Abstract
This work presents results of the research on the occurrence of Coxiella burnetii and Francisella tularensis in the tissues of wild-living animals and ticks collected from Drawsko County, West Pomeranian Voivodeship. The real-time PCR testing for the pathogens comprised 928 samples of animal internal organs and 1551 ticks. The presence of C. burnetii was detected in 3% of wild-living animals and in 0.45–3.45% (dependent on collection areas) of ticks. The genetic sequences of F. tularensis were present in 0.49 % of ticks (only in one location – Drawa) and were not detected in animal tissues. The results indicate respectively low proportion of animals and ticks infected with C. burnetii and F. tularensis.
Key words: Coxiella burnetii, Francisella tularensis, reservoirs, real-time PCR
Wild-living animals play a significant role in epidemiology of zoonoses; they constitute the main source of pathogens dangerous for humans and domestic animals. Number of zoonotic agents carried by wild-living animals increases and still the threat they pose is not well known, particularly for humans with direct contact to animals. The scope of danger may vary and it depends not only on the source of infection but also on transmission routes (Artois 2003; Jones et al. 2008). The presence of vectors in the environment (e.g. ticks) is correlated with the existence of ecological niches inhabited by the hosts and tick-specific environmental conditions (temperature and humidity levels) (Daszak et al. 2000). Ticks are among the most common zoonosis vectors; tick-borne diseases (TBDs) are a significant group of diseases impacting public health.
Some of the zoonosis-related threats are the diseases caused by C. burnetii and F. tularensis. C. burnetii can be isolated from both domestic and wild-living animals such as bears, bisons, red deers, roe deers, boars, rabbits, shrews and marsupials. The infections occur through direct contact with infected animals, by aerosol inhalation, ingestion, direct contact with wounded skin and as a result of tick bites (Norlander 2000; Bossi et al. 2004; Woldehiwet 2004). Arthropods are a significant vector of pathogen transmission; however, multiple other transmission routes exist. Infection can occur after bite of a feeding arthropod, mechanically (e.g. flies), by contact with wounded skin or by inhaling the faeces of parasites (ticks) (Marrie 1990; Anusz 1995; Mediannikow et al. 2010). F. tularensis is found to be important threat in forests and on farmlands; while its main reservoir are wild-living rodents. Arthropods (ticks, mites, mosquitoes, fleas and flies) can also be key vectors of the disease transmission (Dennis et al. 2001; Tarnvik et al. 2003; Oyston et al. 2004; Michelet et al. 2016). Additionally, transfer of the pathogen to humans may occur through direct contact with contaminated animal products (blood, faeces, skin), through inhalation of contaminated air or dust, and by ingesting contaminated food and water (Ohtake et al. 2011).
In order to monitor the presence of C. burnetii and F. tularensis in environmental samples during epidemiological surveillance, PCR method can be implemented (Higgins 2000; Emanuel et al. 2003; Seshadri et al. 2003; Fujita et al. 2006; Klee et al. 2006; Petersen et al. 2009; Bielawska-Drózd et al. 2010).
The goal of this study was to determine the occurrence of C. burnetii and F. tularensis in samples from wild animals and ticks collected from forested areas of Drawsko County including Drawsko military ground (West Pomeranian Voivodeship) using the real-time PCR method.
