/25. PubMed PMID: 21395465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams J.M., Adewunmi A., Schek R.M., Flanagan C.L., Krebsbach P.H., Feinberg S.E. Bone tissue engineering using polycaprolactone scaffolds fabricated via selective laser sintering. Biomaterials. 2005;26(23):4817–4827. doi: 10.1016/j.biomaterials.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 34.Pathria P., Louis T.L., Varner J.A. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40(4):310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Meng X., Li M., Wang X., Wang Y., Ma D. Both CD133+ and CD133− subpopulations of A549 and H446 cells contain cancer-initiating cells. Canc. Sci. 2009;100(6):1040–1046. doi: 10.1111/j.1349-7006.2009.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Held M.A., Curley D.P., Dankort D., McMahon M., Muthusamy V., Bosenberg M.W. Characterization of melanoma cells capable of propagating tumors from a single cell. Canc. Res. 2010;70(1):388. doi: 10.1158/0008-5472.CAN-09-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leone K., Poggiana C., Zamarchi R. The interplay between circulating tumor cells and the immune system: from immune escape to cancer immunotherapy. Diagnostics (Basel) 2018;8(3):59. doi: 10.3390/diagnostics8030059. PubMed PMID: 30200242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joosse Simon A., Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Canc. Cell. 2015;28(5):552–554. doi: 10.1016/j.ccell.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Luan Khoo B., Chin Lee S., Kumar P., Zea Tan T., Ebrahimi Warkiani M., Gw Ow S. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015;6(17) doi: 10.18632/oncotarget.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spindler K.-L.G., Pallisgaard N., Vogelius I., Jakobsen A. Quantitative cell-free DNA, KRAS, BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin. Canc. Res. 2012;18(4):1177. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 41.Khoo B.L., Lee S.C., Kumar P., Tan T.Z., Warkiani M.E., Ow S.G. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015;6(17):15578. doi: 10.18632/oncotarget.3903. PubMed Central PMCID: PMCPMC4558172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoo B.L., Grenci G., Jing T., Lim Y.B., Lee S.C., Thiery J.P. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2016;2(7) doi: 10.1126/sciadv.1600274. PubMed PMID: 27453941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676. doi: 10.1038/nmeth.2019. https://www.nature.com/articles/nmeth.2019#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.