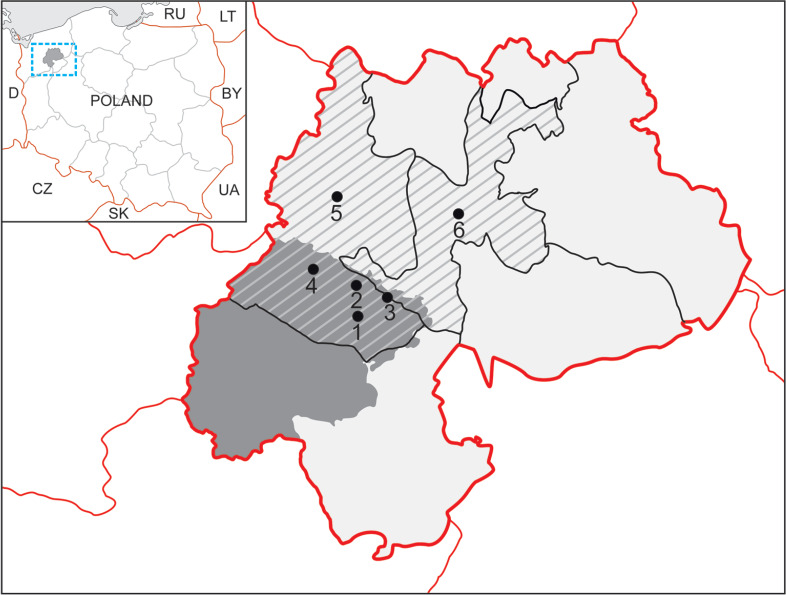
The wild animals to be sampled were hunted in the years 2016–2017 in the areas, which included the hunting of the following clubs “Cyraneczka” – Drawsko Pomorskie; “Żbik” – Cieszyno near Złocieniec; “Bażant” – Stawno near Złocieniec (Fig. 1). For testing, the samples were collected from internal organs (liver, spleen, heart and lungs). In total, 928 tissue samples were collected from 232 animals from the following species: red deer (140 animals), roe deer (40), and boar (52). The samples, which consisted of mixture of tissues (5–10 g) from four organs examined were suspended in saline solution and homogenized (rotor-stator homogenizer – SHM1, Stuart). Subsequently, 1 ml of the homogenizate was transferred to a new 2 ml test tube and centrifuged for 1 min at room temperature at 6300 rpm. The supernatant (1000 µl) was transferred to a new test tube and centrifuged again for 3 minutes at room temperature at 15 000 rpm. The pellet was resuspended in a lysis buffer, then 2 µl RNase and 10 µl proteinase K were added and incubated for 18 hours at 56°C. Subsequent stages were carried out according to the protocol of GeneMatrix Tissue&Bacterial DNA Purification Kit (EURx Ltd., Poland).
Fig. 1.
The locations of sample collection.
Legend: Area marked by the red line: Drawsko County; dark grey slanted striped areas: Drawsko and Złocieniec Districts; grey areas: Drawsko military training ground.
The numbers indicate the areas of sample collections: ticks (1 – Konotop, 2 – Lake Konotop, 3 – Karwice, 4 – Oleszno), and animals (5 – Drawsko-Pomorskie, 6 – Złocieniec).
The research also included 1551 ticks collected from five testing sites that were forested areas of Drawsko military training ground (Konotop Encampment, Oleszno and Karwice, Drawa and Konotop Lake) from April to May 2017 (Fig. 1). These ticks were collected by the flagging-dragging method. Tick species were identified using taxonomic keys; the species collected was found to be Ixodes ricinus. Ticks were pooled according to the collection site and divided according to sex (adults) and development stages (nymphs): ♀ – female (296 ticks), ♂ – male (250), N – nymph (1005).
80 pooled tick samples (pools) were obtained. Among them, 72 pools contained 20 tick imagos or nymphs (10 pools – ♂, 12 pools – ♀, 50 pools – N), 3 pools contained 19 ticks (1 pool – ♂, 1 pool – ♀, 1 pool – N), 1 pool contained 17 ticks – ♀, 1 pool contained 13 ticks – ♂, 1 pool contained 12 ticks – ♀, 1 pool contained 7 ticks – N, 1 pool contained 5 ticks – N. In Drawa location pools were as follows: 211 (♀ – 2 pools/20 individuals, 1 pool/19 individuals; ♂ – 2 pools/20 individuals, 1 pool/12 individuals; N – 5 pools/20 individuals), in Karwice location the following samples were obtained: 460 (♀ – 5 pools/20 individuals; ♂ – 4 pools/20 individuals; N – 14 pools/20 individuals), in Lake Konotop location: 377 (♀ – 4 pools/20 individuals, 1 pool/13 individuals; ♂ – 3 pools/20 individuals, 1 pool/19 individuals; N – 10 pools/20 individuals, 1 pool/5 individuals), in Konotop location: 117 (♀ – 1 pool/17 individuals; ♂ – 1 pool/20 individuals; N – 4 pools/20 individuals), in Oleszno location: 386 (♀ – 1 pool/20 individuals, 1 pool/7 individuals; ♂ – 1 pool/19 individual, N – 17 pools/20 individuals) were collected, respectively. The pooled tick samples were placed in 2 ml test tubes with 300 µl of ethanol (70%) inside and left for 15 minutes (stirred several times). The alcohol was removed, and the ticks were rinsed with deionized water (300 µl). The residues of water were removed with blotting paper and the samples were placed in liquid nitrogen for 10 minutes. The ticks were then homogenized (mechanically, in a mortar) (Halos et al. 2004; Rodriguez et al. 2014; Jose et al. 2017). Following that, 1 ml of deionized water was added, and the samples were frozen at –80°C for further analyses. To isolate the genetic material, 200 µl of the homogenized liquid was used. The material was centrifuged at 15 000 rpm for 3 minutes at room temperature. The pellet was resuspended in lysis buffer LyseT, 2 µl RNase and 10 µl proteinase K were added and the material was incubated for 12 hours at the temperature of 56°C. Subsequent stages were carried out according to the protocol by GeneMatrix Tissue&Bacterial DNA Purification Kit (EURx Ltd., Poland).
Screening tests were performed by the real-time PCR method using a C. burnetii – specific multicopy insertion sequence IS1111+ (transposase gene) and the outer membrane coding sequences: fopA and tul4 for F. tularensis (these sequences confirmed the presence of F. tularensis in ticks). The oligonucleotides used in the reactions are presented in Table I.
Table I.
The oligonucleotides used in real-time PCR.
| C. burnetii | F. tularensis | ||
|---|---|---|---|
| IS1111+ (Klee et al. 2006) | fopA (Emmanuel et al. 2003) | tul4 (Fujita et al. 2006) | |
| Forward primer | 5’-GTCTTAAGGTGGGCTGCGT G-3’ | 5’-AACAATGGCACCTAGTAAT ATTTCTGG-3’ | 5’-ATTACAATGGCAGGCTCC AGA-3’ |
| Reverse primer | 5’-CCCCGAATCTCATTGATC AGC-3’ | 5’-CCACCAAAGAACCATGTT AAACC-3’ | 5’-TGCCCAAGTTTTATCGTTC TTCT-3’ |
| Probe | 5’-FAM-AGCGAACCATTGGTATC GGACGTTT-TAMRA-TATGG-Pho-3’ | 5’-FAM-TGGCAGAGCGGGTACT AACATGATTGGT-TAMRA-3’ | 5’-FAM TCCTAAGTGCCATGAT ACAAGCTTCCCAATTACTAAG-BHQ1-3’ |
The oligonucleotides were synthesized by Genomed S.A. (Poland). The reactions for both pathogens were conducted using LightCycler 2.0 instrument (Roche, Germany) according to the following thermal profile: initial denaturation at 95°C for 10 minutes; 40 cycles (95°C for 15 seconds, 60°C for 30 seconds); 40°C for 30 seconds. DNA extracted from C. burnetii Nine Mile and F. tularensis subsp. holarctica strain Kodar were used as positive controls. The test samples were positive when the cycle threshold Ct was lower than 36.
The prevalence of infected ticks in pools was analyzed based on the number of individuals. Statistical analyses to calculate estimated prevalence for fixed and variable pool sizes were performed with EpiTools (http://www.ausvet.com.au; Andreassen et al. 2012; Ndeereh et al. 2017).
Q fever research in Poland are based on immunological status of both domestic and wild-living animals (Niemczuk et al. 2011). Only few studies involved molecular analyses of clinical or environmental material (arthropods) (Tylewska et al. 1996; Szymańska et al. 2013; Bielawska-Drózd et al. 2014; Bielawska-Drózd et al. 2016). Seven wild-living animals (three boars, three stags and one roe deer) were found positive for IS1111+, characteristic of C. burnetii (3%). In the light of other studies, the positive results obtained in this research are similar or even lower when compared to the results by others: 0.7% in red deers (Smetanova et al. 2016), 4.3% in wild boars 5.1% in roe deers and 9.1% (European hares) (Astobiza et al. 2011).
In this study any characteristic sequences were detected in testing for the presence of F. tularensis DNA in wild-living animals. Despite a large proportion of positive results obtained in ELISA by others: 3.5% (Al Dahouk et al. 2005), 7.5% (Kuehn et al. 2013), 7.4% (Otto et al. 2014) or even 15–30% (Taussing and Landau 2008), in this study the biological agent remains undetected in animal tissues.
Epidemiological situation of Q fever and tularemia in Poland as well as worldwide seems to be stable. Although Q fever and tularemia outbreaks have been registered almost all around the world, the numbers of infections are still low, but Scandinavian countries, Hungary and Czech Republic in relation to tularemia (Bielawska-Drózd et al. 2013; ECDC 2016). Most outbreaks of tularemia and Q-fever have been in wildlife species. In Germany between 2002 and 2016, 10 clusters of tularemia were reported. A serological study in various wildlife species in Brandenburg revelated a total of 101/1353 positive sera (7.5%) of foxes, raccoon dogs, and wild boars (Faber et al. 2018). The reports from Germany between 1992 and 2012 showed that 2.4% of dead wild European rabbits were positive for F. tularensis. In Austria, Bulgaria, Germany, Hungary, Kosovo, Slovakia and Sweden small rodents were examined and the detection rate of F. tularensis varied from 0.7% to 20.8%. In Austria, the bacteria was detected in 1.3% of the hunter red foxes. In Portugal, 212 migratory shore of various species were tested for bacteria, which resulted in identification of F. tularensis (Hestvik et al. 2015). Q fever epidemics and epizooties in 1948–2004 were registered in the region of the Balkan Peninsula (Hukici et al. 2010). Serum samples from 464 wild rabbits were collected and analyzed from European wild rabbits in Spain, Portugal and Chafarinas Islands during the time period 2003–2013. Seroprevalence in wild rabbit populations ranged from 6.7% to 81.3%. European rabbits can also be reservoirs of C. burnetii (González-Barrio et al. 2015).
The estimated prevalence of C. burnetii in pools of ticks ranged from 0.45% to 3.45% for the various locations. The estimated prevalence in the Lake Konotop and Konotop areas showed significantly higher value than in the other sites examined (p-value 0.00348). These results correspond with the results that have been already reported in Poland and in other countries e.g. Senegal, Netherlands, Iran, Slovakia, Hungary, Spain, Germany (Tylewska and Chmielewski 1996; Spitalska et al. 2003; Toledo et al. 2009; Mediannikov et al. 2010; Fard and Khalili 2011; Hilderbrandt et al. 2011; Sprong et al. 2012; Bielawska-Drózd et al. 2016). Moreover, in the current study fopA and tul4 positive results (0.49%) for F. tularensis were found only in one location – Drawa (Table II). Such a small proportion of ticks infected with F. tularensis (0.2% to 1.4%) was also found by other studies conducted in Poland, France, Germany and Portugal (Franke et al. 2010; Lopes de Carvalho et al. 2010; Reis et al. 2011; Wójcik-Fatla et al. 2015). Slightly higher proportions: 1.98% and 3.8% were detected by Zhang et al. (2008) and Tomanović et al. (2009), respectively. However, according to the newest research, the two genes fopA and tul4 can also be present in Francisella-like endosymbionts (FLEs). Screening research conducted in Portugal by Lopes de Carvalho et al. (2010) demonstrated that no less than 32% ticks of Dermacentor reticulatus had the tul4 gene. In subsequent research by Michelet et al. (2016) FLEs were detected in 86% of D. reticulatus as it was shown by the presence of the fopA gene. Therefore, indisputable presence of F. tularensis may be only confirmed when both genes tul4 and fopA are present in the sample tested, while FLEs are supposed when only one of the markers is detected. Due to the highest prevalence of FLEs in D. reticulatus the simultaneous monitoring of the presence of both sequences is necessary to exclude FLEs. Although the research presented here did not encompass D. reticulatus but only I. ricinus species, one has pay attention to D. reticulatus since it can constitute an unquantified FLEs reservoir in Poland. Therefore, future studies ought to comprise separate analyses for this species and consider its specifics and seasonality.
Table II.
The estimated prevalence of positive ticks pools for C. burnetii and F. tularensis.
| Site | Number of pools | PoolSize | Number of positive pools for C. burnetii | Number of positive pools for F. tularensis | Number of individuals | Total number of individuals | Estimated prevalence (%)1 C. burnetii | Estimated prevalence (%)1 F. tularensis |
|---|---|---|---|---|---|---|---|---|
| Drawa | 9 | 20 | 1 | 0 | 59 | 211 | 0.5% | 0.49% |
| 1 | 19 | 0 | 0 | 52 | ||||
| 1 | 12 | 0 | 1 | 100 | ||||
| Karwice | 23 | 20 | 2 | 0 | 460 | 460 | 0.45% | 0 |
| Lake Konotop | 17 | 20 | 8 | 0 | 340 | 377 | 3.1% | 0 |
| 1 | 19 | 0 | 0 | 19 | ||||
| 1 | 13 | 0 | 0 | 13 | ||||
| 1 | 5 | 1 | 0 | 5 | ||||
| Konotop | 5 | 20 | 2 | 0 | 100 | 117 | 3.45% | 0 |
| 1 | 17 | 1 | 0 | 17 | ||||
| Oleszno | 18 | 20 | 2 | 0 | 360 | 386 | 0.55% | 0 |
| 1 | 19 | 0 | 0 | 19 | ||||
| 1 | 7 | 0 | 0 | 7 |
The results obtained in this work correlate with the data found by other researchers. However, despite the results did not present an increase in the proportion of the ticks infected, it is worth to continue studies by Formińska et al. (2015) and Chmielewski et al. (2010) and to examine more numerous tick species and biological agents that can exist in arthropods and constitute a significant threat to human health and lives. It is also advisable to extend the scope of research to include other wild-living animal species as potential reservoirs of zoonotic pathogens.
Additionally, this research demonstrates the useful molecular tool for the detection of F. tularensis and C. burnetii during natural tularemia and Q fever outbreaks.
Acknowledgements
This work was partly supported by the project NPZ (Health National Programme); Assessment of exposure to chemical, biological and physical factors specific to the military with the exception of the Airforce and Navy Service.
The authors are grateful to Paweł Rutyna for preparing Figure 1, Aleksander Michalski and Justyna Wiechetek for participating in sampling of ticks.
Authors’ contributions
ABD and PC designed the study and processed the data. PC, BWS and DZ completed the analysis. PZ and AZ analyzed the reservoirs and biological samples, ABD, PC, PG participated in the manuscript preparation. All authors read, completed and approved the final manuscript.
Conflict of interest
Author does not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Al Dahouk S, Nöckler K, Tomaso H, Splettstoesser WD, Jungersen G, Riber U, Petry T, Hoffmann D, Scholz HC, Hensel A, et al. . 2005. Seroprevalence of brucellosis, tularemia, and yersiniosis in wild boars (sus scrofa) from north-eastern Germany. Vet Med. 52(10):444–455. [DOI] [PubMed] [Google Scholar]
- Andreassen A, Jore S, Cuber P, Dudman S, Tengs T, Isaken K, Hygen HO, Viljugrein H, Anestad G, Ottesen P, et al. . 2012. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Par Vect. 5:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anusz Z. 1995. Q fever in humans and animals Olsztyn (Poland): ART. [Google Scholar]
- Artois M. 2003. Wildlife infectious disease control in Europe. J Mountain Ecol. 7:89–97. [Google Scholar]
- Astobiza I, Barral M, Ruiz-Fons F, Barandika JF, Gerrikagoitia X, Hurtado A, García-Pérez AL. 2011. Molecular investigation of the occurrence of Coxiella burnetii in wildlife and ticks in an endemic area. Vet Microbiol. 147(1):190–194. [DOI] [PubMed] [Google Scholar]
- Bielawska-Drózd A, Cieślik P, Wlizło-Skowronek B, Zięba P, Pitucha G, Gaweł J, Knap J. 2016. Studies on the occurrrence of Coxiell burnetii infection in ticks in selected eastern and central regions of Poland. JMBFS. 5(4):355–357. [Google Scholar]
- Bielawska-Drózd A, Cieślik P, Mirski T, Gaweł J, Michalski A, Niemcewicz M, Bartoszcze M, Żakowska D, Lasocki K, Knap J, et al. . 2014. Prevalence of Coxiella burnetii in environmental samples collected from cattle farms in Eastern and Central Poland (2011– 2012). Vet Microbiol. 174(3–4):600–606. [DOI] [PubMed] [Google Scholar]
- Bielawska-Drózd A, Cieślik P, Mirski T, Bartoszcze M, Knap J, Gaweł J, Żakowska D. 2013. Q fever-selected issues. Ann Agric Environ Med. 20(2):222–232. [PubMed] [Google Scholar]
- Bielawska-Drózd A, Niemcewicz M, Gaweł J, Bartoszcze M, Graniak G, Joniec J, Kołodziej M. 2010. Identyfikacja Francisella tularensis techniką real-time PCR z wykorzystaniem sond hybrydyzujących zaprojektowanych dla fragmentów sekwencji genów fopA i tul4. Med Dosw Mikrobiol. 4(62):351–360 [in Polish]. [PubMed] [Google Scholar]
- Bossi P, Tegnell A, Baka A, Van Loock F, Hendriks J, Werner A, Maidhof H, Gouvras G. 2004. Bichat guidelines for the clinical management of Q fever and bioterrorism-related Q fever. Euro Surveill. 9(12):1–5. [DOI] [PubMed] [Google Scholar]
- Chmielewski T, Tylewska-Wierzbanowska S. 2013. Q fever outbreaks in Poland during 2005–2011. Med Sci Monit. 19:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunnigham AA, Hyatt AD. 2000. Emerging infectious diseases of wildlife-threats of biodiversity and human health. Science. 287(5452):443–449. [DOI] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, et al. . 2001. Tularemia as a biological weapon: medical and public health management. JAMA. 285(21):2763–2773. [DOI] [PubMed] [Google Scholar]
- Emmanuel PA, Bell R, Dang JL, McClanahan R, David JC, Burgess RJ, Thompson J, Collins L, Hadfield T. 2003. Detection of Francisella tularensis within infected mouse tissues by using a handheld PCR thermocycler. J Clin Microbiol. 41(2):689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC 2016. Annual Epidemiological Report 2016 – Tularemia [Internet] Stockholm (Sweden): European Centre for Disease Prevention and Control; [cited 2018 Jul 12]. Available from http://ecdc.europe.eu/en/healthtopics/tularemia/Pages/Annual-epidemiological-report-2016.aspx [Google Scholar]
- Faber M, Heuner K, Jacob D, Grunow R. 2018. Tularemia in Germany – A Re-emerging zoonosis. Front Cell Infect Microbiol. 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fard SRN, Khalili M. 2011. PCR-detection of Coxiella burnetii in ticks collected from sheep and goats in Southeast Iran. Iran J Arthropod Borne Dis. 5(1):1–6. [PMC free article] [PubMed] [Google Scholar]
- Formińska K, Zasada AA, Rastawicki W, Śmietańska K, Bander D, Wawrzynowicz-Syczewska M, Yanushevych M, Niścigórska-Olsen J, Wawszczak M. 2015. Increasing role of arthropod bites in tularemia transmission in Poland – case reports and diagnostic methods. Ann Agric Environ Med. 22(3):443–446. [DOI] [PubMed] [Google Scholar]
- Franke J, Fritzsch J, Tomaso H, Straube E, Dorn W, Hildebrandt A. 2010. Coexistence of pathogens in host-seeking and feeding ticks within a single natural habitat in Central Germany. Appl Environ Microbiol. 76(20):6829–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita O, Tatsumi M, Tanabayashi K, Jamada A. 2006. Development of a real-time PCR assay for detection and quantification of Francisella tularensis. Jpn J Infect Dis. 59(1):46–51. [PubMed] [Google Scholar]
- González-Barrio D, Maio E, Vieira-Pinto M, Ruiz-Fons F. 2015. European rabbits as reservoir for Coxiella burnetii. Emerg Infect Dis. 21(6):1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halos L, Jamal T, Vial L, Maillard R, Suau A, Le Menach A, Boulouis HJ, Vayssier-Taussat M. 2004. Determination of an efficient and reliable method for DNA extraction from ticks. Vet Res. 35(6):709–713. [DOI] [PubMed] [Google Scholar]
- Hestvik G, Warns-Petit E, Smith LA, Fox NJ, Uhlhorn H, Artois M, Hannant D, Hutchings MR, Mattsson R, Yon L, et al. . 2015. The status of tularemia in Europe in a one-health context: a review. Epidemiol Infect. 143:2137–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA, Hubalek Z, Halouzka J, Elkins KL, Sjöstedt A, Shipley M, Ibrahim MS. 2000. Detection of Francisella tularensis in infected mammals and vectors using a probe-based polymerase chain reaction. Am J Trop Med Hyg. 62(2):310–318. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Straube E, Neubauer H, Schmoock G. 2011. Coxiella burnetii and coinfections in Ixodes ricinus ticks in Central Germany. Vector Borne Zoonotic Dis. 11(8):1205–1207. [DOI] [PubMed] [Google Scholar]
- Hukić M, Numanović F, Šiširak M, Moro A, Dervović E, Jakovac S, Salimović Bešić I. 2010. Surveillance of wildlife zoonotic diseases in the Balkans Region. Med. Glas (Zenica). 7(2):96–105. [PubMed] [Google Scholar]
- Jain J, Bindu L, Aravindakshan TV, Hitaishi VN, Binu KM. 2017. Evaluation of DNA extraction protocols from Ixodid ticks. IJEST. 6(3):1912–1917. [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature. 451:990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee SR, Tyczka J, Ellebrok H, Franz T, Linke S, Baljer G, Appel B. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn A, Schulze C, Kutzer P, Probst C, Hlinak A, Ochs A, Grunow R. 2013. Tularaemia seroprevalence of captured and wild animals in Germany: the fox (Vulpes vulpes) as a biological indicator. Epidemiol Infect. 141(4):833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes de Carvalho I, Santos N, Soares T, Ze-Ze L, Nuncio S. 2010. Francisella-like endosymbiont in Dermacentor reticulatus collected in Portugal. Vector Borne Zoonotic Dis. 11(2):185–188. [DOI] [PubMed] [Google Scholar]
- Marrie TJ. 1990. Q fever – the disease Boca Raton (USA): CRC Press. [Google Scholar]
- Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez JF, Sokhna C, Trape JF, Raoult D. 2010. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 4(4):e654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Jancour G, Devillers E, Torina A, Vayssier-Taussat M, Bonnet SI, Moutailler S. 2016. Tick species, tick-borne pathogens and symbionts in an insular environment off the coast of Western France. Ticks Tick Borne Dis. 7(6):1109–1115. [DOI] [PubMed] [Google Scholar]
- Ndeereh D, Muchemi G, Thaiyah A, Otiende M, Angelone-Alasaad S, Jowers MJ. 2017. Molecular survey of Coxiella burnetii in wildlife and ticks at wildlife – livestock interfaces in Kenya. Exp Appl Acarol. 72:277–289. [DOI] [PubMed] [Google Scholar]
- Niemczuk K, Szymańska-Czerwińska M, Zarzecka A, Konarska H. 2011. Q fever in a cattle herd and humans in the Southeastern Poland. Laboratory diagnosis of the disease using serological and molecular methods. Bulletin – Veterinary Institute in Pulawy. 55(4):593–598. [Google Scholar]
- Norlander L. Q-fever epidemiology and pathogenesis. 2000. Microbes Infect. 2(4):417–424. [DOI] [PubMed] [Google Scholar]
- Ohtake S, Martin RA, Saxena A, Lechuga-Ballesteros D, Santiago A, Barry EM, Truong-Le V. 2011. Formulation and stabilization of Francisella tularensis live vaccine strain. J Pharm Sci. 100(8):3076–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto P, Chaignat V, Klimpel D, Diller R, Melzer F, Müller W, Tomaso H. 2014. Serological investigation of wild boars (Sus scrofa) and red foxes (Vulpes vulpes) as indicator animals for circulation of Francisella tularensis in Germany. Vector Borne Zoonotic Dis. 14(1):46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston PCF, Sjöstedt A, Titball RW. 2004. Tularemia: bioterrorism defence renews interest in Francisella tularensis. Nature Rev Microbiol. 2(12):967–978. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Carlson J, Yockey B, Pillai S, Kuske C, Garbalena G, Pottumarthy S, Chalcraft L. 2009. Direct isolation of Francisella spp. from environmental samples. Lett Appl Microbiol. 48(6):663–667. [DOI] [PubMed] [Google Scholar]
- Reis C, Cote M, Paul RE, Bonnet S. 2011. Questing ticks in suburban forest are infected by at least six tick-borne pathogens. Vector Borne Zoonotic Dis. 11(7):907–916. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Fraga J, Noda AA, Mayet M, Duarte Y, Echevarria E, Fernández C. 2014. An alternative and rapid method for the extraction of nucleic acids from Ixodid ticks by potassium acetate procedure. Braz Arch Biol Technol. 57(4):542–547. [Google Scholar]
- Seshadri R, Paulsen TI, Eisen JA, Read TD, Nelson KE, Nelson WC, Ward NL, Tattelin H, Davidsen TM, Beanan MJ, et al. . 2003. Complete genome sequence of the Q fever pathogen, Coxiella burnetii. Proc Natl Acad Sci. 100(9):5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetanova K, Schwarzova K, Kocianova E. 2006. Detection of Anaplasma phagocytophilum, Coxiella burnetii, Rickettsia spp., and Borrelia burgdorferi s.I in ticks, and wild-living animals in western and middle Slovakia. Ann N Y Acad Sci. 1078(1):312–315. [DOI] [PubMed] [Google Scholar]
- Spitalska E, Kocianova E. Detection of Coxiella burnetii in ticks collected in Slovakia and Hungary. 2003. Eur J Epidemiol. 18:263–266. [DOI] [PubMed] [Google Scholar]
- Sprong H, Tijsse-Klasen E, Langelar E, De Bruin M, Fonville M, Gassner F, Takken W, Van Wieren S, Nijhof A, Jongejan F, et al. . 2012. Prevalence of Coxiella burnetii in ticks after a large outbreak of Q fever. Zoonoses Public Health. 59(1):69–75. [DOI] [PubMed] [Google Scholar]
- Szymańska-Czerwińska M, Galińska EM, Niemczuk K, Zasepa M. 2013. Prevalence of Coxiella burnetii infection in foresters and ticks in south-eastern Poland and comparison of diagnostic methods. Ann Agric Environ Med. 20(4):699–704. [PubMed] [Google Scholar]
- Tarnvik A, Berglund L. 2003. Tularemia. Eur Respir J. 21(2):361–373. [DOI] [PubMed] [Google Scholar]
- Taussing L, Landau L. 2008. Pediatric Respiratory Medicine 2nd Edition. Philadelphia (USA): Mosby Elsevier. [Google Scholar]
- Toledo A, Jado I, Olmeda AS, Casado-Nistal MA, Gil H, Escudero R, Anda P. 2009. Detection of Coxiella burnetii in ticks collected from Central Spain. Vector Borne Zoonotic Dis. 9(5):465–468. [DOI] [PubMed] [Google Scholar]
- Tomanowić S, Radulowić Z, Masuzawa T, Milutinović M. 2010. Coexistence of emerging bacterial pathogens in Ixodes ricinus ticks in Serbia. Parasite. 17(3):211–217. [DOI] [PubMed] [Google Scholar]
- Tylewska-Wierzbanowska S, Kruszewska D, Chmielewski T, Zukowski K, Zabicka J. 1996. Ticks as a reservoir of Borrelia burgdorferi and Coxiella burnetii on Polish terrain. Przegl Epidemiol. 50(3):245–251. [PubMed] [Google Scholar]
- Woldehiwet Z. 2004. Q fever (coxiellosis): epidemiology and pathogenesis. Res Vet Sci. 77(2):93–100. [DOI] [PubMed] [Google Scholar]
- Wójcik-Fatla A, Zając V, Sawczyn A, Cisak E, Sroka J, Dutkiewicz J. 2015. Occurrence of Francisella spp. in Dermacentor reticulatus and Ixodes ricinus ticks collected in eastern Poland. Ticks Tick Borne Dis. 6(3):253–257. [DOI] [PubMed] [Google Scholar]
- Zhang F, Liu W, Wu XM, Xin ZT, Zhao QM, Yang H, Cao WC. 2008. Detection of Francisella tularensis in ticks and identification of their genotypes using multiple-locus variable-number tandem repeat analysis. BMC Microbiol. 8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]